Live Attenuated Influenza Vaccines by Computer-Aided Rational Design (original) (raw)
. Author manuscript; available in PMC: 2011 Jan 1.
Published in final edited form as: Nat Biotechnol. 2010 Jun 13;28(7):723–726. doi: 10.1038/nbt.1636
Abstract
Influenza claims 250,000 - 500,000 lives annually worldwide. Despite existing vaccines and enormous efforts in biomedical research, these staggering numbers have not changed significantly over the last two decades1, motivating the search for new, more effective, vaccines that can be rapidly designed and easily produced. Using influenza virus strain A/PR/8/34, we describe a systematic, rational approach, termed Synthetic Attenuated Virus Engineering (SAVE), to develop new, efficacious live attenuated influenza virus vaccine candidates through genome-scale changes in codon pair bias. Attenuation is based on many hundreds of nucleotide changes across the viral genome, offering high genetic stability and a wide margin of safety. The method can be applied rapidly to any emerging influenza virus in its entirety, an advantage that is significant for dealing with seasonal epidemics and pandemic threats, such as H5N1- or 2009-H1N1 influenza.
Influenza viruses are negative stranded, enveloped orthomyxoviruses with eight gene segments, each encoding one or two proteins2. The signature antigenicity of the A and B types of influenza viruses is determined by the glycoproteins hemagglutinin (HA) and neuraminidase (NA). Antigenicity undergoes yearly genetic drift by point mutations, which is the basis for seasonal epidemics1-3. Swapping of gene segments by reassortment between viruses of aquatic birds, swine and humans produces new type A influenza viruses (genetic shift) with novel antigenicity that may cause devastating pandemics1-3.
The capacity of influenza viruses for immune escape demands annual updating of vaccine strains to reflect changes in the HA and NA genes within the impending seasonal strains. Two types of vaccines are currently used: a chemically inactivated virus delivered by injection, and a live attenuated influenza vaccine (LAIV) of cold-adapted virus4, delivered as a nasal-spray (“FluMist”) (CDC; http://www.cdc.gov/flu/protect/keyfacts.htm). Either vaccine comes with limitations. While cell-mediated responses are being recognized as a major determinant of influenza immunity5-8, the traditional, killed vaccines act mainly by inducing neutralizing antibodies. Unfortunately, this vaccine appears less effective than hoped in the elderly population (>65 yr)9. LAIV induces both humoral and cellular immunity but its administration is as yet restricted to healthy children, adolescents and adults (non-pregnant females), age 2 – 49. LAIV works relatively better in immunologically naive young children than in adults10, 11.
Here we illustrate a new technology we call **S**ynthetic **A**ttenuated **V**irus **E**ngineering (SAVE)12 as a rational design approach for building live attenuated influenza vaccines, as well as vaccines to other diseases.
The central idea of SAVE is to recode and synthesize a viral genome13 in a way that perfectly preserves the wild-type amino acid sequence, but that re-arranges existing synonymous codons to create a sub-optimal arrangement of pairs of codons12. For reasons that are not understood, some pairs of codons occur more frequently while others occur less frequently than expected (codon pair bias)14. Every examined species has a statistically significant codon pair bias15. This bias evolves slowly; yeast and humans have a radically different codon pair bias, but all mammals share essentially the same codon pair bias (unpublished results).
Codon pair bias is independent of codon bias. For example, consider the amino acid pair Arg-Glu. Since there are six codons for Arg and two for Glu, there are 12 encodings for this pair of amino acids. Taking into account the frequency of the two contributing codons (codon bias), the pair CGC-GAA is expected 2397 times in the annotated human ORFeome, but in fact is observed only 268 times (observed/expected = 0.11). This is an infrequently used codon pair. In contrast, the Arg-Glu encoding AGA-GAA is expected 2644 times, but is observed 4195 times (observed/expected = 1.59); this is a frequently used codon pair. By whole genome synthesis13,16 we previously recoded poliovirus so as to contain “poor” (i.e., infrequently used) codon pairs, and found that this dramatically attenuated the virus12. Although the mechanism of attenuation is unclear, preliminary evidence suggests that translation is affected12. Attenuation can be “titrated” by adjusting the extent of codon pair deoptimization12. Because codon pair deoptimization is due to miniscule effects at each of hundreds or thousands of nucleotide mutations (without changing amino acid sequences), reversion to virulence is extremely unlikely12. Aided by computer algorithms12, codon pair-deoptimized viral genomes can be rapidly designed and synthesized, and live virus can be generated by reverse genetics.
To attenuate influenza virus, we redesigned large parts of the coding regions of the PB1, NP, and HA genes of influenza virus A/PR/8/34 (“PR8”), using our deoptimization computer program12. These genes play important roles in genome replication and assembly (although other genes may also have served). Without altering either amino acid sequence or codon bias, the program re-arranged existing synonymous codons to deoptimize codon pairs. This resulted in hundreds of silent mutations per genome segment without any amino acid changes. The characteristics of the synthetic genome segments and their changes in Codon Pair Bias (CPB) are summarized in Table 1 and in Figure S1.
Table 1.
Characteristics of deoptimized Influenza Genome Segments
| Gene Segment | Deoptimized Coding Regiona | CPB of wt Segmentb | CPB of deoptimized Segmentc | Number of silent Mutations |
|---|---|---|---|---|
| NPMin | 125-1426 | 0.012 | -0.421 | 314 |
| PB1Min | 519-1494 | 0.007 | -0.386 | 236 |
| HAMin | 157-1654 | 0.019 | -0.420 | 353 |
The deoptimized segments were synthesized de novo and cloned into a standard ambisense, 8-plasmid system17,18. To generate influenza viruses carrying one or more deoptimized segments, the plasmids carrying the recoded, synthetic segments, together with the complement of the remaining PR8 wt plasmids, were transfected into susceptible cells (Materials and Methods). Each deoptimized segment PB1Min, NPMin, and HAMin in the background of the complementing 7 wt segments yielded a viable virus, as did any combination thereof, including the virus simultaneously containing all three deoptimized segments (PR8-PB1/NP/HAMin, abbreviated “PR83F”) (Fig. 1; and data not shown).
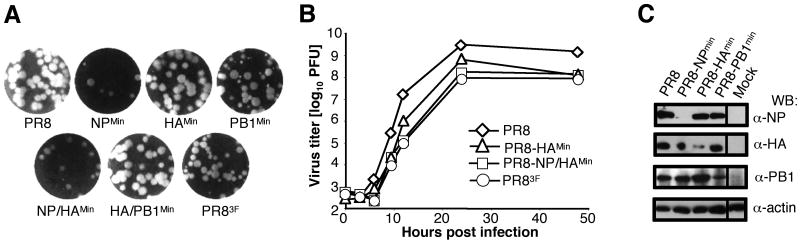
Fig. 1. Characterization in tissue culture of synthetic codon pair-deoptimized Influenza viruses.
(A) Plaque phenotypes on MDCK cells of PR8 wild type virus and synthetic PR8 derivatives, carrying one (NPMin, HAMin, PB1Min), two (NP/HAMin; HA/PB1Min) or three (PR83F) deoptimized gene segments. (B) Growth kinetics of PR8 wild type virus and three synthetic PR8 derivatives in MDCK cells after infection with 0.001 MOI of the indicated viruses. All combinations grew within log 8-9 although only PR8-HAMin, PR8-NP/HAMin and PR83F are shown as compared to wild type. (C) Analysis of Influenza virus protein expression in infected cells. MDCK cells were infected with PR8 wild type virus or synthetic PR8 derivatives, carrying one deoptimized gene segment each (NPMin, HAMin, PB1Min), as indicated. Western blot (WB) analysis of proteins extracted from whole cell lysates was carried out with PB1, NP, HA, or actin antibodies. Actin was used to indicate equal protein loading.
Several of these new synthetic viruses were analyzed for their in vitro growth characteristics in MDCK cells. All mutant viruses formed plaques that were either indistinguishable from, or only slightly smaller than that of the wt virus (Fig. 1A). The mutant viruses grew less well than wt, but typically only to about ten-fold lower titers (Fig. 1B). The properties of viruses carrying combinations of synthetic segments other than depicted in Fig. 1 fall in between the curves or plaque phenotypes of PR8 and PR83F (data not shown). The deoptimized virus PR83F had about the same response to changes in temperature (to either 33°C or 39.5°C) as did the wild type virus (Fig. S3).
Previously, we found that codon or codon pair-deoptimized polioviruses had a reduced specific infectivity12,19. This was not the case for deoptimized influenza viruses, as their ratio of PFU to HA units was nearly identical to wt (data not shown).
Our working hypothesis postulates that deoptimization of viral ORFs results in reduced protein synthesis, which in turn brings forth a virus that is attenuated12. Therefore we tested protein synthesis driven from the deoptimized gene segments in PR8-NPMin, PR8-HAMin, and PR8-PB1Min by Western blotting of infected whole cell lysates (Fig. 1C). In all three viruses the deoptimized viral gene product was specifically reduced compared to other proteins from the same virus, or to the same protein in wild type-infected cells, or to the actin control in the same cells (Fig. 1C). Protein synthesis from the wild type segments in a deoptimized virus was apparently not affected. The exact molecular mechanisms responsible for the reduced protein production remain to be determined.
Despite their reasonably robust growth, codon pair-deoptimized influenza viruses proved to be remarkably attenuated in mice (Table 2). Each individual deoptimized segment had a demonstrable attenuating effect, leading to a reduction in LD50 of about 10, 30, and 500 fold, for PR8-NPMin, PR8-HAMin, and PR8-PB1Min, respectively. Combinations of two deoptimized genes have not been tested, but combining all three attenuating genes into one virus (PR83F) led to a cumulative attenuation of about 13,000 fold (Table 2).
Table 2.
Lethal Dose 50 and Protective Dose 50 of deoptimized Influenza Viruses
| Virus | LD50 (PFU)a | PD50 (PFU)b |
|---|---|---|
| PR8 (wt) | 6.1 × 101 | ∼1.0 × 100c |
| PR8-NPMin | 5.0 × 102 | n.d. d |
| PR8-PB1Min | 3.2 × 104 | n.d. |
| PR8-HAMin | 1.7 × 103 | n.d. |
| PR8-NP/HA/PB1Min (PR83F) | 7.9 × 105 | 1.3 × 101 |
To test the codon pair-deoptimized viruses in animals, BALB/c mice were infected intranasally with 104 PFU of PR83F or PR8, and monitored for disease symptoms (ruffled fur, lethargy, weight loss, death). At this dose, mice infected with wild-type PR8 developed severe symptoms with rapid weight loss and did not survive beyond day 5 (Fig. 2A). Mice infected with PR83F, on the other hand, experienced no observable symptoms or weight loss, save for a small, transient delay in weight gain as compared to mock-infected animals (Fig. 2A).
Fig. 2. Attenuation of codon pair-deoptimized Influenza virus PR83F in BALB/c mice.
(A) Body weight curve following intranasal infection with 104 PFU of PR8 wild type (triangles), 104 PFU of deoptimized PR83F (diamonds), or mock-infected (saline; squares). The average of 5 mice per time point and standard deviations are indicated. Wild type-infected mice did not survive beyond day 5 (indicated by a cross). (B) Virus titer in whole lung homogenate after infection with either 103 PFU PR8 wild type (squares), or deoptimized PR83F (circles). Average of three mice per time point. * On day 9 post infection, PR83F was no longer detectable (below 40 PFU/lung).
Live attenuated virus vaccines depend on a limited, yet safe, degree of replication within the host to stimulate the immune system. To assess the replicative potential of these viruses in an immune competent host, we monitored viral load in lungs of BALB/c mice infected intranasally with either 103 PFU of PR83F or PR8 wt virus. Within 24 hours, wt-infected mice were marked by 3000 fold higher viral load in their lungs compared to PR83F, leading to death in under 6 days (Fig. 2B). Conversely, in PR83F-infected animals, amplification of the vaccine virus progressed slowly and peaked at a lower viral load than the wild type virus, resulting in a controlled infection with no overt disease symptoms, and virus clearance after nine days (Fig. 2B).
Infection by a sub-lethal dose of wild type virus can in principle provoke protective immunity, which is the usual course of natural human infections. The Chinese scholar Li Shizhen described the art of inoculating humans with live smallpox in his Compendium of Materia Medica (1593)18. This method of smallpox vaccination, practiced in China for centuries20, was very dangerous because the difference between the lethal dose (LD) and the immunizing protective dose (PD) of wt smallpox is very small. To address the issue of safety margin quantitatively with our influenza viruses, we determined the LD50 and the protective dose 50 (PD50, the dose providing protective immunity to half the animals) for wild type PR8 and also for the attenuated strain, PR83F (Fig. 3). PR8 had a very low PD50 of 1 PFU (due to robust replication). (1 PFU of PR8, titred on MDCK cells, is about 40 virus particles21). The LD50 of PR8 was 61 PFU, resulting in an LD50/PD50 ratio of about 60. This ratio between the LD50 dose and the PD50 dose is the “safety margin” of a given virus if it were to be used as a vaccine. The safety margin of the wt (LD50/PD50 = 60) is narrow--hence it is inadequate as a vaccine. In contrast, the attenuated virus PR83F had a PD50 of 13 PFU, higher than the PD50 of the wild-type, but still very low. Strikingly, the attenuated PR83F had an LD50 of 790,000 PFU and, thus, an LD50/PD50 ratio (safety margin) of 60,000, which is 1000-fold better than the wild-type virus (Fig. 3A versus Fig. 3B, shaded areas under the curve). Thus, it is easy to determine a dose of the attenuated virus PR83F that is both safe to administer and effective in provoking immunity (see also Fig. S2).
Fig. 3. Immune responses and protection.
(A, B) Vaccine Margin of Safety for PR8 wild type and deoptimized PR83F viruses. The left ordinate indicates the percentage of animals surviving the primary inoculation (black squares) with (A) PR83F or (B) wt PR8, at doses ranging between 100 to 106 PFU. After 28 days, the surviving, vaccinated animals were challenged with a single 1000 × lethal dose 50 of PR8 wild type virus. Disease and survival were monitored (right ordinate; open circles) for (A) PR83F- and (B) PR8-vaccinated mice. (C) Virus load in mouse lungs following wild type challenge of PR83F-vaccinated animals. 28 days following a single intranasal vaccination with 104 PFU PR83F mice were challenged with 1000 × LD50 of PR8 wt virus. Three days thereafter the level of challenge virus in lung homogenates was determined. (D) ELSIA determination of influenza-specific serum antibodies. 28 days after a primary infection, serum was collected, and anti-influenza IgG serum antibody titers were determined from animals that had received a primary inoculation of 0.01 × LD50 (black diamonds) or 0.001 × LD50 of PR83F (black circles), 0.01 × LD50 of PR8 (white squares), or saline (black triangles). ELISA antibody titer against PR8 virus antigen is expressed as the lowest reciprocal serum dilution that resulted in a positive ELISA signal (5 standard deviation above background).
To characterize the protective immunity in more detail, we immunized mice with a single intranasal vaccination of 104 PFU of PR83F. After 28 days, these mice were challenged with 1000 × LD50 of wt PR8. Three days after challenge, PR8 titres in lung homogenates were determined. In 80% of the mice, challenge virus was below the level of detection (suppressed at least 106-fold) (Fig. 3C; open circles), whereas titers of ∼107 PFU were found in lungs of mock-vaccinated animals (Fig. 3C; open squares).
The mean anti-influenza serum IgG-antibody titer in mice immunized with 0.01 × LD50 of the respective viruses was 312,500 for PR83F and 27,540 for PR8 (Fig. 4C). At an even lower and, thus, safer vaccine dose of 0.001 × LD50 the immune response toward PR83F was nearly unchanged with an antibody titer of 237,500 (Fig. 4C). Thus, at identical doses relative to their respective LD50, PR83F is a more potent inducer of influenza-specific antibodies than the wild type. These findings attest to the strong immunizing potential of a low-grade influenza virus infection in general, and to the safety profile of codon pair-deoptimized influenza viruses in particular.
Vaccines created in this way encode all viral proteins with the exact wild-type amino acid sequences. Thus the viruses express the entire wild-type repertoire of antigenic sites and would have the maximum chance of inducing both cellular and humoral immunity against all epitopes. Since attenuation results from hundreds or thousands of nucleotide changes the probability of reversion to virulence is extremely low, an advantage over other approaches to making live vaccines. Co-infection of the vaccine recipient with a naturally circulating wt virus could lead to gene reassortment, but this is unlikely to produce variants more virulent than the co-infecting virus. The deoptimized segments of the vaccine strain would “poison” any such reassortant.
In spite of striking genetic dissimilarities, codon pair deoptimization has given similar results with both influenza virus and poliovirus12. However, poliovirus is a plus-stranded RNA virus whose genome encodes a single polyprotein. Codon pair-deoptimization in the 5′-region of the poliovirus genome disturbs the synthesis of all viral proteins with drastic effects on replication12. In contrast, influenza virus, a minus-stranded RNA virus, has a genome divided into eight segments and its proteins are expressed independently from individual mRNAs. The fact that two such extremely different viruses each are highly sensitive to codon pair deoptimization suggests that the strategy may operate quite generally.
As with any new technology, there are issues to resolve before applying this approach for human use. These include our incomplete knowledge of the molecular mechanism(s) involved in attenuation, an in-depth assessment of the genetic stability of the attenuation phenotype, and a need to find the right balance between attenuation (for provoking an immune response) and robust replication (for purposes of production). For seasonal epidemics, SAVE may allow a different strategy than the existing live vaccines, such as FluMist™. For FluMist™, the attenuating mutations (obtained by lengthy selection procedures) map to six viral “backbone” genes, which therefore must be kept constant every year, with only HA and NA being updated annually (“6:2 recombinants”). Individuals immunized yearly could develop immunity against “backbone” proteins, limiting the replication and thus the efficacy of the vaccine (possibly helping to explain the relatively poor efficacy of FluMist in previously-immunized army personnel22). In contrast, using SAVE one could attenuate the entire impending seasonal or pandemic strain's genome, providing a perfect antigenic match between the vaccine and target virus. However, this approach would require an appropriate regulatory environment that might, for instance, approve a method of attenuating each new strain to a certain standardized degree with a standardized change in codon pair bias (just as the current inactivated virus vaccine uses a standardized method of inactivation). To attenuate in a standardized and predictable way will require many more experiments to exhaustively explore the variables.
The recoded influenza viruses described here present a novel paradigm for vaccines, since attenuation is the result of hundreds or thousands of nucleotide changes without change of a single amino acid. The attenuated phenotype results from large-scale rearrangements of existing synonymous codons, producing under-represented pairs of codons12. Combined with (i) the expected high genetic stability of the attenuating genetic changes (“attenuation by thousand cuts”23), (ii) the possibility of systematically designing such viruses, the favorable growth kinetics in tissue culture (108 PFU/ml), (iii) the small protective dose of “deoptimized” influenza viruses, (iv) the efficacy of PR83F as revealed in challenge experiments, and, particularly, (v) the wide safety margin, the SAVE technology sets the stage for making efficient live attenuated influenza vaccines. While it is difficult to extrapolate to human use, in our system 10 milliliter of culture supernatant would appear to contain enough virus to vaccinate and protect approximately 1 million mice with a single vaccination of 100 PD50 doses of PR83F (Fig. 3A, Fig. S2).
Vaccines based on changes in codon pair bias could be generated within weeks for any emerging influenza virus once its genome sequence is known, although of course a further period of testing would be required before the vaccine could be used. The margin of safety appears to be unusually high. This new strategy will be applicable to the rapid development of human vaccines against seasonal flu epidemics and pandemics.
Materials and Methods
Cell lines
293T and Madin Darby Canine Kidney cells (MDCK) cells were obtained from the American Type Culture Collection (ATCC). Cells were grown in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum (HyClone) and penicillin-streptomycin (Invitrogen).
Reverse Genetics and Generation of Synthetic Influenza Viruses
All synthetic influenza viruses used here are based on the strain A/PR/8/34 (Mt. Sinai variant, short PR8). The reference sequences of the 8 gene segments for this strain are available under genbank accession numbers AF389115, AF389116, AF389117, AF389118, AF389119, AF389120, AF389121, AF389122). An 8-plasmid ambisense system for this strain cloned in the vector pDZ 24 was kindly made available by Peter Palese and Adolfo García-Sastre (Mt. Sinai School of Medicine).
Codon pair-deoptimized genome segments were designed using a computer algorithm previously described12. Coding regions of the segments PB1, HA, and NP were targeted to be recoded. A minimum of 120 nucleotides at either segment terminus were left unaltered since these sequences contain packaging signals2. All of the codon changes were synonymous; i.e., none of them introduced any amino acid changes. The precise extent of recoded sequence for each target segment and the resulting changes in codon pair bias are summarized in Table 1.
The recoded gene segments were synthesized de novo (Mr. Gene GmbH, Regensburg, Germany) and introduced into wt plasmids to replace the respective wt counterpart sequence. Codon pair-deoptimized viruses were derived by DNA transfection of either one, or combinations of two, or three deoptimized segments together with the remaining wt plasmids. For this purpose a total of 2μg plasmid DNA (250ng of each of 8 plasmids) was transfected into co-cultures of 293T and MDCK cells in 35mm dishes using Lipofectamine 2000 (Invitrogen) according to manufacturers recommendations. After 6 hours of incubation at 37 °C, the serum free Opti-MEM containing the transfection mix was replaced with DMEM containing 0.2% Bovine Serum Albumin (BSA). After a further 24 hours of incubation, 1 μg/ml TPCK-Trypsin was added to the dishes. Two days thereafter virus containing cell supernatants were collected and amplified on MDCK cells.
In vitro Growth characteristics and titration of synthetic Influenza viruses
The growth characteristics of codon pair-deoptimized synthetic viruses were analyzed by infecting confluent monolayers of MDCK cells in 100 mm dishes with 0.001 multiplicities of infection (MOI). Infected cells were incubated at 37°C in DMEM, containing 0.2% Bovine Serum Albumin (BSA) and 2μg/ml TPCK-Trypsin (Pierce, Rockford, IL). At the given time points 200 μl of supernatant was removed and stored at -80°C until titration. Viral titers and plaque phenotypes were determined by plaque assay on confluent monolayers of MDCK cells in 35mm six well plates using a semisolid overlay of 0.6% tragacanth gum (Sigma-Aldrich) in minimal Eagle medium (MEM) containing 0.2% Bovine Serum Albumin (BSA) and 4μg/ml TPCK-Trypsin. After 72 hours of incubation at 37°C plaques were visualized by staining the wells with crystal violet.
Mouse pathogenicity, in vivo virus replication, and vaccination
A minimum of 5 BALB/c mice (5-6 weeks old) per group were infected once by intranasal inoculation with doses ranging from 100 to 106 PFU of PR83F or of wt PR8. Inoculum virus was diluted in 25μl PBS and administered evenly into both nostrils. A control group of 5 mice was inoclulated with PBS only (mock). Venous blood from the tail vein was collected from all animals prior to initial infection for subsequent determination of pre-vaccination antibody titers.
Morbidity and mortality (weight loss, reduced activity, death) was monitored. The Lethal Dose 50, LD50, of the wt virus and the vaccine candidates was calculated by the method of Reed and Muench 25. Mice experiencing severe disease symptoms (rapid, excessive weight loss over 25%) were euthanized and scored as a lethal outcome.
For vaccination experiments mice were infected as above. 28 days following the initial infection (vaccination), venous blood from the tail vein was drawn for subsequent determination of post-vaccination antibody titers.
The mice were then challenged with 105 PFU of the wt virus PR8 corresponding to more than 1000 times the LD50. Mortality and morbidity (weight loss, reduced activity, death) was monitored. The Protective Dose 50 (PD50) of codon pair-deoptimized PR8-3F versus that of the PR8 was determined as the dose required to protect 50% of mice from a challenge with 1000 × LD50 of the wt virus, 28 days after a single inoculation with the vaccine virus.
To assess virus replication in the lungs of infected animals, BALB/c mice were infected intranasally with 103 PFU of either PR8 or PR83F. At 1, 3, 5, 7, and 9 days post infection the lungs of three mice each were collected (wt infected mice did not survive beyond day 6). Lungs were homogenized in 1 ml of PBS and the virus load per organ was determined by plaque assay on MDCK cells, as described above. Similarly, replication of PR8 challenge virus in lungs of PR83F-vaccinated mice were determined. 28 days following a single intranasal inoculation with 104 PFU of PR83F or saline, 5 animals each were challenged with 1000 × LD50 of PR8. 3 days after challenge (the usual peak of viral replication), lungs were processed and viral load per organ determined as described above.
Western Blot analysis of Influenza virus proteins in infected cells
MDCK cells were infected with virus at a multiplicity of infection of 5 and incubated for 4 hours at 37°C. Subsequently, the cells were harvested in Laemmli buffer. The proteins were resolved by SDS-PAGE, analyzed by Western blotting with α-HA, α-PB1 and α-actin antibodies (Santa Cruz Biotechnology), and α-NP antibody (generous gift from Dr. Peter Palese), and HRP-conjugated secondary antibodies, and detected by autoradiography.
ELISA Determination of Influenza-specific serum IgG-antibodies after vaccination
Nunc Maxisorp ELISA 96 well plates were coated over night with 100 ng purified Influenza PR8 virus in 100μl PBS followed by blocking with 100 μl 1% BSA in PBS. Serial 5-fold dilutions in PBS/1% BSA of mouse sera obtained prior to and 28 days after a single intranasal vaccination were incubated for 2 hours at room temperature. Mice were previously vaccinated with approximately 0.01 or 0.001 × LD50 of PR83F (103 PFU or 104 PFU, respectively), 0.01 × LD50 of PR8 wt (100 PFU) or mock vaccinated. After 4 washes with PBS the wells were incubated with 1:500 of anti mouse-alkaline phosphatase conjugated secondary anti-mouse IgG antibody (Santa Cruz) for another 2 hours at room temperature. Following 4 washes with PBS and brief rinsing with distilled water 100 μl of a chromatogenic substrate solution containing 9mg/ml p-nitrophenylphosphate in 200 mM diethanolamine, 1 mM MgCl2, pH 9.8 was added. The color reaction was stopped by addition of an equal volume of 500 mM NaOH. Absorbance at 405 nm was read using a Molecular Devices ELISA reader. The endpoint antibody titer was defined as the highest dilution of serum that gave a signal greater than 5 standard deviations above background. Background level was determined from wells processed identically to experimental samples, in the absence of any mouse serum.
Supplementary Material
1
2
3
Acknowledgments
The authors are indebted to Adolfo García-Sastre and Peter Palese for sharing with us their 8-plasmid system for PR8, antibodies, and information. We thank Aniko Paul and Jeronimo Cello for comments on the manuscript. All authors declare they have a patent pending relating to certain aspects of this work. Supported by NIH grants AI075219 and AI15122 (EW) and a TRO-Fusion Award by Stony Brook University (SM and SS).
References
- 1.Salomon R, Webster RG. The influenza virus enigma. Cell. 2009;136:402–410. doi: 10.1016/j.cell.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palese P, Shaw ML. In: Field's Virology. Knipe DM, et al., editors. Vol. 2. Lippincott Williams & Wilkins (LWW); Philadelphia: 2007. pp. 1647–1689. [Google Scholar]
- 3.Steinhauer D, Skehel JJ. Genetics of influenza viruses. Annu Rev Genet. 2002;36:305–332. doi: 10.1146/annurev.genet.36.052402.152757. [DOI] [PubMed] [Google Scholar]
- 4.Maassab HF. Adaptation and growth characteristics of influenza virus at 25 degrees c. Nature. 1967;213:612–614. doi: 10.1038/213612a0. [DOI] [PubMed] [Google Scholar]
- 5.Rimmelzwaan GF, Fouchier RA, Osterhaus AD. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol. 2007;18:529–536. doi: 10.1016/j.copbio.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Sant AJ, et al. Immunodominance in CD4 T-cell responses: implications for immune responses to influenza virus and for vaccine design. Expert Rev Vaccines. 2007;6:357–368. doi: 10.1586/14760584.6.3.357. [DOI] [PubMed] [Google Scholar]
- 7.Hikono H, et al. T-cell memory and recall responses to respiratory virus infections. Immunol Rev. 2006;211:119–132. doi: 10.1111/j.0105-2896.2006.00385.x. [DOI] [PubMed] [Google Scholar]
- 8.Swain SL, et al. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunol Rev. 2006;211:8–22. doi: 10.1111/j.0105-2896.2006.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simonsen L, et al. Impact of influenza vaccination on seasonal mortality in the US elderly population. Arch Intern Med. 2005;165:265–272. doi: 10.1001/archinte.165.3.265. [DOI] [PubMed] [Google Scholar]
- 10.Belshe RB, Van Voris LP, Bartram J, Crookshanks FK. Live attenuated influenza A virus vaccines in children: results of a field trial. J Infect Dis. 1984;150:834–840. doi: 10.1093/infdis/150.6.834. [DOI] [PubMed] [Google Scholar]
- 11.Belshe RB, et al. The efficacy of live attenuated, cold-adapted, trivalent, intranasal influenzavirus vaccine in children. N Engl J Med. 1998;338:1405–1412. doi: 10.1056/NEJM199805143382002. [DOI] [PubMed] [Google Scholar]
- 12.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 14.Gutman GA, Hatfield GW. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moura G, et al. Large scale comparative codon-pair context analysis unveils general rules that fine-tune evolution of mRNA primary structure. PLoS ONE. 2007;2:e847. doi: 10.1371/journal.pone.0000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wimmer E, Mueller S, Tumpey TM, Taubenberger JK. Synthetic viruses: a new opportunity to understand and prevent viral disease. Nat Biotechnol. 2009;27:1163–1172. doi: 10.1038/nbt.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schickli JH, et al. Plasmid-only rescue of influenza A virus vaccine candidates. Philos Trans R Soc Lond B Biol Sci. 2001;356:1965–1973. doi: 10.1098/rstb.2001.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shizhen L. Bencao Gangmu: Compendium of Materia Medica. 1593 [Google Scholar]
- 21.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J Virol. 2008;82:11869–11879. doi: 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Tobler S, Roayaei J, Eick A. Live attenuated or inactivated influenza vaccines and medical encounters for respiratory illnesses among US military personnel. JAMA. 2009;301:945–953. doi: 10.1001/jama.2009.265. [DOI] [PubMed] [Google Scholar]
- 23.Coffin JM. Attenuation by a thousand cuts. N Engl J Med. 2008;359:2283–2285. doi: 10.1056/NEJMcibr0805820. [DOI] [PubMed] [Google Scholar]
- 24.Quinlivan M, et al. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol. 2005;79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed LJ, Muench M. A simple method for estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3