RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage (original) (raw)
Abstract
Dnmt2 proteins are the most conserved members of the DNA methyltransferase enzyme family, but their substrate specificity and biological functions have been a subject of controversy. We show here that, in addition to tRNAAsp-GTC, tRNAVal-AAC and tRNAGly-GCC are also methylated by Dnmt2. Drosophila Dnmt2 mutants showed reduced viability under stress conditions, and Dnmt2 relocalized to stress granules following heat shock. Strikingly, stress-induced cleavage of tRNAs was Dnmt2-dependent, and Dnmt2-mediated methylation protected tRNAs against ribonuclease cleavage. These results uncover a novel biological function of Dnmt2-mediated tRNA methylation, and suggest a role for Dnmt2 enzymes during the biogenesis of tRNA-derived small RNAs.
Keywords: Cytosine methylation, Dnmt2, tRNA, angiogenin
The covalent modification of nucleic acids plays an important role in regulating the functions of DNA and RNA. DNA modifications have been analyzed in considerable detail, and the characterization of (cytosine-5) DNA methylation has been crucial for understanding the molecular basis of epigenetic gene regulation (Klose and Bird 2006). (Cytosine-5) methylation has also been documented in various RNA species, including tRNA, but the function of RNA methylation has not been firmly established yet (Motorin et al. 2010).
Dnmt2 proteins were originally assigned to the DNA methyltransferase family, because of their strong sequence conservation of catalytic DNA methyltransferase motifs (Okano et al. 1998; Yoder and Bestor 1998). A recent study has suggested that Dnmt2-mediated DNA methylation is important for transposon silencing in Drosophila (Phalke et al. 2009). However, only a weak and distributive DNA methylation activity has been reported in various systems (Jeltsch et al. 2006).
The ambiguities associated with the DNA methyltransferase activity of Dnmt2 have also prompted the search for alternative enzyme substrates, and resulted in the discovery of a tRNA methyltransferase activity of Dnmt2 (Goll et al. 2006). Purified recombinant human Dnmt2 methylated RNA preparations from Dnmt2 mutant mice, flies, and plants. Further experiments identified C38 in the anti-codon loop of tRNAAsp as the methylation target site of Dnmt2 (Goll et al. 2006). However, the functional relevance of the tRNA methyltransferase activity of Dnmt2 remains to be established. Dnmt2 mutant mice, flies, and plants were reported to be viable and fertile (Goll et al. 2006) under standard laboratory conditions. A distinct Dnmt2 mutant phenotype, caused by morpholino knockdown experiments, has so far been reported only in zebrafish, leading to lethal differentiation defects in the retina, liver, and brain (Rai et al. 2007). In addition, two studies have indicated increased stress tolerance in Dnmt2-overexpressing flies and amoebas (Lin et al. 2005; Fisher et al. 2006). However, the underlying molecular mechanisms have not been investigated yet.
Results and Discussion
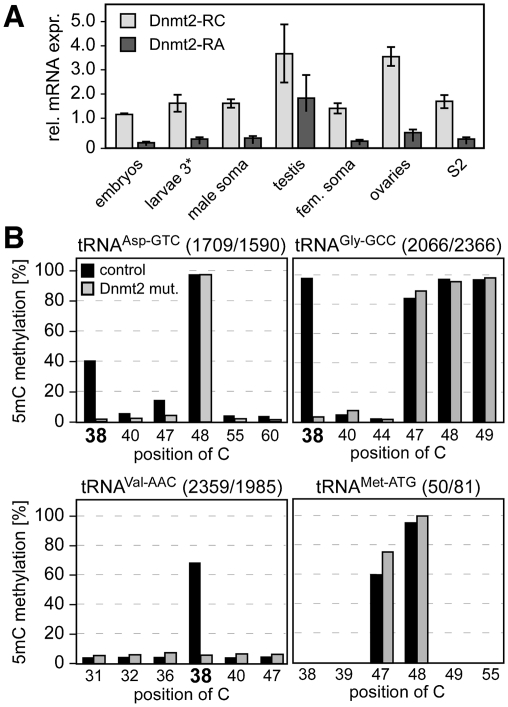
The Drosophila Dnmt2 locus (chromosome 2, 33C4) gives rise to two transcripts: Dnmt2-RC and Dnmt2-RA. The predicted protein products Dnmt2-PC and Dnmt2-PA differ in their N termini by a peptide stretch of 21 amino acids (Supplemental Fig. S1A), which eliminates the catalytic DNA methyltransferase motif I from the putative Dnmt2-PA protein. Both transcripts were detectable during all stages of development and in adult flies (Fig. 1A), suggesting a general function throughout development.
Figure 1.
Dnmt2 is a multisubstrate tRNA methyltransferase. (A) Quantitative PCR analysis of Dnmt2 mRNA expression in Drosophila tissues. Expression was normalized to rp49 levels. (B) Identification of novel Dnmt2 substrate tRNAs. Deep bisulfite sequencing analysis of various tRNAs in wild-type (control) and Dnmt2 mutant (Dnmt2 mut.) flies. Numbers of sequence reads are indicated at the top of each panel. Diagrams show fractions of nondeaminated cytosines, thus revealing the pattern of m5C modification in individual tRNAs. Cytosines that were found to be methylated in a Dnmt2-dependent manner are C38 in tRNAAsp-GTC, tRNAVal-AAC, and tRNAGly-GCC (highlighted by bold numbers).
In order to establish a mutant allele that causes the loss of both transcripts, we generated a deletion in the genomic Dnmt2 locus by mobilizing the P-element EP(2)GE15695, which is located upstream of the Dnmt2 coding sequence (Jurkowski et al. 2008). Since the P-element insertion mapped to the promoter region of both Dnmt2 and the adjacent CG6712 locus, we performed an imprecise excision screen for deletions that allowed the expression of CG6712, but deleted parts of the coding region of Dnmt2. One of the excision alleles generated (Dnmt299) contained a proximal deletion of the EP element and fused the remnant of the EP element to the 3′ half of the Dnmt2 coding region, thereby deleting large parts of the Dnmt2 gene, including motifs I–VII and part of motif VIII (Supplemental Fig. S1B). Quantitative PCR analysis showed that both transcripts generated from the Dnmt2 locus were undetectable in homozygous Dnmt299 mutants (Supplemental Fig. S1C). Dnmt2 protein could not be detected in embryonic protein extracts from homozygous Dnmt299 mutants, but was detectable as a 40-kDa polypeptide in wild-type embryos using Dnmt2-specific antibodies (Supplemental Fig. S1D). These results strongly suggested that Dnmt299 represents a null allele suitable for further analysis.
To investigate whether Dnmt2 methylates additional tRNAs, we used RNA bisulfite sequencing, a recently established method that allows the direct analysis of RNA methylation patterns in their native sequence context (Schaefer et al. 2009). Obvious candidate substrates were tRNAs that also contain a cytosine residue at position 38, assuming that substrate recognition might be similar to tRNAAsp. Among these, tRNAGly and tRNAVal warranted particular consideration, because these RNAs have been reported to be methylated at C38 (Garel and Keith 1977; Addison et al. 1982).
Using next-generation sequencing of PCR amplicons derived from bisulfite-treated total RNA, we obtained sequence information for tRNAs from adult Dnmt2 wild-type and Dnmt299 mutant animals. The analysis revealed robust methylation of C38 in tRNAAsp-GTC from control flies (Dnmt2rev). In contrast, only background methylation levels were observed in tRNAAsp-GTC from Dnmt299 mutant flies (Fig. 1B). These results confirmed previous RNA bisulfite sequencing data for tRNAAsp-GTC (Schaefer et al. 2009). Similarly, C38 of tRNAGly-GCC and tRNAVal-AAC were strongly methylated in control flies, and methylation was close to background levels in Dnmt299 mutants (Fig. 1B). Dnmt2-dependent tRNAVal methylation was also confirmed by site-specific, DNAzyme-mediated cleavage, and analysis by thin-layer chromatography (Hengesbach et al. 2008), as well as by a biochemical methylation assay using purified recombinant human Dnmt2 (Supplemental Fig. S2). Together, these results demonstrate that the tRNA methyltransferase activity of Dnmt2 is not limited to C38 of tRNAAsp-GTC, and that Dnmt2 can methylate additional tRNAs both in vitro and in vivo.
Methylation changes in Dnmt299 mutants were observed only at C38, and the methylation status of other cytosine residues in the tRNA fragments investigated did not become detectably altered in Dnmt299 mutants (Fig. 1B). A certain level of enzyme specificity was also supported by our findings that other C38-containing tRNAs—including tRNAMet-ATG, tRNAGlu-CTC, and tRNAHis-GTG—were not detectably methylated in a Dnmt2-dependent manner (Fig. 1B; data not shown).
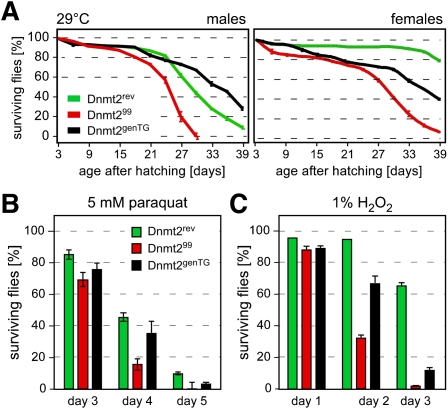
Dnmt299 mutant flies were viable and fertile under standard laboratory conditions, as described for other mutant alleles (Goll et al. 2006; Jurkowski et al. 2008). Because a previous study suggested that Dnmt2-overexpressing flies showed increased stress tolerance (Lin et al. 2005), the viability of Dnmt299 animals was analyzed under thermal stress conditions. When flies were raised continuously at 29°C (4°C above standard temperature), Dnmt299 mutants showed a reduced mean lifespan (25 d) when compared with Dnmt2rev controls (30 d) (Fig; 2A). Importantly, transgenic flies carrying a genomic copy of the Dnmt2 locus in the Dnmt299 background (Dnmt2genTG) showed a partially or fully restored lifespan (Fig. 2A), indicating that this phenotype is Dnmt2-dependent.
Figure 2.
Loss of Dnmt2 causes increased stress sensitivity. (A) Viability plot of Dnmt299 mutant, control (Dnmt2rev), and transgenic rescue (Dnmt2genTG) flies reared at elevated temperature (29°C). For each genotype and sex, the survival of 20 flies (3 d old) was counted over a period of 39 d. Error bars represent standard deviations from five independent experiments. (B) Dnmt2 mutant flies show elevated sensitivity to paraquat. For each genotype, 20 male or female adult flies were exposed to paraquat, and live flies were counted for 5 d. Error bars represent standard deviations from five independent experiments. (C) Dnmt2 mutant flies show elevated sensitivity to H2O2. Twenty male or female adult flies were exposed to 1% H2O2, and live flies were counted for 3 d. Error bars represent standard deviations from five independent experiments.
In order to examine a potential function of Dnmt2 in oxidative stress responses, adult male flies were exposed to paraquat, and surviving flies were counted daily. The results showed that the paraquat sensitivity of Dnmt299 mutant flies was substantially higher than that of control flies (Dnmt2rev) after exposure to 5 mM paraquat (Fig. 2B). Dnmt2genTG showed a robust rescue effect (Fig. 2B), confirming that the increased sensitivity to paraquat is Dnmt2-dependent. Exposing flies to another oxidizing agent, H2O2, showed that Dnmt299 mutant flies were also more sensitive to H2O2 than control flies (Dnmt2rev). The increased sensitivity to H2O2 was partially rescued when Dnmt2genTG flies were analyzed (Fig. 2C), confirming that the response to H2O2 is mediated by Dnmt2. The appearance of Dnmt2 mutant phenotypes under stress conditions implies that Dnmt2 is associated with Drosophila stress response pathways.
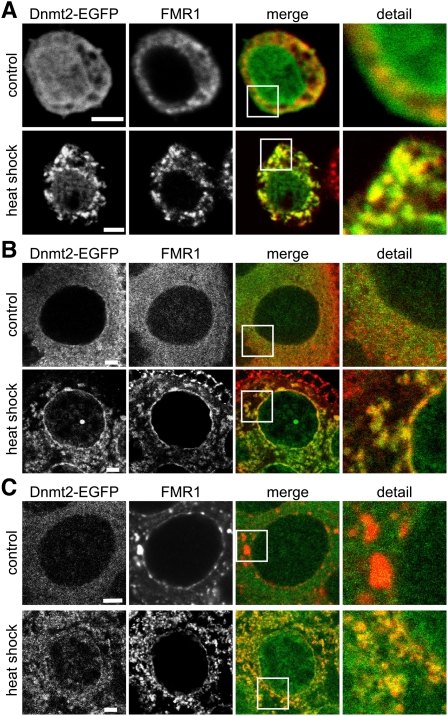
Having identified a role of Dnmt2 in stress responses, we analyzed the association of the protein with cellular stress compartments. To analyze the subcellular localization of Dnmt2 under stress conditions, we used flies expressing EGFP-tagged Dnmt2 from a genomic construct in the Dnmt299 background (Dnmt2genTG-EGFP). Activity of the tagged Dnmt2 construct was confirmed by bisulfite sequencing of tRNA from adult males, which showed that C38 methylation of tRNAAsp-GTC and tRNAGly-GCC was restored to wild-type levels (Supplemental Fig. S3). Methylation was not rescued for tRNAVal-AAC (Supplemental Fig. S3), which indicates that the transgenic construct does not completely restore Dnmt2 expression or functionality in all tissues or developmental stages. These findings are consistent with the incomplete rescue of stress phenotypes observed with tagged genomic constructs (see Fig. 2). Ovaries from Dnmt2genTG-EGFP flies showed a homogeneous, mostly cytoplasmic distribution of Dnmt2-EGFP, which aggregated into discernable granular loci after heat shock. This increase in granular Dnmt2-EGFP signal was observed in both live and fixed ovarian tissue (Supplemental Fig. S4), thus excluding fixation artifacts. Similar results were also obtained with Drosophila S2 cells stably expressing Dnmt2-EGFP, and pronounced Dnmt2-EGFP-positive granules became visible after heat shock (Fig. 3A).
Figure 3.
Dnmt2 is associated with stress compartments. (A) Immunofluorescence analysis of FMR1 in S2 cells expressing Dnmt2-EGFP. FMR1 localizes to the cytoplasm, whereas Dnmt2-EGFP is localized ubiquitiously to the cytoplasm and nucleus in control cells. Upon heat shock (1 h at 40°C), FMR1 concentrates in large cytoplasmic granules where the signal colocalizes with granular Dnmt2-EGFP. (B) Similar observations were made in ovaries after heat shock. (C) Immunofluorescence analysis of the P-body-associated protein ME31B on fixed Dnmt2-EGFP ovaries. After heat shock, the number of ME31B foci increased, and colocalization with Dnmt2-EGFP structures could be observed. Bars: A, 10 μM; B,C, 1 μM.
To confirm the association of Dnmt2 with stress-related subcellular compartments, we costained heat-shocked S2 cells for Dnmt2-EGFP and fragile X mental retardation 1 (FMR1), a ubiquitious RNA-binding protein that relocalizes to stress granules in Drosophila (Farny et al. 2009). The results showed that part of the Dnmt2-EGFP signal clearly colocalized with FMR1 loci (Fig. 3A). A distinct colocalization between Dnmt2-EGFP and FMR1 after heat shock was also observed in ovaries (Fig. 3B). Staining of Dnmt2genTG-EGFP ovaries with an antibody to ME31B, a Drosophila RNA processing body component in the germline (Lin et al. 2006), also showed processing body signals after heat shock with strong colocalization of Dnmt2 and ME31B (Fig. 3C). Together, these data illustrate the ability of Dnmt2 to localize to stress granules and RNA processing bodies following heat shock, and thus further illustrate the association between the subcellular localization of Dnmt2 and stress response pathways.
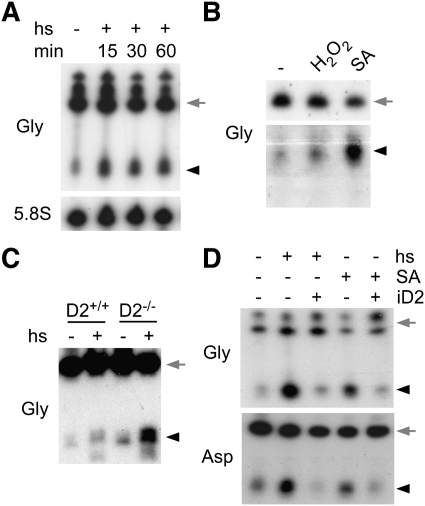
Stress granules and processing bodies contain high numbers of RNA molecules (Anderson and Kedersha 2009), and heat-shock stress responses have been linked to changes in RNA methylation in bacteria (Bugl et al. 2000). Interestingly, tRNA cleavage in the anti-codon loop has been identified recently as a molecular mechanism associated with eukaryotic stress responses (Thompson et al. 2008; Yamasaki et al. 2009). We therefore established whether tRNA cleavage under thermal and oxidative stress conditions is conserved for Dnmt2 substrates and in Drosophila. Using Drosophila S2 cells, we investigated the cleavage of tRNAGly-GCC as a representative Dnmt2 substrate. Northern analysis showed the rapid appearance of tRNAGly-GCC fragments following heat shock (Fig. 4A), and also indicated tRNAGly-GCC cleavage following oxidative stress by H2O2 and arsenite, respectively (Fig. 4B). A slight, heat-shock-dependent increase of tRNAGly-GCC fragments was also observed by Northern analysis of RNA prepared from ovaries (Fig. 4C). Notably, this effect became much more pronounced when tRNAGly-GCC cleavage was analyzed in ovaries from aged Dnmt299 mutant females (Fig. 4C), thus suggesting a role of Dnmt2 in the regulation of tRNA cleavage. To confirm these findings, we used transgenic S2 cells that allow inducible overexpression of Dnmt2. When these cells were subjected to heat shock or oxidative stress, Northern blot analysis showed the stress-dependent appearance of tRNAGly-GCC fragments (Fig. 4D). Ectopic expression of Dnmt2 substantially decreased the amount of detectable cleavage products (Fig. 4D). Essentially similar results were also obtained when analyzing tRNAAsp-GTC (Fig. 4D). Together, these findings indicate that either Dnmt2 binding to tRNAs or ectopic Dnmt2-mediated tRNA methylation protected tRNAGly-GCC and tRNAAsp-GTC molecules from cleavage.
Figure 4.
Stress-induced cleavage of Dnmt2 substrate tRNAs. (A) Northern analysis of tRNAGly-GCC cleavage in S2 cells that were heat-shocked (1 h at 40°C) for the times (in minutes) indicated. 5.8S rRNA was used as a loading control. (B) Cleavage of tRNAGly-GCC in S2 cells that were treated with H2O2 (1 mM H2O2, 60 min) or sodium arsenite (0.5 mM SA, 90 min). (C) Cleavage of tRNAGly-GCC in wild-type (D2+/+) or Dnmt2 mutant (D2−/−) ovaries (resected 35 d after hatching) that were heat-shocked (+) in medium at 40°C. Controls (−) show results from parallel experiments with ovary incubation at 25°C. (D) Northern analysis of tRNAGly-GCC and tRNAAsp-GTC cleavage in S2 cells that allow inducible overexpression of Dnmt2 (iD2). Full-length tRNAs are marked by gray arrows, and tRNA fragments are marked by black arrowheads.
The RNase A-like ribonuclease angiogenin has been shown recently to mediate stress-induced tRNA cleavage in human cells (Yamasaki et al. 2009). Since the endonuclease targeting the anti-codon loop of Drosophila tRNAs has not been identified yet, we treated Drosophila S2 cells with recombinant human angiogenin and analyzed cleavage of tRNAAsp-GTC and tRNAGly-GCC by Northern blot analysis. The results clearly showed that exogenous angiogenin induces the generation of small tRNA fragments (Fig. 5A). Importantly, Dnmt2 overexpression protected Dnmt2 substrate tRNAs from angiogenin cleavage, while tRNAMet-ATG, which is not a substrate of Dnmt2, remained unaffected by Dnmt2 overexpression (Fig. 5A). These experiments indicated that Dnmt2 functions to protect against angiogenin-mediated tRNA cleavage.
Figure 5.
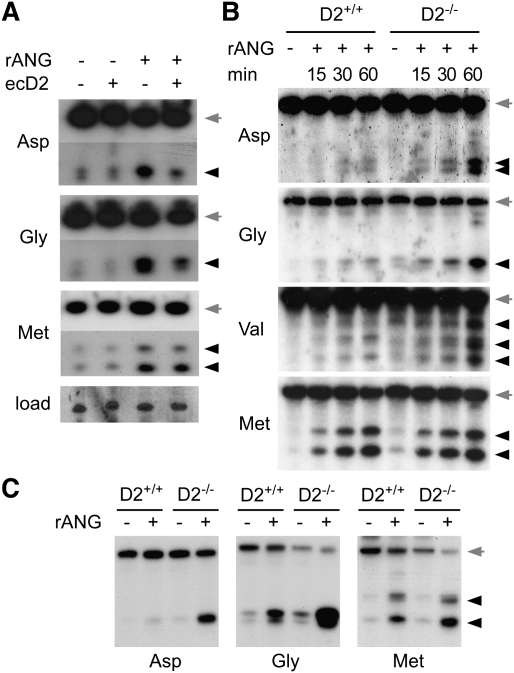
Dnmt2-mediated tRNA methylation inhibits endonucleolytic cleavage by angiogenin. (A) Dnmt2 overexpression (ecD2) inhibits angiogenin-induced tRNA cleavage. tRNA cleavage was induced in S2 cells by the addition of recombinant angiogenin (rANG) to the medium (1 μg/mL), and was analyzed by Northern blotting. Full-length tRNAs (gray arrows) and cleavage products (black arrowheads) are shown from the same original blot. (B) Dnmt2 substrate tRNAs isolated from wild-type embryos (D2+/+) are protected against angiogenin cleavage in vitro. No protection effect was observed with Dnmt2 substrate tRNAs isolated from Dnmt2 mutant embryos (D2−/−), or with tRNAMet-ATG, which is not a Dnmt2 substrate. (C) Dnmt2-mediated protection against angiogenin cleavage is conserved in mice. tRNA was purified from wild-type (D2+/+) or Dnmt2 mutant (D2−/−) MEFs, incubated with recombinant angiogenin (1 μM, 60 min), and analyzed by Northern blotting. Full-length tRNAs are marked by gray arrows, and tRNA fragments are marked by black arrowheads.
To test whether Dnmt2-mediated tRNA methylation at C38 inhibits angiogenin-catalyzed tRNA cleavage, we purified tRNAs from wild-type and Dnmt2 mutant embryos, and incubated these RNAs with recombinant angiogenin in a biochemical assay. Northern blot analysis showed very low levels of angiogenin cleavage of tRNAAsp-GTC, tRNAGly-GCC, and tRNAVal-AAC when the tRNAs were isolated from wild-type embryos (Fig. 5B). In contrast, all three Dnmt2 substrates were cleaved efficiently when the tRNAs were purified from Dnmt2 mutant embryos (Fig. 5B). The cleavage kinetics on tRNAMet-ATG was not affected by the genotype of the RNA preparation (Fig. 5B), which confirmed that C38 methylation functions to protect Dnmt2 substrate tRNAs from endonucleolytic cleavage. While small RNA sequencing studies have indicated cleavage sites in the vicinity of the Dnmt2 target nucleotide C38 (Kawaji et al. 2008; Fu et al. 2009), angiogenin is also known to be a ribonuclease with comparably low specificity (Rybak and Vallee 1988). We therefore determined the cleavage sites by sequencing of 5′-tRNA fragments from tRNAAsp-GTC, tRNAGly-GCC, and tRNAVal-AAC. The results showed cleavage at various positions in the anti-codon stem–loop (Supplemental Fig. S5), suggesting an effect of C38 methylation on the structure and endonuclease accessibility of this tRNA region.
Because of the remarkable evolutionary conservation of Dnmt2, we sought to confirm the function of Dnmt2 in endonuclease cleavage protection in a different model system. To this end, we derived embryonic fibroblasts from wild-type and Dnmt2 homozygous mutant mice (Goll et al. 2006). tRNAs were isolated from both cell lines, and Dnmt2-dependent tRNAAsp-GTC methylation at C38 was confirmed by RNA bisulfite sequencing (Supplemental Fig. S6). Purified tRNAs from wild-type and Dnmt2 mutant mouse embryonic fibroblasts (MEFs) were incubated with recombinant angiogenin, and tRNA cleavage of tRNAAsp-GTC, tRNAGly-GCC, and tRNAMet-ATG was analyzed by Northern blot. The results showed a clear protection against angiogenin cleavage for the Dnmt2 substrates tRNAAsp-GTC and tRNAGly-GCC if derived from Dnmt2 mutant MEFs, but not for tRNAMet-ATG (Fig. 5C). These data provide important confirmation for a role of Dnmt2-mediated tRNA methylation in protection from endonucleolytic cleavage.
The lack of phenotypic and molecular data has been a major obstacle in defining the biological function of Dnmt2. The distributive Dnmt2-mediated DNA methylation that was reported in several studies (Jeltsch et al. 2006) has been difficult to interpret as biologically relevant. Similarly, the methylation of a single cytosine residue in a single tRNA made it difficult to envisage a fundamental biological role for Dnmt2 enzymes. However, the initial assumption that Dnmt2 activity is restricted to tRNAAsp was based largely on a low-resolution analysis of cellular RNAs (Goll et al. 2006). The recent development of RNA bisulfite sequencing (Schaefer et al. 2009), and the use of methods for the purification and site-specific methylation analysis of defined tRNAs (Hengesbach et al. 2008) allowed us to identify additional tRNAs that were methylated by Dnmt2 in Drosophila, thus providing evidence for multisubstrate methyltransferase activity of Dnmt2. Multisubstrate specificity has been described for other RNA modification enzymes, including pseudouridine synthase 1 (Behm-Ansmant et al. 2006), and, significantly, TRM4, the only other characterized cytosine-5 tRNA methyltransferase (Motorin and Grosjean 1999). Considering that TRM4 methylates minisubstrates as small as 40 nucleotides, it remains a possibility that other unidentified substrate RNAs (e.g., small regulatory RNAs) might be methylated by RNA methyltransferases such as Dnmt2. Furthermore, since we were unable to reproduce Dnmt2-dependent DNA methylation of Invader4 transposons in Drosophila, it remains possible that the reported role of Dnmt2 in transposon silencing is mediated by the RNA methyltransferase activity of the enzyme (M Schaefer and F Lyko, in prep.).
It has been established recently that ribonucleases play an important role in the stress-induced cleavage of tRNAs (Yamasaki et al. 2009), a mechanism that promotes the assembly of stress granules (Emara et al. 2010). Our results suggest that endonucleolytic cleavage of Dnmt2 substrate tRNAs can be inhibited by the methylation mark at C38. While it is known that the mcm5s2 modification of U34 can promote the cleavage of certain yeast tRNAs by the Kluveromyces lactics γ-toxin endonuclease (Lu et al. 2008), our findings represent the first example for the inhibition of ribonucleases by cytosine-5 RNA methylation.
Defined tRNA fragments have been identified through small RNA cloning approaches (Kawaji et al. 2008; Cole et al. 2009; Haussecker et al. 2010), suggesting a physiological role for tRNA fragments. Interestingly, tRNA fragments can be processed to smaller RNAs by the Dicer RNase, indicating a potential interaction of tRNA-derived sequences and canonical small RNA processing pathways (Cole et al. 2009). Indeed, a recent study provided evidence that tRNA-derived small RNAs act to down-regulate target mRNAs, and affect different RNA silencing pathways by associating with effector core components (Haussecker et al. 2010). This suggests that altered tRNA cleavage (e.g., under stress conditions) could have phenotypic consequences.
Materials and methods
tRNA bisulfite sequencing
Total RNA isolation and bisulfite conversions were carried out as described before (Schaefer et al. 2009). Primer sequences are indicated in Supplemental Table S1. Amplicons were analyzed on a GS FLX 454 sequencer (Roche). Bioinformatic analysis was performed using a mapping script for individual bar-coded reads developed in Biopython.
Phenotype analysis
For survival assays, 20 flies were maintained on standard medium at 25°C or 29°C, 60% humidity, under a 12-h light–dark cycle, and were transferred to new medium every 3 d. Mortality was scored at transfer times. Twenty flies were tested for each genotype in five biological replicates. Paraquat (methyl viologen) and H2O2 toxicity assays were performed on 2-d-old animals placed in vials (20 per vial) with food media made of 1.3% agar containing 1% sucrose and 5 mM paraquat (Sigma) or 1% H2O2, respectively. Surviving animals were scored daily. Twenty flies were tested for each genotype in five biological replicates.
tRNA cleavage assays
S2 cells containing pRmHA3-Dnmt2-Flag constructs were induced with 0.7 mM CuSO4, chased with medium without copper for 5 h, and treated with 1 μg/mL angiogenin (R&D Systems) for 1 h at 25°C. RNA was extracted and separated on 15% urea-PAGE, transferred to Nytran SuperCharge membranes (Schleicher and Schuell Bioscience), and hybridized overnight at 47°C with 32P-end-labeled oligonucleotides (detecting both 5′ and 3′ ends of tRNAs) in hybridization solution (5× SSC, 20 mM Na2HPO4 at pH 7.4, 7% SDS, 1× Denhardt's). After washing with 3× SSC/5% SDS (15 min at 47°C) and with 1× SSC/1% SDS (15 min at room temperature), membranes were exposed to film at −80°C. For in vitro cleavage assays, 1.3 μg of urea-PAGE-purified tRNA was heated in water for 5 min at 80°C, followed by addition of cleavage buffer (30 mM Hepes at pH 6.8, 30 mM NaCl, 10 mM MgCl2). tRNA was allowed to renature for 15 min to room temperature. BSA (to 0.001%) and angiogenin (to 1 μM) were added, and the reaction was incubated for the indicated times at 37°C, followed by separation on 15% urea-PAGE, blotting, and hybridization as described above.
MEFs
MEFs were derived from day 13.5 isogenic Dnmt2−/− (Goll et al. 2006) and wild-type embryos. Single embryos were genotyped, dispersed, and trypsinized in a 10-cm-diameter dish. Cells were maintained in DMEM supplemented with 10% fetal bovine serum and a commercial cocktail of antibiotics (Invitrogen), and were passaged serially according to the 3T3 protocol (Todaro and Green 1963) until spontaneus immortalization.
See the Supplemental Material for additional details.
Acknowledgments
We thank Akira Nakamura (RIKEN Center for Developmental Biology, Japan) for providing the anti-ME31B antibody. This work was supported by grants from the Deutsche Forschungsgemeinschaft to M.H. (HE 3397/6-1), M.S., and F.L. (Priority Programme Epigenetics, FOR1082).
Footnotes
References
- Addison WR, Gillam IC, Tener GM 1982. The nucleotide sequence of tRNA4Val of Drosophila melanogaster. Chloroacetaldehyde modification as an aid to RNA sequencing. J Biol Chem 257: 674–677 [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2009. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Massenet S, Immel F, Patton JR, Motorin Y, Branlant C 2006. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA 12: 1583–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U 2000. RNA methylation under heat shock control. Mol Cell 6: 349–360 [DOI] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P 2010. Angiogenin-induced tiRNAs promote stress-induced stress granule assembly. J Biol Chem 285: 10959–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farny NG, Kedersha NL, Silver PA 2009. Metazoan stress granule assembly is mediated by P-eIF2α-dependent and -independent mechanisms. RNA 15: 1814–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher O, Siman-Tov R, Ankri S 2006. Pleiotropic phenotype in Entamoeba histolytica overexpressing DNA methyltransferase (Ehmeth). Mol Biochem Parasitol 147: 48–54 [DOI] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583: 437–442 [DOI] [PubMed] [Google Scholar]
- Garel JP, Keith G 1977. Nucleotide sequence of Bombyx mori L. tRNA1Gly. Nature 269: 350–352 [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398 [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16: 673–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengesbach M, Meusburger M, Lyko F, Helm M 2008. Use of DNAzymes for site-specific analysis of ribonucleotide modifications. RNA 14: 180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A, Nellen W, Lyko F 2006. Two substrates are better than one: Dual specificities for Dnmt2 methyltransferases. Trends Biochem Sci 31: 306–308 [DOI] [PubMed] [Google Scholar]
- Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A 2008. Human DNMT2 methylates tRNAAsp molecules using a DNA methyltransferase-like catalytic mechanism. RNA 14: 1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. 2008. Hidden layers of human small RNAs. BMC Genomics 9: 157 doi: 10.1186/1471-2164-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP 2006. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Lin MJ, Tang LY, Reddy MN, Shen CK 2005. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J Biol Chem 280: 861–864 [DOI] [PubMed] [Google Scholar]
- Lin MD, Fan SJ, Hsu WS, Chou TB 2006. Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev Cell 10: 601–613 [DOI] [PubMed] [Google Scholar]
- Lu J, Esberg A, Huang B, Bystrom AS 2008. Kluyveromyces lactis γ-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res 36: 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Grosjean H 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA 5: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Lyko F, Helm M 2010. 5-methylcytosine in RNA: Detection, enzymatic formation and biological functions. Nucleic Acids Res 38: 1415–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220 [DOI] [PubMed] [Google Scholar]
- Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G 2009. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine 5 methyltransferase DNMT2. Nat Genet 41: 696–702 [DOI] [PubMed] [Google Scholar]
- Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR 2007. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev 21: 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak SM, Vallee BL 1988. Base cleavage specificity of angiogenin with Saccharomyces cerevisiae and Escherichia coli 5S RNAs. Biochemistry 27: 2288–2294 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Lyko F 2009. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37: e12 doi: 10.1093/nar/gkn954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, Green H 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 17: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Bestor TH 1998. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum Mol Genet 7: 279–284 [DOI] [PubMed] [Google Scholar]