Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements (original) (raw)
. Author manuscript; available in PMC: 2011 Aug 27.
Summary
Chromosome replication initiates at multiple replicons and terminates when forks converge. In E. coli, the Tus-TER complex mediates polar fork converging at the terminator region and aberrant termination events challenge chromosome integrity and segregation. Since in eukaryotes termination is less characterized, we used budding yeast to identify the factors assisting fork fusion at replicating chromosomes.
Using genomic and mechanistic studies we have identified and characterized 71 chromosomal termination regions (TERs). TERs contain fork pausing elements that influence fork progression and merging. The Rrm3 DNA helicase assists fork progression across TERs counteracting the accumulation of X-shaped structures. The Top2 DNA topoisomerase associates at TERs in S-phase and G2/M facilitates fork fusion and prevents DNA breaks and genome rearrangements at TERs.
We propose that in eukaryotes replication fork barriers, Rrm3 and Top2 coordinate replication fork progression and fusion at termination regions thus counteracting abnormal genomic transitions.
Keywords: Replication termination, Top2, Rrm3, Replication pausing
Introduction
Chromosome replication initiates at multiple origins that fire throughout S-phase. Following origin firing, the replication forks move bi-directionally until they fuse with forks coming from adjacent origins (Edenberg and Huberman, 1975). In E. coli, chromosome termination takes place within a broad region containing several Tus/TER complexes, specialized polar fork barriers confining fork fusion to a site of 270 kb (Duggin et al., 2008). In eukaryotes, replication termination appears to occur randomly within a 4 kb zone (Greenfeder and Newlon, 1992a; Zhu et al., 1992). Two of the three termination regions identified in yeast contain fork pausing elements (Greenfeder and Newlon, 1992a). Certain loci, such as the RTS1 region and the rDNA locus exhibit specific termination sites (Brewer and Fangman, 1988; Dalgaard and Klar, 2000). Within these regions, specialized Fork Barriers (RFBs) mediate termination in an orientation-dependent manner arresting one of the two forks. Fork pausing can destabilize the fork and RFBs can be associated with chromosome breakage and genomic rearrangements (Kobayashi, 2006; Lambert et al., 2005). Replication forks frequently stall also at centromeres (Greenfeder and Newlon, 1992b), Replication Slow Zones (RSZs) (Cha and Kleckner, 2002), tRNA genes or Ty elements (Admire et al., 2006; Lemoine et al., 2005) and regions where collision of transcription and replication occurs (Azvolinsky et al., 2009; Deshpande and Newlon, 1996; Tuduri et al., 2009). The helicase Rrm3, a component of the replisome, facilitates fork progression through non-histone protein-DNA complexes (Ivessa et al., 2003).
Catenated intertwines can arise when two replicons fuse together (Fields-Berry and DePamphilis, 1989; Wang, 2002). In vivo and in vitro studies have implicated both type IA (Top3) and type II (Top2) topoisomerases in replication termination (Baxter and Diffley, 2008; Cuvier et al., 2008; DiNardo et al., 1984; Suski and Marians, 2008; Wang, 2002). Top3 has been involved in the resolution of sister chromatid junctions which have been also related to termination structures (Branzei et al., 2006; Chan et al., 2009). Top2 associates with chromosomal regions during S-phase (Bermejo et al., 2007) and localizes at centromeres in metaphase (Bachant et al., 2002). Cells lacking Top2 experience DNA breakage upon cell division (Holm et al., 1989).
We investigated whether in eukaryotes termination occurs at specific chromosomal loci. To identify the chromosomal termination regions, we used genomic approaches to monitor replication fork progression and fusion. We identified 71 termination regions (TERs) with an average length of 5 kb. TERs contain fork pausing elements. Rrm3 assists fork progression across TERs, and in rrm3Δ cells X-shaped intermediates accumulate at TERs. Top2, but not Top3, facilitates fork fusion and the resolution of the topological constrains at TERs. In top2 mutants, TERs accumulate breaks and rearrangements.
Altogether our results contribute to elucidate the mechanisms coordinating chromosome replication termination in eukaryotes and those cellular pathways that control the integrity of termination regions.
Results
Genomic approaches to identify termination regions (TERs)
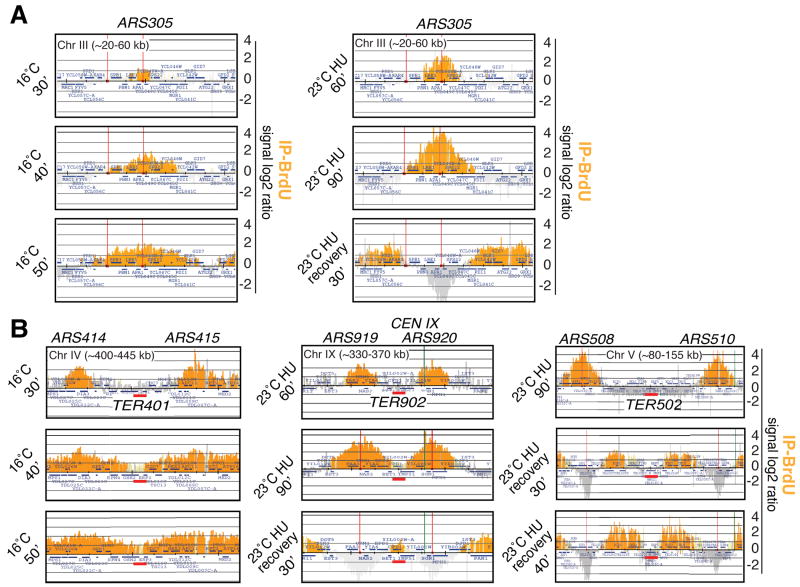
We used ChIP-chip and Bromodeoxyuridine (BrdU) incorporation (Katou et al., 2003) to monitor with time the movement of the BrdU peaks arising from origins of replication and progressively invading adjacent chromosomal regions. With this approach we were able to identify those chromosomal areas where two fork-related BrdU peaks converged. We defined as termination zones (TERs) the minimal un-replicated regions flanked by BrdU peaks arising from adjacent origins of replication. It is expected that the fork fusion sites would lie somewhere within TERs. To maximize cells synchronization we performed our experiments at low temperature or in the presence of hydroxyurea (HU) to slow down fork progression. Three sets of experiments were performed (Figure 1A): 1) wild type (wt) (Table S1) G1 cells were released in BrdU at 16°C and samples were taken ever 10 min for 1 hour. 2) G1 cells were released in BrdU and HU at 23°C and samples were collected every 30′ for 3 hour. 3) G1 cells were released in HU for 90 min and then in fresh medium with BrdU at 23°C. Samples were taken every 10 min for 90 min. Under these conditions, we specifically monitored termination of those forks arising from late origins.
Figure 1. Replication fork dynamics and fusion.
wt (sy2201) cells were arrested in G1 and released in S-phase with BrdU in 3 different sets of experiments (untreated 16°C, HU and HU recovery). Orange histogram bars (BrdU) in the Y-axis represent the average signal ratio of loci significantly enriched in the immunoprecipitated fraction (IP) along the indicated regions in log2 scale (detection p-value and change p-value are <0.001. Light orange bars should contain at least 10 contiguous probes with a p-value <0.001). The X-axis shows chromosomal coordinates. ARS elements are indicated (red lines) and the blue bars mark the ORFs. (A) Examples of fork movement monitored by BrdU incorporation at ARS305 with method 1, 2 and 3. See Table S2 for the list of the origin-related BrdU peaks (B) Visualization of 3 termination regions using the 3 methods. Red bars indicate TERs. Replication origins and experimental conditions are shown. The green line indicates the centromere. See Figure S1 and Table S3 for TERs size and position.
Consistent with previous analyses (Raghuraman et al., 2001; Yabuki et al., 2002) (http://www.oridb.org/index.php), we identified 146 BrdU peaks, corresponding to early origins and 83 to late origins (Table S2). We also identified 71 _TER_s with an average length of 5 kb (Figure S1 and Table S3). We excluded from our analysis the regions containing BrdU peaks close to telomeres and those termination areas that were either too large or not well defined. Some TERs were previously described or inferred from previous analysis (Greenfeder and Newlon, 1992a; Raghuraman et al., 2001; Zhu et al., 1992).
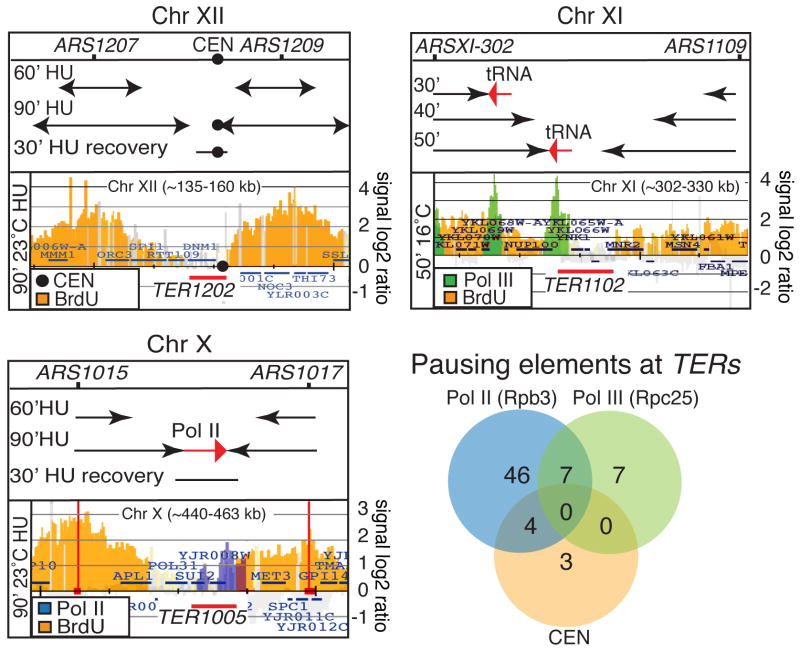
We then investigated whether fork termination at the 71 TERs correlated with loci or events that could potentially interfere with fork progression. Since Pol II and Pol III-mediated transcription interferes with replication (Azvolinsky et al., 2009; Deshpande and Newlon, 1996; Olavarrieta et al., 2002), we performed in S-phase ChIP-chip analysis of Rpb3 and Rpc25, which are subunits of RNA polymerase II and III respectively. The S-phase enrichment of Rpb3 at mRNA genes or of Rpc25 at tRNA genes and LTR (long terminal repeats), besides revealing transcription activity, may also mark potential fork pausing regions. We included in our analysis also those pausing elements that have been previous annotated (such as centomeres, RSZs and non-coding RNA genes) (Cha and Kleckner, 2002; Deshpande and Newlon, 1996; Greenfeder and Newlon, 1992b). Almost all TERs contain one or more potential replication pausing elements (examples in Figure 2 and Table S4). In fact in 64/71 cases, the TER zones contained transcription clusters and in 7/71 cases centromeres were located within TERs. We did not detect obvious features in 4/71 TERs, although in these cases transcription clusters were within a range of 1-3kb away from the TER zones (* in Table S4). Overall, 67/71 TERs contained one or more pausing elements that might affect fork progression (Table S4). The association between pausing elements and TERs is greater than random (p= 0.00021, Table S5).
Figure 2. Identification of pausing elements at TERs.
TER1202 (CHRXII), TER1102 (CHRXI) and TER1005 (CHRX) are shown as examples. BrdU-labeled forks are indicated in orange color and derive from the analysis carried out in strain sy2201 using the conditions for experiments 1 (TER1102) or experiment 2 and 3 (TER1202 and 1005) respectively. In each panel, the top part shows the extension of fork movements (black arrows) at the indicated time points based on BrdU data. In each panel, the bottom part represents a selected time point when forks reach the TER area. The black circle within TER1202 indicates the centromere. The green peaks in the bottom part of the TER1102 panel indicate the S-phase clusters of the Pol III subunit Rpc25 using strain cy8735. The blue peaks in the bottom part of the TER1005 panel indicate the S-phase clusters of the Pol II subunit Rpb3 using strain cy8519 (see methods section for details). Red arrows indicate transcription direction. Red bars mark the TER zones. The Venn diagram shows the relative number of TERs containing centromere (orange), Pol II (blue) and Pol III (green). See also Tables S4 for list of TERs containing pausing elements.
Yeast replication pause sites have been identified by mapping the high-occupancy sites of DNA polymerase ε (Polε) in wild type and rrm3Δ cells (Azvolinsky et al., 2009). We found that 47/71 TERs correlate with high occupancy Polε sites observed in wt and/or in rrm3Δ mutants, further suggesting that the replisome physiologically stalls at TERs (Figure S2 and Table S4).
At TERs, in most of the cases, transcription was on a head-on orientation with only one of the two converging forks even at those TERs that contained more than one transcription clusters. Even if we cannot always predict which of the two converging replication fork is slowed down we notice that in 62/71 cases, the pausing elements could slow either the left or the right forks but not both (Figure 2 and Table S4). This includes the 4 TERs in which the pausing elements were adjacent (* in Table S4) and 5/7 _CEN_-containing TERs where one of the two forks reaches the CEN before the other. Out of the 9 remaining TERs, in 2 cases (TER704 and TER1604), the right and left forks seemed to converge at CENs simultaneously; in 1 (TER1503) the polarity was dubious and in 6 cases (TER304, 702, 801, 1101, 1601 and 1602) termination was associated with two divergent Pol III transcribed units that potentially paused both converging forks.
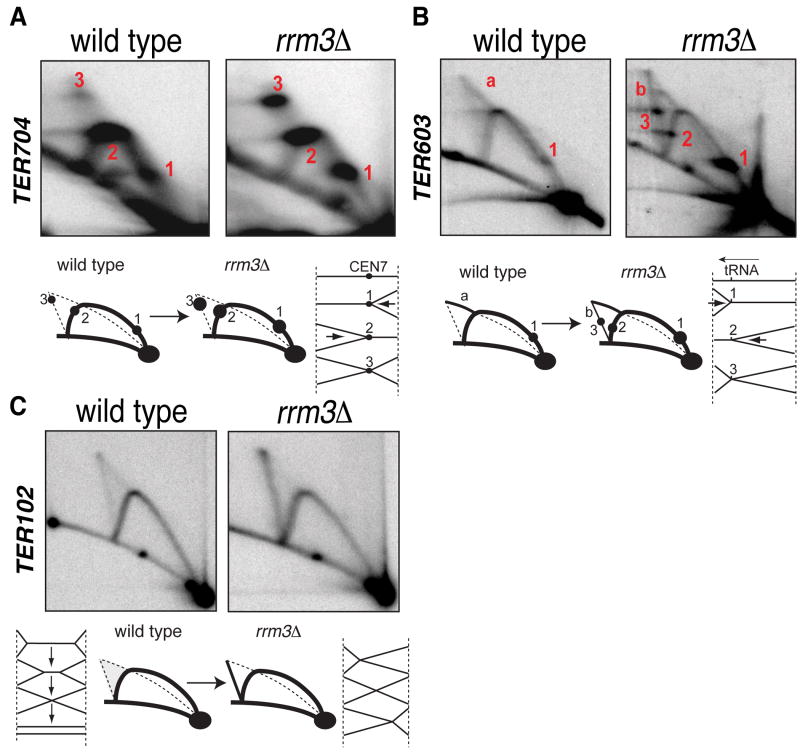
Rrm3 is required for fork progression across TERs
We used 2D gels to visualize replication intermediates at TERs in wt and rrm3 cells (Figures 3 and S3, Table S4 and data not shown). The visualization of replication termination intermediates is hampered by their fast turnover and by fork velocity. We found that the best approach to visualize termination structures is the 2D gel technique coupled with psoralen-crosslinking treatment. This procedures maximizes the visualization of the intermediates resulting form the converging of the two forks while it selectively resolves fork-related cruciform intermediates (Lopes et al., 2003)(Lopes and Foiani, unpublished observation) which are unrelated to replication termination and might interfere with the visualization of termination structures. We focused on two classes of TERs, those with (21/71) Rrm3-dependent pause sites such as CENs and tRNA genes and those without (46/71) which correlate with the presence of Pol II clusters (Azvolinsky et al., 2009).
Figure 3. Rrm3 contributes to fork progression across TERs.
(A-C) wt (sy2209) and rrm3Δ (cy6807) cells were pre-synchronized in G2 with Nocodazole and released in α-Factor. Cells were then released in S-phase at 23°C and samples collected at 40′. Genomic DNA was analyzed by 2D gels. Schematic representations of the different fork pausing and termination signals are shown. The red letters in panel b indicate double Y and Xs respectively. The red numbers indicate pausing sites (see text for details). Relative BrdU maps, restriction digestion strategy and 2D gels quantification are shown in Figure S3.
In wt cells, at TER704, two spots (1 and 2 in Figure 3A) appeared on the Y arc reflecting fork pausing at CEN7. We also observed a diffuse termination cone signal with a defined X-spot (3) likely reflecting delayed termination at CEN7. rrm3Δ cells exhibited an increase in the intensity of Y- and X-spots consistent with its role in facilitating replication across pause sites (Ivessa et al., 2003). We obtained analogous results in other _CEN_-associated TERs (TER402, 1504 and 1604) (data not shown).
TER603 contains a tRNA gene and wt cells accumulated a pausing signal on the Y arc (1 in Figure 3B) (Deshpande and Newlon, 1996) and termination intermediates (a). In rrm3Δ cells, the intensity of the Y spot increased and another pause signal appeared (2) because Rrm3 facilitates fork progression even at tRNA genes transcribed co-directionally with the fork (Ivessa et al., 2003). rrm3Δ cells also exhibited a transition of the termination intermediates from a double Y conformation (a) to an X conformation (b) (Figure 3B). Moreover, an asymmetric X-spot accumulated (3) due to termination at the tRNA site. We obtained analogous results with TER1102 and 1503 (data not shown).
The accumulation of X-shaped converging forks in rrm3Δ cells may result from slowing down of one of the two forks at a pause site, which is then more likely to become a termination site as the other converging fork approaches. However this does not rule out that Rrm3 might also directly assist fork fusion later at termination.
The majority of TERs, including TER102, contains a Pol II transcribed gene that slows down forks independently of Rrm3 (Azvolinsky et al., 2009). Fork pausing throughout highly transcribed RNA polymerase II genes is not confined to specific sites and occurs over the entire ORF regions (Azvolinsky et al., 2009; Bermejo et al., 2009), thus it does not always generate obvious discrete spots o the Y arc of the 2D gel. While wt cells accumulated at TER102 a cone signal due to random termination (Figure 3C) (Greenfeder and Newlon, 1992a), rrm3Δ mutants accumulated Xs. A possible interpretation, although not exclusive, is that these X-shaped molecules result from the impaired fusion of converging forks. Indeed, partially replicated double Y termination intermediates are progressively converted into fully replicated Xs and then into replicated linear molecules (Figure 3C). While in wt cells, the conversion of Xs into linear intermediates is likely very fast as X molecules do not accumulate, rrm3Δ cells might be delayed in this termination step since these unresolved termination structures accumulate and persist during S-phase. Similar results were seen for TER101, 202, 301, 502, 601, 902, 1002, 1005, 1303 and 1608 (data not shown).
X-shaped structures can also arise as a result of recombination (Liberi et al., 2005; Schwacha and Kleckner, 1994). We failed to observe a significant difference between the level of Xs at TERs in rrm3Δ and rrm3Δ rad51Δ mutants, thus suggesting that these X-structures did not arise from recombination (data not shown). Moreover the X-shaped structures were detected at TERs but not at _TER_-flanking regions (data not shown) further suggesting that they are related to termination events.
In conclusion we analyzed by 2D gel 20 TERs corresponding to the three classes of TER. In all of them termination intermediates were visualized, thus validating our genomic approaches. Moreover, in all 20 cases, termination signals were enhanced in the absence of Rrm3, even at those TERs that do not contain obvious Rrm3-dependent pausing elements.
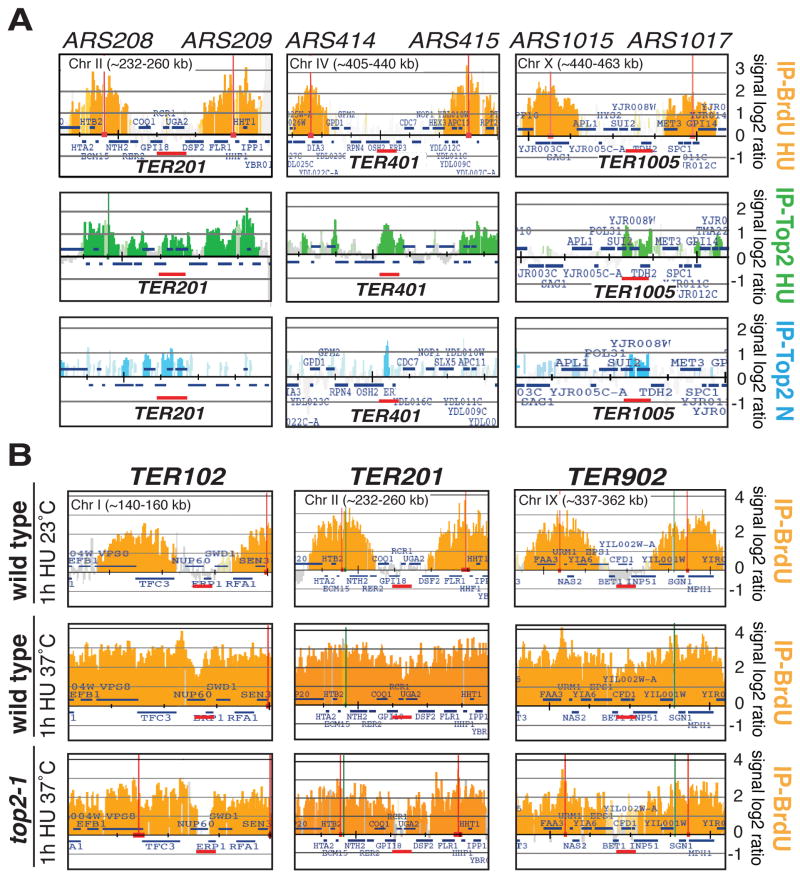
Top2 is recruited at TERs and facilitates replication termination
Top1, Top2 and Top3 move with forks (Bermejo et al., 2007) (data not shown). Topoisomerases might approach TERs by traveling with the forks or associate with TERs before or after the arrival of converging forks. The presence of topoisomerases at TERs may not be confined to S-phase as, topological constrains could persist after S-phase (Fields-Berry and DePamphilis, 1989; Holm et al., 1985). We investigated by ChIP-chip the presence of Top2 and Top3 at TERs, both in S and in G2/M cells.
Cells were released from G1 in HU or nocodazole. No enrichment was observed for Top3 at TERs under both conditions (data not shown). Top2 clusters were observed in S-phase and G2/M but not in G1 (Bermejo et al., 2007). The majority of S-phase Top2 clusters are related to fork associated Top2 and S-phase transcribed genes (Bermejo et al., 2009). We found that Top2 associates with 51/71 TERs in HU (p=0,00047) and in 55/71 in nocodazole (p=0,0065) even at those TERs that do not contain transcription units (Figure 4A, Tables S4 and S5). We obtained similar results when S-phase cells were grown with a different carbon source (53/71, p=0,0000056) (Tables S4 and S5). We failed to visualize Top2 in 4/71 TERs. Hence, Top2 associates with the majority of TERs before fork arrival and persists in G2/M.
Figure 4. Top2 is required for efficient replication termination.
The ChIP-chip data are described as in Figure 1. (A) BrdU-labeled forks are indicated in orange. Top2-10Flag (cy7315) cells were arrested in G1 with α-factor and released at 23°C in the presence of HU for 1 hour or in the presence of nocodazole for 3 hours. Samples were collected at the indicated time points and processed for ChIP-chip analysis. As a control we show the BrdU-maps (in orange) that correspond to forks that have experienced 1h in HU. Green histogram bars represent the Top2 clusters in HU (IP-Top2 HU) and the blue ones indicate the Top2 clusters in nocodazole (IP-Top2 N). Red bars indicate the TER zones. (B) wt (sy2201) and top2-1 (cy7421) cells were released from α-factor in YPD with BrdU and HU at 37°C for 1 hour. BrdU maps of wt cells experiencing HU treatment at 23°C is also shown. Red bars indicate the TER zones. List of TERs containing Top2 clusters are shown in table S4 and the relative statistical analysis in table S5.
We then investigated whether fork fusion at TERs was affected in top2 mutants. We analyzed the convergence of the BrdU-labeled forks in wt and top2 cells released from G1 into HU at the restrictive temperature for one hour. Only TERs within an inter-origin spacing of ≤20Kb could be considered for this analysis. While wt cells efficiently completed replication at TER102, 103, 201, 403, 404, 902, 1005, 1202, 1302, 1401 and 1604 (Figure 4B and data not shown), in top2 mutants the same TERs exhibited un-replicated regions with an approximate size of 1kb. Since in top2 mutants the timing of origin firing is not delayed compared to wt cells (Bermejo et al., 2007), this result suggests that the replication of the last 1kb at TERs is somewhat limiting in top2 cells, perhaps due to the topological constrains generated at the point where forks converge. In support of this conclusion, kinetics analysis showed that, within the same replicon, specifically the fork experiencing termination was delayed but not the other one (data not shown). This observation further confirms previous findings indicating that sister replication forks can be uncoupled (Doksani et al., 2009; Wang et al., 2008). Replication termination at TERs was delayed but not prevented in top2 mutants as the forks converged later on (data not shown).
In top2 mutants at the restrictive temperature the chromosomes remain entangled and undergo breakage during cell division as shown by Pulse Field Gel Electrophoresis (PFGE) (Figure 5A). Conversely, we failed to detect obvious differences between wt and top3 mutants. We then investigated in top2 cells by PFGE a 109 kb EagI fragment of CHR III that includes two TERs between ARS305 and ARS307. In wt cells the genomic fragment was fully replicated by one hour, while in top2 mutants, it remained in the wells even at 4 hours and later accumulated DNA breaks (Figure 5B). DNA breaks appearance correlated with the decrease of the signal in the wells. We note that in top2 mutants at 37°C the nocodazole block persists for no more than 3 hours (Figure S4A). Again we failed to visualize entangled chromosomes and DSBs formation in top3 mutant. To address whether at least a fraction of DNA breaks in top2 mutants may be related to abnormal termination, we deleted ARS305 and ARS306 to prevent fork fusion in the EagI fragment. DSB formation in top2-1 ars305Δ ars306Δ, compared to top2-1 mutants was reduced about 3 fold at the EagI fragment but not at other regions (Figure S4B and data not shown). The residual breaks are likely due to faulty coordination between replication and transcription (Bermejo et al., 2009) and/or to rare termination events perhaps resulting from firing of the dormant ARS302-303-320 origins cluster (Wang et al., 2001), although we failed to detect by 2D gel any obvious bubble structure under our conditions.
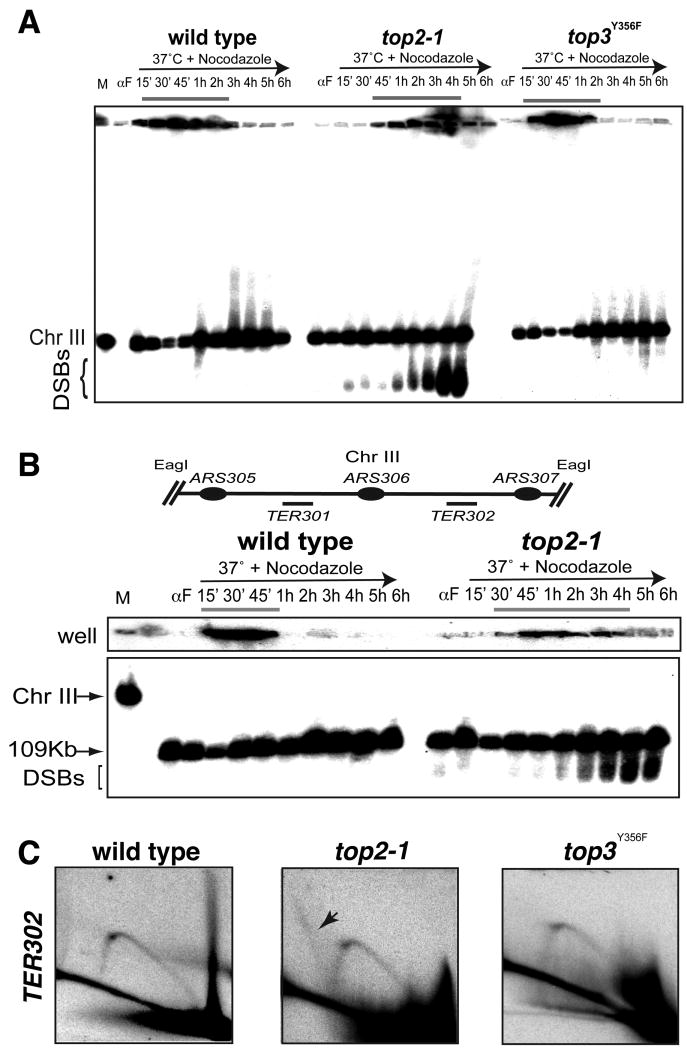
Figure 5. Top2 is required for chromosome resolution.
(A-B) wt (cy7627), top2-1 (cy7671) and top3Y356F (cy 7629) cells were arrested in G1 with α-Factor and released in S-phase in YP+ Gal at 37°C in the presence of Nocodazole. Genomic DNA was extracted in agarose plugs at the indicated time points. Yeast chromosomes were separated by PFGE and analyzed by Southern blotting with the TER302 probe. M indicates the chromosome marker. DSBs indicate double strand breaks. (B) Agarose plugs were digested with _Eag_I. Schematic representation of the analyzed region is shown. (C) wt (cy7627), top2-1 (cy7671) and top3Y356F (cy 7629) were released in S-phase at 37°C and different samples (30′-40′-50′) were pulled together to increase the chance to visualized the replication intermediates. 30 μg of DNA were digested with _Hind_III and _Pst_I and analyzed by 2D gels using TER302 probes. FACS, PFGE and 2D gels are also shown in Figure S4.
We then analyzed the replication intermediates at TER302 in wt, top3 and top2 cells at the restrictive temperature (Figure 5C). wt cells exhibited Ys but no obvious termination structures, perhaps because of their fast turn over at 37°C. We note that termination structures can be seen in the same region in wt cells at 23°C (data not shown). top3 mutants exhibited 2D gel profiles similar to wt. Conversely, top2-1 mutants accumulated additional fully duplicated X-intermediates only at TERs (Figure 5C) but not at other genomic locations (Figure S4C). These structures likely represent X-shaped entangled precatenane derivatives resulting from aberrant termination (Bermejo et al., 2007). We obtained analogous results for TER704 and TER1504 (data not shown). We conclude that Top2 and not Top3 plays a major role in the resolution of S-phase chromosomes and that genetic defects affecting the resolution process correlate with DNA breaks formation.
Top2 protects the integrity of TERs
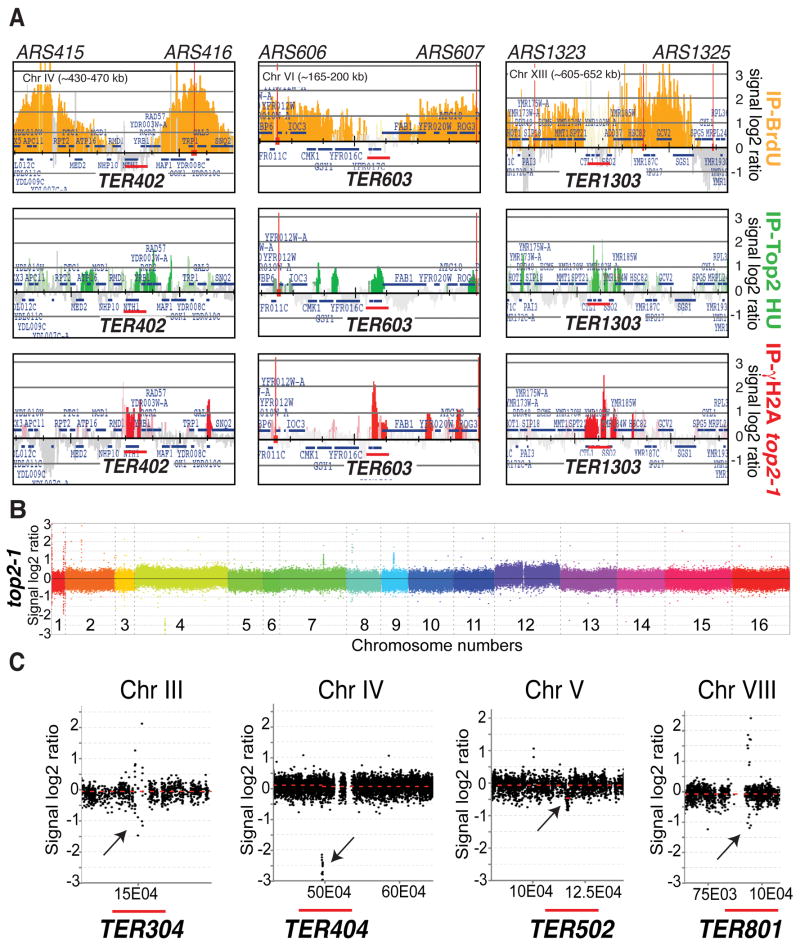
Top2 prevents the expression of fragile sites and, in top2 mutants, aberrant S-phase events cause DNA break formation during cell division (Baxter and Diffley, 2008; Bermejo et al., 2009; Bermejo et al., 2007; Holm et al., 1985). Hence, we investigated whether Top2 prevents abnormal transitions at TERs. Histone H2A phosphorylation on Ser129 (γH2A) marks nicks/gaps and DNA breaks (Lydall and Whitehall, 2005; Vidanes et al., 2005). We analyzed by ChIP-chip the γH2A clusters in top2-1 cells at the restrictive temperature following cell division. γH2A clusters significantly accumulate throughout the genome at Top2-bound regions (Bermejo et al., 2009). Accordingly, we found γH2A peaks also at 37/67 Top2-bound TER regions (Figure 6A and Table S4).
Figure 6. Top2 prevents fragility at TERs.
(A) top2-1 (cy8423) cells were released from G1 in S phase at 37°C. The sample was collected after 150 min (following cell division) and processed for ChIP-chip with antibodies against γH2A. The red histogram bars represent the γH2A clusters. BrdU-labeled forks (orange, IP-BrdU) and Top2 peaks (green, IP-Top2 HU) obtained from independent experiments are also shown. The red bars mark the TERs. See also Table S4 (B) top2-1 (cy7671) cells were released from G1 in S-phase at 25°C or 37°C in the presence of nocodazole to compare the relative genomes within one cell cycle. Samples were collected after 2,5 hour and processed for CGH analysis. The plot of the log2 ratio value on Y-axis shows DNA copy number changes between test-DNA and reference-DNA. The different colors represent all 16 chromosomes and the corresponding number is indicated. (C) SignalMap ver1.9 (NimbleGen) magnification of 4 regions detected by CGH. Plot of the log2 ratio value on Y-axis shows DNA copy number changes. The X-axis shows chromosomal coordinates. Black arrows indicate sites of genomic instability. Red bars indicate position of TER sites. Genome instability regions are also shown in Figure S1 and Table S6.
Hence, TERs like other genomic loci (Bermejo et al., 2009), express DNA fragility during cell division. To visualize potential chromosomal instability at TERs owing to top2 mutations before chromosome segregation, we performed comparative genome-wide analysis in top2 mutants experiencing one round of DNA synthesis. Comparative Genome Hybridization (CGH) was performed in wt and top2 cells released from G1 in S-phase at 25°C (reference-DNA sample) or 37°C (test-DNA sample) with nocodazole. This approach allows us to measure those genomic locations where test DNA is present in an equal, reduced or increased amount compared to the reference DNA.
13 loci exhibited deletions and/or amplifications in top2 mutants (Figure 6B, C and Table S6). These include 4 TERs (TER304, 404, 502 and 801), 3 hypothetical TERs (our analysis did not allow us to define a clear TER in these regions), 3 Ty elements, the left sub-telomeric region and the right telomere of CHR I, and partially the rDNA locus. (The majority of the rDNA locus, as well as other repetitive sequences, is not present in the array). We note that TER304 is a known genome instability site (Lemoine et al., 2005) and that rDNA instability was already described in top2 mutants (Christman et al., 1988; Holm et al., 1989). Hence, within a cell population lacking a functional Top2 activity, there are specific chromosome regions that are more subject than others to chromosome instability and that 1/3 of these loci are TERs. Moreover, these data indicate that in top2 mutants, a fraction of TERs already exhibited abnormalities at the end of S-phase, while the majority of TERs accumulated γH2A, later on, during cell division.
Discussion
We showed that eukaryotic replication termination occurs at TERs containing fork barriers. There are intriguing analogies with prokaryotes where specific termination sites and polar pausing elements influence termination. It is possible that fork barriers have passively localized through evolution in proximity of TERs, because if replication forks have to pause, it is least disadvantageous when this occurs at a site where forks are converging. Alternatively, evolution has brought fork barriers at TERs to influence fork fusion. Intriguingly, we note that deleting an efficient origin causes the re-localization of fork fusion from the original TER to another pausing element (data not shown), thus suggesting that the site of termination is influenced by the presence of pause sites.
Our findings also suggest that the polarity of fork barriers had an evolutionary impact on chromosome replication and on TERs integrity. Indeed using the yeast comparative genomics database we notice that in 5/6 TERs (TER304, 702, 801, 1601 and 1602) containing two divergent Pol III-dependent pause sites (tRNA/LTR), one of them is totally or partially not conserved (Figure S5 and Ted Weinert personal communication). On the other hand, those 58 TERs that contain polar barriers have conserved the pause sites in other yeasts. We excluded from the analysis the 7 _TERs_-containing centromeres as CENs are known to rapidly diverge in evolution (Henikoff et al., 2001) (and on the other side represent bipolar pausing elements). This correlation (p= 0.00000465) further suggests the existence of an evolutionary pressure against _TER_-containing pause sites on both strands perhaps to avoid genome instability events. In this view, we note that TER502 (the remaining un-conserved TER), 304 and 801 are unstable in top2 mutants as shown by CGH analysis (Figure 6C), TER304 and TER702 are hot spots for genome rearrangements (Admire et al., 2006; Lemoine et al., 2005), and γH2A accumulates in TER304, 502, 702 and 1601 (Table S4). It will be of interest to address how replication termination is achieved when transcription is dispensable as in the frog embryonic cell cycle. We also note that TERs seem to correlate with low nucleosome regions (p=0,07) (Table S5).
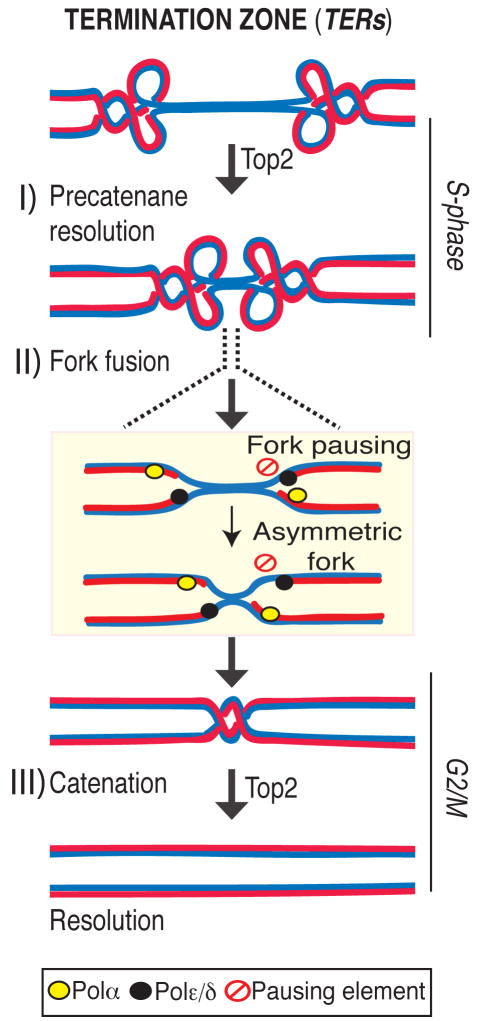
Based on in vivo and in vitro studies, both Top2 and Top3 have been suggested to play a role in replication termination (Baxter and Diffley, 2008; Branzei et al., 2006; Chan et al., 2009; Cuvier et al., 2008; DiNardo et al., 1984; Suski and Marians, 2008; Wang, 2002). Our data argue against a major contribution for Top3 at replication termination at the chromosomal level, rather they pinpoint the importance of Top2 in mediating topological transitions at TERs. Although alternative possibilities could be envisaged we propose the following III steps model (Figure 7).
Figure 7. A model for replication termination.
I) Precatenane resolution: Top2 mediates fork progression at the TER zone by resolving precatenanes behind the forks. II) Fork fusion: the right fork stalls at a pausing site (pausing element, red symbol) and emerges with an asymmetric conformation. The leading polymerase (black oval) and the lagging apparatus (yellow oval) are shown. III) Catenation: Top2 then resolves the last catenation at TERs before DNA segregation allowing chromosomes resolution.
- Rrm3, Top1 and a fraction of Top2 travel with the fork (Azvolinsky et al., 2006; Bermejo et al., 2007). Rrm3 facilitates forks progression across pausing sites (Ivessa et al., 2003) while Top1 and Top2 are both needed to resolve the torsional stress ahead of the fork generated during fork progression: while Top1 resolves positive supercoiling ahead of the fork (Wang, 2002), also contributing to prevent interference between replication and transcription (Tuduri et al., 2009), Top2 likely acts behind the fork to resolve precatenanes (Lucas et al., 2001; Wang, 2002). When forks approach the termination zone, the topological constrains at converging forks can no longer be resolved by Top1 (Fields-Berry and DePamphilis, 1989) and therefore the only option for fork progression is to rely on Top2 activity. This is consistent with the observation that top2 mutants are selectively delayed in completing the last portion of replication but not the bulk of DNA synthesis. However, we cannot rule out that the topological architecture of the termination zone (e.g. chromosome loops) needs specifically Top2 activity for resolution. Indeed a subpopulation of Top2 is also bound to TERs in early S-phase, perhaps due to the affinity of Top2 for nucleosome-free regions (p= 2.10E-58). Moreover, other S-phase Top2 clusters have been recently suggested to correlate with the formation of chromosome loops (Bermejo et al., 2009). We found that the Top2 clusters at TERs are established already at the cdc7 dependent step and are not influenced by origin firing (data not shown), thus suggesting that TERs represent CIS chromosomal elements that undergo topological transitions requiring Top2 activity.
- When fork fusion occurs, the lagging polymerase encounters the leading strand polymerase from the opposite fork, thus physically occupying the remaining un-replicated region (Sundin and Varshavsky, 1981). It is still unclear how the replisome is dismantled and how fork fusion occurs. Perhaps the presence of polar fork barriers may guarantee that the two forks do not converge simultaneously thus ensuring that at least one of the two forks emerges from the pausing region with asymmetric leading and lagging strands before fusing with the other fork. This is consistent with the findings that stalled forks exhibit an asymmetric configuration (Gruber et al., 2000; Sogo et al., 2002). Rrm3 could simply facilitate fork progression at the pause sites located within the TERs. However, we cannot exclude the possibility that Rrm3 actively participates at fork fusion as suggested by the finding that unresolved termination structures accumulate even at those TERs that do not contain obvious Rrm3-dependent pause elements.
Considering that i) the termination context might be ideal for fork reversal as topological constrains accumulate and the replisome must be dismantled (Postow et al., 2001) ii) the Mec1-Rad53 checkpoint pathway prevents fork reversal when forks stall (Sogo et al., 2002) iii) checkpoint factors have been implicated in mediating termination at the rDNA locus (Mohanty et al., 2006), it is tantalizing to speculate that the Mec1 checkpoint pathway somewhat prevents aberrant fork transitions, such as fork reversal, at termination zones. - Fork fusion then gives rise to catenated sister chromatid junctions that have to be resolved before segregation. We propose that this last step is mediated by a sub-population of pre-assembled _TER_-associated Top2 that can persist even after S-phase. It is also possible that Top2, at least in a fraction of TERs, is loaded at the beginning of mitosis. Given that the catenated junction might be mobile and spread along the chromosomes (Spell and Holm, 1994), the presence of preassembled Top2 might be needed to confine and coordinate its resolution at the TER loci, perhaps through SUMO-mediated regulation (Bachant et al., 2002; Dawlaty et al., 2008).
According to the model proposed, the transient accumulation of topological constrains might facilitate abnormal transitions (Hiasa and Marians, 1994) that could lead to amplification or deletion of TER sites. Moreover, the proper resolution of catenated sister chromatids would be impaired in top2 cells and, following cell division, DNA breaks, and aberrant segregation will be expected (Baxter and Diffley, 2008; Bermejo et al., 2007; DiNardo et al., 1984; Holm et al., 1989).
Altogether our data provide a framework for understanding the eukaryotic molecular mechanisms that control replication termination and coordinate replication with transcription and topological dynamics.
Experimental Procedures
Yeast Strains and growing conditions
All strains (Table S1) are isogenic derivatives of W303-1A. All epitope tags (10Flag and 6PK) were fused to the c-terminus of the protein of interest. Strains were grown in YPD and cells were arrested in G1 by α–factor (2 μg/ml) or in G2/M by nocodazole (10 μg/ml). HU was added at 0.2M. Over-expression of the dominant negative version of Top3 was induced for 3 hour by Galactose 2% in YP + Raffinose 2% media. BrdU was added as previously described (Katou et al., 2003). Rpc25 and Rpb3 subunits were analyzed by ChIP-chip following 1 hour in HU.
Pulse Field Gel Electrophoresis (PFGE)
DNA plugs were prepared as described (Lengronne et al., 2001). Yeast chromosomes were separated by PFGE (Gene Navigator System, Amersham) and electrophoresis was performed for 15 h at 200V with 90s pulses, followed by 9 h with 125s pulses, in TBE 0.5× at 14°C. Plugs digestion was performed in according to New England BioLabs and previously described (Azvolinsky et al., 2006).
Psoralen-crosslinking, DNA extraction, 2D-gel technique
Genomic DNA extraction was performed according the “QIAGEN genomic DNA Handbook”. DNA psoralen-crosslinking and 2D-gel procedure were described (Doksani et al., 2009). Quantifications were done using ImageQuant 5.2 (Molecular Dynamics).
Probes are obtained by PCR using the following oligos: TER102: Fw TCTGCGCCAAGCAAAGATTC, Rv TTTCCTTGCGTCTGATTCGG. TER603: Fw GAATGCCCGAGCCCTAAAAA, Rv ATGTGAGCCATCTGGAAAGG. TER704: Fw TGTGCACATCTTGCCCATTA, Rv GCCTCTATCACTGCAAAGTG.
TER302: Fw GAAGGTTCAACATCAATTGATTGATTCTGCCGCCATGATC, Rv GCTTCCCTAGAACCTTCTTATGTTTTACATGCGCTGGGTA
ChIP-chip analysis
S.cerevisiae oligonucleotide microarrays were provided by Affymetrix (S.cerevisiae Tiling 1.0R, P/N 900645). BrdU and proteins ChIP-chip analyses were carried out as described (Bermejo et al., 2009). Pol2 (Polε) ChIP-chip analysis was performed as described (Azvolinsky et al., 2009).
Comparative Genome Hybridization (CGH)
Roche-Nimblegen 385K Yeast Whole Genome Tiling arrays were used to perform CGH analysis. Experimental processing was performed accordingly to Roche-Nimblegen protocol, data elaboration using the NimbleScan v2.4 software (Roche-Nimblegen) and the analysis using the embedded packages DNAcopy and segMNT.
Statistical methods
Evaluation of the significance of the presence of protein binding peaks and pausing elements within TERs (Table S5) was performed by confrontation against a null hypothesis model generated with a Montecarlo-like simulation.
For each dataset (binding clusters of a specific protein or set of pausing elements) we produced 1000 randomizations of the positions of the features, maintaining unchanged the number and size of the genomic areas covered within each chromosome; the number of peaks and features with random positions within the TERs was then counted and taken as score for each iteration. The distribution of these random scores was validated to be approximately normal (|Skew| < 0.25 and |Kurtosis excess| < 0.25) and then the average and standard deviation for this distribution was taken as null hypothesis.
The increase or decrease ratios for the scores of the actual positions with respect to the expected value for the null hypothesis (defined as the average score of random attempts) was then calculated, and the P-values for the drift were estimated as Standard Normal CDF of |actual−mean|deviation.
Evaluation of significance of overlaps in sets (i.e. for the number of non-conserved TERs versus the TERs containing divergent pausing elements) was performed by means of the Exact Fisher Test.
Supplementary Material
01
Acknowledgments
We thank A. Verreault and E. Schwob for reagents, Ted Weinert for communicating unpublished results, D. Branzei, M. Lopes and Y. Doksani for suggestions and critical reading of the manuscript, and all members of our laboratories for discussions. We thank M. Cesaroni for TER sequence analysis, M. Saponaro for technical advice and F. Ciccarelli for suggestion on evolution analysis. Work in the M.F. laboratory is supported by grants from Italian Association for Cancer Research, Italian Foundation for Cancer Research, Telethon-Italy, European Community, Italian Ministry of Health. D.F. was supported by a AIRC fellowship. Work in KS's laboratory is supported by a grant of the Genome Network Project and Grant-in-Aid for Scientific Research (S) from the MEXT, Japan. YK is a GCOE research associate. Work in V.A.Z.'s laboratory is supported by NIH grant R37 029638.
Footnotes
Accession Numbers: Experimental data are available on Gene Expression Omnibus database with accession number GSE19061.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Admire A, Shanks L, Danzl N, Wang M, Weier U, Stevens W, Hunt E, Weinert T. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 2006;20:159–173. doi: 10.1101/gad.1392506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K, Foiani M. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell. 2009;138:870–884. doi: 10.1016/j.cell.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Fink GR. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- Cuvier O, Stanojcic S, Lemaitre JM, Mechali M. A topoisomerase II-dependent mechanism for resetting replicons at the S-M-phase transition. Genes Dev. 2008;22:860–865. doi: 10.1101/gad.445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 2009;137:247–258. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggin IG, Wake RG, Bell SD, Hill TM. The replication fork trap and termination of chromosome replication. Mol Microbiol. 2008;70:1323–1333. doi: 10.1111/j.1365-2958.2008.06500.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Huberman JA. Eukaryotic chromosome replication. Annu Rev Genet. 1975;9:245–284. doi: 10.1146/annurev.ge.09.120175.001333. [DOI] [PubMed] [Google Scholar]
- Fields-Berry SC, DePamphilis ML. Sequences that promote formation of catenated intertwines during termination of DNA replication. Nucleic Acids Res. 1989;17:3261–3273. doi: 10.1093/nar/17.8.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder SA, Newlon CS. A replication map of a 61-kb circular derivative of Saccharomyces cerevisiae chromosome III. Mol Biol Cell. 1992a;3:999–1013. doi: 10.1091/mbc.3.9.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder SA, Newlon CS. Replication forks pause at yeast centromeres. Mol Cell Biol. 1992b;12:4056–4066. doi: 10.1128/mcb.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber M, Wellinger RE, Sogo JM. Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol Cell Biol. 2000;20:5777–5787. doi: 10.1128/mcb.20.15.5777-5787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Hiasa H, Marians KJ. Tus prevents overreplication of oriC plasmid DNA. J Biol Chem. 1994;269:26959–26968. [PubMed] [Google Scholar]
- Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. Strategies to maintain the stability of the ribosomal RNA gene repeats--collaboration of recombination, cohesion, and condensation. Genes Genet Syst. 2006;81:155–161. doi: 10.1266/ggs.81.155. [DOI] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Liberi G, Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol Cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- Lucas I, Germe T, Chevrier-Miller M, Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. Embo J. 2001;20:6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Whitehall S. Chromatin and the DNA damage response. DNA Repair (Amst) 2005;4:1195–1207. doi: 10.1016/j.dnarep.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olavarrieta L, Hernandez P, Krimer DB, Schvartzman JB. DNA knotting caused by head-on collision of transcription and replication. J Mol Biol. 2002;322:1–6. doi: 10.1016/s0022-2836(02)00740-4. [DOI] [PubMed] [Google Scholar]
- Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Spell RM, Holm C. Nature and distribution of chromosomal intertwinings in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1465–1476. doi: 10.1128/mcb.14.2.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wang X, Reyes-Lamothe R, Sherratt DJ. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 2008;22:2426–2433. doi: 10.1101/gad.487508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Vujcic M, Kowalski D. DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol Cell Biol. 2001;21:4938–4948. doi: 10.1128/MCB.21.15.4938-4948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- Zhu J, Newlon CS, Huberman JA. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01