Protein Interaction Domain Mapping of Human Kinetochore Protein Blinkin Reveals a Consensus Motif for Binding of Spindle Assembly Checkpoint Proteins Bub1 and BubR1 (original) (raw)
Abstract
The kinetochore is a supramolecular structure essential for microtubule attachment and the mitotic checkpoint. Human blinkin/human Spc105 (hSpc105)/hKNL1 was identified originally as a mixed-lineage leukemia (MLL) fusion partner and later as a kinetochore component. Blinkin directly binds to several structural and regulatory proteins, but the precise binding sites have not been defined. Here, we report distinct and essential binding domains for Bub1 and BubR1 (here designated Bubs) at the N terminus of blinkin and for Zwint-1 and hMis14/hNsl1 at the C terminus. The minimal binding sites for Bub1 and BubR1 are separate but contain a consensus KI motif, KI(D/N)XXXF(L/I)XXLK. RNA interference (RNAi)-mediated replacement with mutant blinkin reveals that the Bubs-binding domain is functionally important for chromosome alignment and segregation. We also provide evidence that hMis14 mediates hNdc80 binding to blinkin at the kinetochore. The C-terminal fragment of blinkin locates at kinetochores in a dominant-negative fashion by displacing endogenous blinkin from kinetochores. This negative dominance is relieved by mutations of the hMis14 binding PPSS motif on the C terminus of blinkin or by fusion of the N sequence that binds to Bub1 and BubR1. Taken together, these results indicate that blinkin functions to connect Bub1 and BubR1 with the hMis12, Ndc80, and Zwint-1 complexes, and disruption of this connection may lead to tumorigenesis.
The kinetochore is formed on centromeric DNA and is essential for microtubule attachment and mitotic checkpoint signaling during mitosis (3, 6, 23, 27, 34). Recent genetic and mass spectrometric analyses have identified more than 80 kinetochore components in fungi, nematodes, insects, and mammalian cells and have revealed both conserved core components and species-specific kinetochore proteins (4, 14, 22, 25, 30). These proteins form several subcomplexes and assemble and disassemble in an ordered fashion to build functional kinetochores during mitosis.
The Mis12 complex is a key conserved kinetochore subcomplex which consists of four proteins, Mis12, Mis13/Dsn1, Mis14/Nsl1, and Nnf1/PMF1 (4, 10, 11, 18, 25). The human Mis12 (hMis12) complex interacts with heterochromatin protein 1 (HP1) during interphase and dissociates from HP1 during mitosis to form the inner centromere structure between sister kinetochores (16). During mitosis, the hMis12 complex associates with the microtubule-binding protein blinkin (alternatively called hSpc105, hKNL1, CASC5, and D40) and the Ndc80/Hec1 complex (2, 5, 33), as well as the mitotic checkpoint-related proteins Zwint-1 and Bub1 and BubR1 (Bub1-related protein) (17). Thus, the hMis12 complex plays a key role in inner centromere and kinetochore assembly as well as kinetochore-microtubule attachment and mitotic checkpoint signaling in human cells.
Yeast two-hybrid (Y2H) mapping, localization dependency, and RNA interference (RNAi) rescue experiments have suggested that the hMis12 complex directly interacts with the C terminus of blinkin and that the N terminus of blinkin directly recruits Bub1 and BubR1 (here designated Bubs) to kinetochores (17). The molecular mechanisms of these interactions are elusive but are indispensable for understanding the mechanism of kinetochore assembly, checkpoint signaling, and regulation. In this study, we performed extensive Y2H analyses of blinkin and identified its minimal binding sites for Bub1, BubR1, Zwint-1, and hMis14. Deletion constructs or substitution mutants of blinkin suggest a critical functional importance for these domains. Furthermore, we show that hMis14 links blinkin to the Ndc80 complex at kinetochores during mitosis. We discuss the conserved and species-specific function of the Mis12 complex.
MATERIALS AND METHODS
Strains and media.
HeLa cells and strains that stably express the green fluorescent protein (GFP)-hMis14 wild type (WT) or GFP-hMis14 m2E mutant were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM; Kohjin Bio) supplemented with 10% fetal bovine serum (FBS; Biowest), 1% penicillin-streptomycin, and 1% antibiotic and antimycotic (Gibco). Stable T-Rex HeLa and Flip-in TRex 293 cells were constructed and cultured according to the manufacturer's protocol (Invitrogen). To induce transgenes, cells were incubated with 1 μg/ml tetracycline (tet; MP Biomedicals). Cells were treated with 100 ng/ml nocodazole (MP Biomedicals) or 150 ng/ml TN16 (Wako) for 15 to 18 h to obtain the mitotic arrest extract.
Plasmids and siRNA transfection.
The nucleotide sequences of all blinkin mutations were verified by DNA sequencing. Plasmid DNAs purified using an EndoFree Maxi kit (Qiagen) were transfected using an Effectene transfection kit (Qiagen) to obtain the stable cell lines. The conventional 21-nucleotide small interfering RNA (siRNA) for blinkin (siRNA 4) or stealth siRNA 3 (designed for the middle region of blinkin) (17) was transfected with Oligofectamine (Invitrogen) or RNAi Max (Invitrogen), respectively. The cell culture and transfection of siRNA were performed according to the manufacturer's instructions.
Yeast two-hybrid analysis.
The two-hybrid analysis was performed according to the procedures described in the two-hybrid analysis kit (Matchmaker; Clontech) using plasmids pGBT9 and pGAD424 (17). The β-galactosidase filter assay was performed using the SFY526 strain that carried the GAL1-lacZ reporter, as described in the manufacturer's instructions.
Antibodies.
Immunoblots and immunofluorescence experiments were performed using the following antibodies: hMis12 (anti-rabbit Ig, 1:30) (10), hMis13 (anti-rabbit Ig, 1:1,000) (25), hMis14 (anti-mouse Ig, 1:20) (16), hNnf1 (anti-mouse Ig, 1:20; A2), HP1α (anti-mouse Ig, 1:500; Millipore), CENP-A (mouse, 1:100), CENP-C (anti-guinea pig Ig, 1:1,000), blinkin (anti-mouse 31F2, 1:20) (17) and blinkin(M) (anti-rabbit C5, 1:1,000), Zwint-1 (anti-mouse Ig, 1:20) (17), Bub1 (anti-sheep Ig, 1:1,000), BubR1 (anti-sheep Ig, 1:1,000) (32), anti-histone H3S10ph (anti-rabbit Ig, 1:1,000, catalog number ab5176; Abcam), GFP (anti-mouse Ig, 1:500; Roche), and tubulin (anti-mouse Ig, 1:500, DM1A; Sigma). Rabbit polyclonal antibody against BLKC (C3) and BLKM (C5) and mouse monoclonal antibody against hNnf1 were produced using the C-terminal 25 amino acids (aa 2145 to 2169), the central 23 amino acids (aa 1757 to 1779), and the N-terminal 19 amino acids, respectively, as the antigen.
Live-cell imaging and immunofluorescence.
Live-cell analysis was performed as previously described (12). Cells grown on glass-based dishes (Iwaki, Tokyo, Japan) were supplemented with 20 mM HEPES (pH 7.4) and observed using a Delta Vision RT system (Applied Precision, Inc.) at a temperature of 37°C. For DNA staining, HeLa cells were cultured in the presence of 50 ng/ml Hoechst 33342 stain, and the live images were generated at 5-min intervals with a LiveUV filter. Immunofluorescence microscopy was performed as previously described (17) with the antibodies indicated in Fig. 1, 3, 4, 5, 6, and 9. DNA was counterstained using Hoechst 33342. For fixed cells, a z series of 20 to 40 images was captured at 0.2-μm intervals and processed using constrained iterative deconvolution. Deconvolved image stacks were projected.
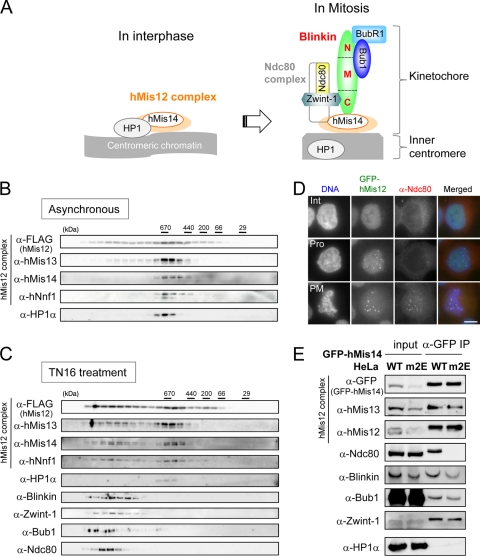
FIG. 1.
Mitotic associations between the hMis12 complex and Ndc80 were abolished by the mutation of the HP1-binding site on hMis14. (A) Summary cartoon of hMis12 complex-interacting proteins in interphase and mitosis (17). (B and C) FLAG-hMis12 was immunoprecipitated from asynchronous (B) and TN16-treated (C) mitotic extracts and was subsequently dissociated by a 3× FLAG peptide. The solubilized protein complexes, which coprecipitated with FLAG-hMis12, were fractionated by a Superose 6 HR 10/30 size-exclusion chromatography column and immunoblotted by the antibodies indicated. The migrations of the molecular mass marker are indicated above the blots (in kilodaltons). (D) GFP-hMis12-expressing cells (green) were fixed and stained with anti-Ndc80 antibody (red) and Hoechst 33342 (blue). Interphase (Int), prophase (Pro), and prometaphase (PM) cells are shown. (E) Immunoprecipitation of GFP-tagged hMis14 wild-type (WT) and hMis14 m2E mutant proteins. Cultures of HeLa cells that stably expressed GFP-tagged hMis14 WT and m2E were used. The expression of GFP-hMis14 m2E was repressed in a subpopulation of the cells. Equal amounts of GFP-hMis14 WT and m2E were applied in the immunoprecipitation lanes. Cells were treated with nocodazole for 18 h. Input and immunoprecipitates (IP) were immunoblotted using the antibodies indicated.
Immunopurification.
Cell extracts were prepared with modified CSK buffer (0.1% TX-100 mCSK), consisting of 500 mM NaCl, 10 mM PIPES [piperazine-N,_N_′-bis(2-ethanesulfonic acid), pH 7.0], 300 mM sucrose, 0.1% Triton X-100 (Triton), 1 mM MgCl2, 1 mM EGTA, 0.1 mM phenylmethylsulfonyl fluoride, and 2 μg/ml leupeptin. The extracts were incubated with anti-FLAG (M2; Sigma-Aldrich)- or anti-GFP (clone RQ2; MBL)-conjugated beads for 2 h. The beads were washed twice with 0.1% TX-500 mCSK, and elutes were obtained with 0.1 M glycine (pH 2.0) for the anti-GFP antibody.
Fluorescence-activated cell sorter (FACS) analysis.
Cells were fixed with cold ethanol at 4°C overnight. After RNase treatment for 30 min, cells were stained with propidium iodide (PI; 50 μg/ml) for more than 30 min and analyzed by flow cytometry (Becton Dickinson). The G2/M population was measured by counting 20,000 cells.
RESULTS
The assembly of blinkin-hMis12-Ndc80 complexes occurs in M phase.
The tetrameric hMis12 complex binds to HP1 during interphase and interacts with blinkin, Zwint-1, the Ndc80 complex, and Bubs during mitosis (16, 17, 25) (Fig. 1A). To examine cell cycle-dependent complex formation more directly, immunoprecipitation against FLAG-hMis12 and a subsequent gel size-fractionation analysis was performed using interphase and mitotic extracts. FLAG-hMis12 was immunoprecipitated with anti-FLAG antibody-conjugated beads and was subsequently dissociated from the beads by a 3× FLAG peptide. The resulting solubilized FLAG-hMis12 protein complexes were fractionated using a Superose 6 size-exclusion chromatography column and analyzed by immunoblotting. As shown in Fig. 1B, the proteins of hMis12, hMis13, Mis14, and hNnf1 (the subunits of the hMis12 complex) and HP1α formed a peak (around 670 kDa) in the asynchronous culture that was composed of mostly interphase cells. All subunits of the hMis12 complex shifted to the larger peak fractions when cells were treated with the microtubule-destabilizing drug TN16 to cause cell cycle arrest in mitosis (Fig. 1C). In contrast, HP1α was not found in the bigger peak fractions. These results suggest that the hMis12 complex shifts from the interphase complex that is bound to HP1 to a larger supramolecular complex with Ndc80, Zwint-1, blinkin, and Bub1 after the entry into mitosis. Separate from the other components, HP1α forms the inner centromere (16).
We next examined the intracellular localization of hMis12 and Ndc80 in the interphase and mitotic nucleus. Consistent with the above results, the signals of hMis12 (GFP) and Ndc80 (antibody) were distinct in the interphase (Int) cells but were indistinguishable in the prometaphase (PM) cells (Fig. 1D). Basically the same result was found for the localization of hMis12 and blinkin (data not shown and reference 17).
hMis14 links the Ndc80 complex to blinkin at the kinetochores.
hMis14 is a trident-binding protein that associates with blinkin, hMis13, and HP1, and the binding motif sequences for these three proteins in hMis14 do not overlap (16). We employed the hMis14 m2E mutant that has two glutamate substitutions for the bulky hydrophobic amino acid residues V and L in the HP-binding PXVXL motif. This mutant specifically abolishes the interaction between hMis14 and HP1 in interphase, leading to the impaired associations of hMis14 with the kinetochore and HP1 with the inner centromere during mitosis (16). To examine whether the m2E mutation might change complex formation in the kinetochore assembly, immunoprecipitation against the GFP-hMis14 WT and hMis14 m2E mutant was performed using mitotic extracts of cells arrested with nocodazole. As expected, the GFP-hMis14 WT coprecipitated with hMis12, hMis13, Ndc80, blinkin, Bub1, and Zwint-1, while the GFP-hMis14 m2E mutant precipitated with each protein except Ndc80 (Fig. 1E). These results suggested that the proper kinetochore localization of hMis14 may be required to link the Ndc80 complex to the hMis12 complex, blinkin, Zwint-1, and Bubs in mitosis (see Discussion).
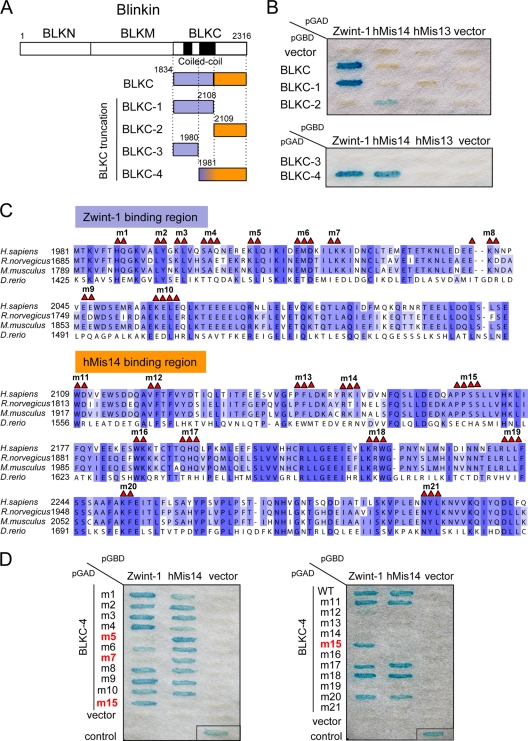
Identification of Zwint-1- and hMis14-binding regions on blinkin.
Our previous two- and three-hybrid analyses indicated that the C-terminal fragment of blinkin, BLKC (aa 1833 to 2316), interacts with Zwint-1 and hMis14 (16, 17). To examine the nature of these binding regions, we generated four truncation fragments of BLKC, BLKC-1 to BLKC-4, as shown in Fig. 2A. BLKC-4 interacts with both Zwint-1 and hMis14, while BLKC-1 interacts with only Zwint-1, and BLKC-2 shows a weak interaction with only hMis14 (Fig. 2B). BLKC-3 did not interact with either Zwint-1 or hMis14. Curiously, BLKC did not interact with hMis14, and the reason for this is unclear. Taken together, these results indicate that the extreme C terminus of blinkin that is conserved in vertebrates (Fig. 2C) is important for binding to both Zwint-1 and hMis14. This is consistent with other results obtained while using purified proteins (26). The binding sites may be distinct but may overlap, and the small region of BLKC from aa 1981 to 2108 (BLKC1981-2108) might be important for the specific interaction with Zwint-1.
FIG. 2.
Identification of the Zwint-1- and hMis14-binding regions in blinkin. (A) The BLKC mutant constructs are shown. The Zwint-1- and hMis14-binding regions are shown in dark purple and orange, respectively. (B) Y2H interaction between BLKC mutants and Zwint-1, hMis14, or hMis13. Plasmids pGBD and pGAD carried the DNA-binding and -activating domains of yeast GAL4, respectively. (C) Amino acid sequences of BLKC-4 (aa 1981 to 2316). Highly conserved amino acids and similar amino acids are boxed in dark and light purple, respectively. Amino acids indicated by red triangles are replaced by alanine in BLKC-4 mutants. (D) Y2H interaction between BLKC-4 mutants and Zwint-1 or hMis14.
To identify the essential amino acid residues in blinkin that are necessary for the interaction with Zwint-1 and hMis14, we next generated 21 alanine mutants of BLKC-4, namely, BLKC-4 m1 to m21 (Fig. 2C shows the sites of the mutations). Y2H analysis revealed that the BLKC-4 m5 and m7 mutants specifically abolished the interaction with Zwint-1, while the m15 mutant failed to interact with hMis14 (Fig. 2D). Several mutations, such as those for m12 to m14, m16, m19, and m21, abolished the interaction of both Zwint-1 and hMis14, suggesting that these mutations might alter the conformation necessary for interactions with both Zwint-1 and hMis14.
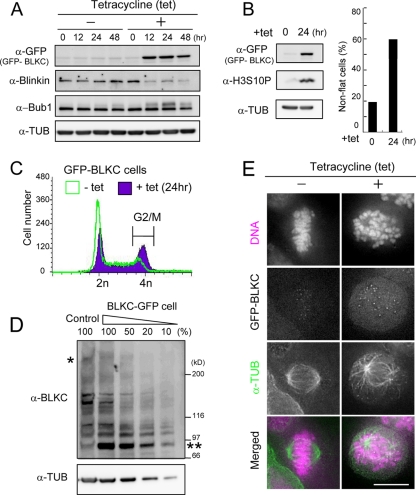
Dominant-negative effect of BLKC.
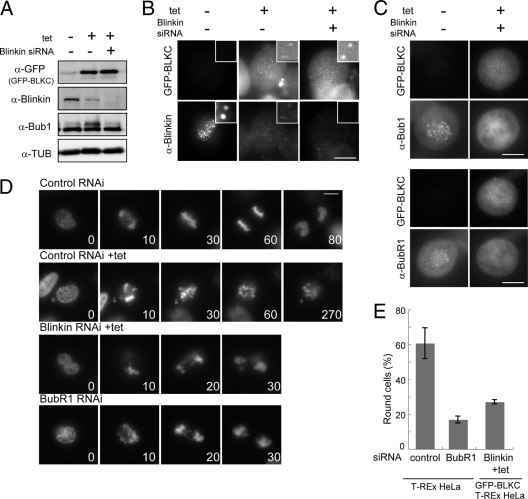
To assess the functional significance of the results presented above, we first examined the effect of expressing blinkin fragments in HeLa cells. The expression of GFP-BLKC under the cytomegalovirus (CMV) promoter caused chromosome misalignment, while the expression of GFP-tagged N-terminal fragments of blinkin had no apparent effect on mitosis (data not shown). To substantiate the effect of GFP-BLKC expression, we generated a stable HeLa cell line that conditionally expressed GFP-BLKC under the control of the tet promoter. GFP-BLKC was expressed 12 h after the addition of tet (Fig. 3A, +), and round, presumably mitotic cells accumulated within 12 to 48 h (Fig. 3B, right). The phosphorylation of histone H3S10 was intense at 24 h after the addition of tet (Fig. 3B) and the G2/M population, determined by FACS analysis, was increased from 14.6% to 38.9% (Fig. 3C), suggesting that GFP-BLKC expression caused mitotic arrest. It is known that Bub1 exhibits a phosphorylation-dependent band shift in SDS-PAGE gels when cells are arrested in mitosis by microtubule toxins (32). The intensity of band-shifted Bub1 peaked at 24 h (Fig. 3A). Interestingly, the band intensity of endogenous blinkin decreased when GFP-BLKC was expressed, while the level of Bub1 remained the same at 12 and 24 h (Fig. 3A). Similar results were found when GFP was fused to the C terminus of BLKC (see Fig. 6B and C). The expression of GFP-tagged BLKC fragments (Fig. 3D, double asterisks) was approximately 10 times higher than that of endogenous blinkin, as determined by serial dilution (single asterisk).
FIG. 3.
The overexpression of the BLKC fragment causes chromosome misalignment and mitotic arrest. (A) TRex-HeLa chromosomally integrated GFP-BLKC was treated with (+) or without (−) tetracycline (tet), and extracts were immunoblotted using the antibodies indicated. The loading control was tubulin (TUB). (B) Immunoblot of GFP, histone H3S10ph, and tubulin after treatment with tet (left). Frequency of nonflat cells determined by phase-contrast microscopy was measured (n > 1,000 cells, right). (C) TRex-HeLa chromosomally integrated GFP-BLKC was treated with tet (+ tet) or without (− tet) for 24 h and fixed with cold ethanol. After RNase treatment, cells were stained with PI (50 μg/ml) and analyzed by flow cytometry. The G2/M population was measured by counting 20,000 cells. (D) Immunoblot of BLKC and TUB with serial dilutions of BLKC-GFP extract. Antibody against the C-terminal region of blinkin was used to detect both endogenous blinkin and BLKC-GFP. One and two asterisks indicate endogenous blinkin and BLKC-GFP, respectively. (E) Cells were incubated for 24 h with (+) or without (−) tetracycline (tet) and stained with the antibodies indicated. Cells without tet treatment were stained as the control.
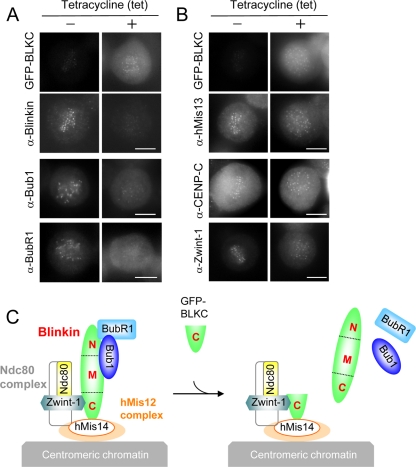
In mitotic cells that expressed GFP-BLKC (Fig. 3E, +), chromosomes were scattered throughout the spindle and failed to align. In the control cells (−), GFP-BLKC was hardly expressed and mitosis was normal. Immunostaining of endogenous blinkin (using an antibody directed against its N terminus), Bub1, and BubR1 revealed faint kinetochore signals in the cells that expressed GFP-BLKC (Fig. 4A, +). However, the signals of the hMis13 subunit of the hMis12 complex, CENP-C (a kinetochore marker), and Zwint-1 were localized normally after induction with tet (Fig. 4B). These results are consistent with the idea that GFP-BLKC associates with Zwint-1 and the hMis14 subunit of the hMis12 complex and prevents endogenous blinkin from functioning normally in kinetochore assembly, as illustrated in Fig. 4C.
FIG. 4.
Dominant-negative effect of BLKC. (A and B) TRex-HeLa chromosomally integrated GFP-BLKC was incubated for 24 h with (+) or without (−) tetracycline (tet) and stained with the antibodies indicated. Antiblinkin antibody recognizes the N terminus of blinkin; thus, it stains endogenous blinkin but not BLKC. Bars, 10 μm. (C) Model of the dominant-negative effect by BLKC. See details in the text.
Blinkin RNAi abolished mitotic arrest caused by GFP-BLKC.
We hypothesized that residual endogenous blinkin might activate the checkpoint and arrest GFP-BLKC-expressing cells in mitosis. To test this, we depleted blinkin by using RNAi while overexpressing GFP-BLKC. The amount of blinkin protein detected using antiblinkin antibodies (α-blinkin) was negligible (Fig. 5A, ++), and blinkin did not localize to the kinetochores of GFP-BLKC-expressing cells after RNAi treatment (Fig. 5B, ++). The kinetochore localization of Bubs was also diminished in cells treated with both tet and blinkin RNAi (Fig. 5C). Consistent with these results, cells that expressed GFP-BLKC showed accelerated mitosis and were not arrested by nocodazole treatment when blinkin RNAi was also included (Fig. 5D and E). Taken together, these results indicate that the presence of the residual endogenous blinkin was required for the mitotic arrest in cells that expressed GFP-BLKC.
FIG. 5.
Blinkin RNAi abolishes the mitotic arrest by BLKC expression. (A) TRex-HeLa chromosomally integrated GFP-BLKC was treated with tet and blinkin siRNA as indicated. Extracts were obtained 48 h after treatment and immunoblotted using the antibodies indicated. The loading control was tubulin. (B and C) Cells were fixed 48 h after the treatment of tet and blinkin siRNA and stained with antibodies as indicated. (D) Time-lapse micrographs of GFP-BLKC-expressing TRex-HeLa cells after control and blinkin RNAi treatment. TRex-HeLa without integration was treated with the control and BubR1 RNAi without the addition of tet. DNA was stained with Hoechst 33342. Numbers indicates the time (in minutes) after nuclear envelope breakdown (NEBD). Bars, 10 μm. (E) Frequency of round cells determined by phase-contrast microscopy was measured 18 h after nocodazole treatment. Each cell line was treated by siRNA and tet as indicated. Bars indicate standard deviation. Control, 60.6 ± 8.8 (n = 2,060 cells); BubR1, 16.8 ± 2.0 (n = 1,484); Blinkin+tet, 27.0 ± 1.3 (n = 1,801).
A BLKC mutant deficient in hMis14 binding abolishes negative dominance.
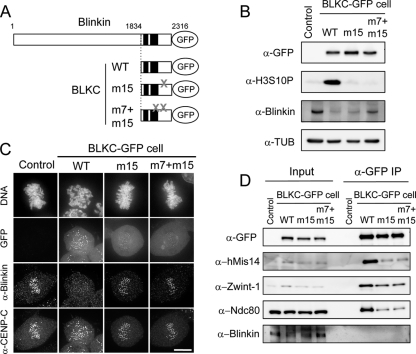
To examine whether BLKC mutants exerted a dominant-negative effect, we used the tet-inducible Flip-In system (Invitrogen) and generated stable cell lines that express the BLKC WT or the mutant protein BLKC-m15-GFP or BLKC-m7+m15-GFP (Fig. 6A). Twenty-four hours after the addition of tet, essentially equal amounts of GFP-tagged proteins were expressed in each cell line (Fig. 6B). The histone H3S10P immunoblot signal (a molecular marker of mitotic cells) was abundant only in BLKC WT-expressing cells (Fig. 6B, 2nd lane), and was reduced in cells expressing BLKC-m15 or BLKC-m7+m15 (Fig. 6B, 3rd or 4th lane, respectively). As shown in Fig. 6C, wild-type BLKC-GFP was robustly localized at kinetochores, while both BLKC mutants were barely recruited at kinetochores. On the other hand, little endogenous blinkin was detected at kinetochores in BLKC WT-expressing cells, but it was abundant at the kinetochores in cells expressing BLKC mutant proteins (Fig. 6C, α-blinkin; α-CENP-C is the control) when chromosome misalignment was also suppressed (Fig. 6C, DNA). These results suggested that endogenous blinkin became unstable in BLKC WT-expressing cells, as free endogenous blinkin displaced from kinetochores was susceptible to degradation.
FIG. 6.
The BLKC mutant deficient in hMis14 binding suppresses the dominant-negative effect by BLKC. (A) The BLKC mutant constructs chromosomally integrated in Flip-In TRex 293 cells are shown. A gray X indicates m7 and m15 mutation sites. (B) Each cell line was treated with tet to express GFP-tagged BLKC constructs. Extracts were obtained 24 h after tet treatment and immunoblotted by antibodies as indicated. Nonintegrated cells were used as controls. (C) Each cell line was fixed and stained by Hoechst 33342 and antiblinkin and anti-CENP-C antibodies 24 h after tet treatment. Bar, 10 μm. (D) Each cell line was cultured for 12 h with tet and for an additional 18 h after the addition of nocodazole. Extracts were immunoprecipitated with anti-GFP antibodies. Input and immunoprecipitates were immunoblotted using the antibodies indicated.
BLKC WT coimmunoprecipitated with hMis14, Zwint-1, and Ndc80/Hec1 (Fig. 6D, α-GFP IP), but BLKC m15 and m7+m15 mutants reduced the interactions with these three kinetochore proteins. None of the BLKC constructs coimmunoprecipitated with endogenous blinkin (Fig. 6D). The BLKC-m15 mutation, which contained the substitutions at 2166PPSS, specifically abolished the interaction with hMis14 in the Y2H assay, affecting the interaction between BLKC, Zwint-1, and Ndc80 in vivo, because blinkin might effectively interact with Zwint-1 and Ndc80 at the kinetochores after it formed the complex with hMis14. Our interpretation of the results is that BLKC expression caused a dominant-negative effect through direct interaction with hMis14, which subsequently interacted with the other subunits of the hMis12 complex, as well as with the Ndc80 and Zwint-1 complexes.
Identification of minimal Bub1- and BubR1-interacting domains in blinkin.
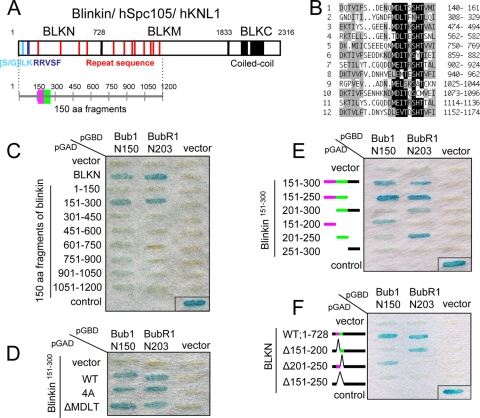
The results suggest that the N-terminal region of blinkin is essential for the proper function of blinkin. Our previous yeast two-hybrid (Y2H) analysis demonstrated that the N terminus of human blinkin, BLKN (aa 1 to 728), interacts strongly with the N-terminal regions of Bub1 and BubR1, while the middle region of blinkin, BLKM (aa 729 to 1833), has a weak interaction with Bub1 (Fig. 7A) (17). Human blinkin/hSpc105/hKNL1 contains two conserved motifs at the extreme N terminus [(S/G)ILK and RRVSF] and a series of M(D/E)(I/L)(S/T) repeats throughout the N-terminal half of the protein (Fig. 7A, red vertical lines, and Fig. 7B) (4, 8, 20, 24, 28). To examine whether any of these motifs play a role in the interactions with the Bubs kinases, we generated a series of 150-aa fragments derived from blinkin1-1200 and examined interactions by using a Y2H assay (Y2H interactions). We found that the blinkin151-300 fragment specifically interacted with Bubs (Fig. 7C). This blinkin151-300 fragment does not contain the (S/G)ILK and RRVSF motifs but does contain one MDLT motif, which was examined to determine if it necessary for the interaction. The deletion or the mutation of the MDLT motif had no influence on the Y2H interaction with Bubs (Fig. 7D), suggesting that the previously reported conserved motifs are not involved in the interaction with Bub1 and BubR1.
FIG. 7.
Identification of minimal Bub1- and BubR1-binding sites in blinkin. (A) Blinkin contains the conserved motifs [S/G]ILK (light blue), RRVSF (blue), MELT repeat (red), and coiled coil (black). Three regions of blinkin, BLKN (aa 1 to 728), BLKM (aa 729 to 1833), and BLKC (aa 1834 to 2316), and 150-aa fragments are shown in the schematic. (B) Repeat sequences of human blinkin. Highly conserved amino acids and similar amino acids are boxed in black and gray, respectively. Number indicates the amino acid position of blinkin. (C) Yeast two-hybrid interaction between 150-aa fragments of blinkin and Bubs. For the control, p53 and simian virus 40 (SV40) T antigen were used. (D) Y2H interaction between blinkin151-300 mutants and Bub1 or BubR1. A MDLT motif on blinkin151-300 was replaced by alanine (4A) or deleted (ΔMDLT). (E) Y2H analysis between 50-aa fragments of blinkin151-300 and Bubs. Bub1- and BubR1-binding fragments are shown in purple and green, respectively. (F) Y2H assay between BLKN deletion mutants and Bub1 or BubR1.
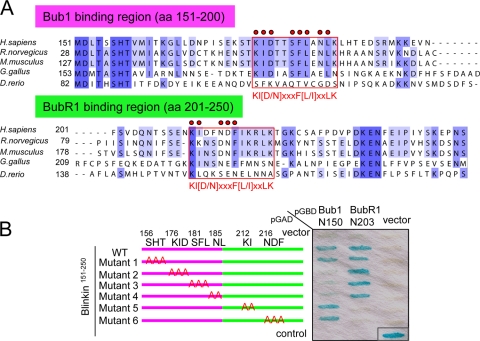
A series of 50-aa fragments of blinkin151-300 were generated to further define its interaction with Bubs. Interestingly, despite the presence of similar TPR domains in Bub1 and BubR1 (7), two distinct fragments, blinkin151-200 and blinkin201-250, were necessary and sufficient for the Y2H interaction with Bub1 and BubR1, respectively (Fig. 7E and F). The sequence of blinkin151-250 is conserved among vertebrates (Fig. 8A). Mutants of blinkin151-250 with an alanine substitution as described for Fig. 8B were generated and tested by Y2H assay, and it was found that 176KID,181SFL, and 185NL were essential for the interaction of blinkin with Bub1 and that 212KI and 216NDF were essential for the interaction of blinkin and BubR1 (Fig. 8B). Interestingly, this analysis revealed a novel amino acid motif common to both interactions, which we name the KI motif [KI(D/N)XXXF(L/I)XXLK; indicated in the red frames in Fig. 8A, where the red circles indicate the alanine-substituted amino acid residues as a possible direct, specific, and essential site for Bub1 and BubR1 binding]. Taken together, the two-hybrid analyses suggest that Bub1 and BubR1 bind to these two closely situated but distinct domains near the N terminus of blinkin.
FIG. 8.
Identification of the Bubs-binding consensus motif in blinkin. (A) Amino acid sequences of Bub1- and BubR1-binding regions in putative blinkin family members in Homo sapiens (GenBank accession no. NP_653091), Rattus norvegicus (XP_230465.2), Mus musculus (NM_029617), Gallus gallus (XM_420938), and Danio rerio (XM_001921844). Conserved amino acids and similar amino acids are highlighted in dark and light purple, respectively. Essential amino acids for the interaction with Bubs identified by a Y2H assay are indicated by red circles. The Bubs recognition KI motif, KI(D/N)XXXF(L/I)XXLK, is enclosed in a red rectangle. (B) Y2H analysis between blinkin151-250 mutants and Bub1 or BubR1. Amino acids replaced by alanine are indicated. The 50-aa fragments of blinkin151-200 and blinkin201-250 are shown in purple and green, respectively.
Adding BLKN abolishes the negative dominance of BLKC.
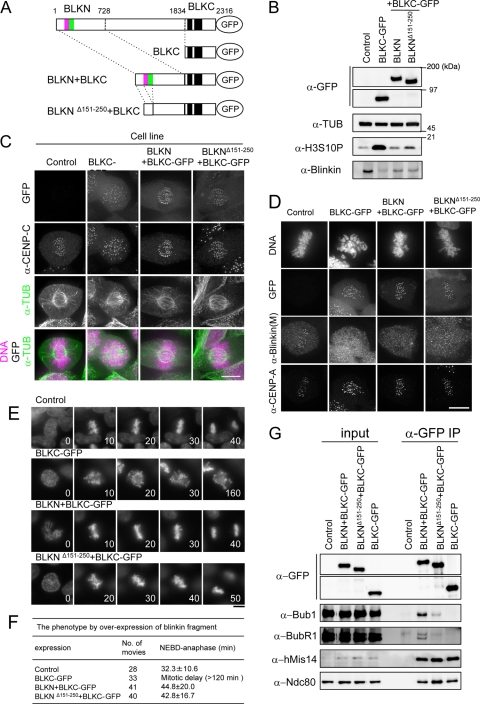
To obtain information about the effect of BLKN on the dominant-negative effect of the BLKC fragment, we constructed stable cell lines that expressed BLKN fused to BLKC-GFP (BLKN+BLKC) or BLKNΔ151-250 fused to BLKC-GFP (BLKNΔ151-250+BLKC) (Fig. 9A). Twenty-four hours after the addition of tet, essentially equal amounts of GFP-tagged proteins were expressed in each cell line (Fig. 9B). The phosphorylated histone H3S10 (mitotic marker) was abundant only in BLKC-GFP-expressing cells and was much reduced when BLKN+BLKC-GFP or BLKNΔ151-250+BLKC was expressed (Fig. 9B, 3rd and 4th lanes). BLKN+BLKC-GFP and BLKNΔ151-250+BLKC were localized at kinetochores (Fig. 9C) and diminished the amount of endogenous blinkin bound to kinetochores (indicated by the antibody against the M region that specifically stained endogenous blinkin but not BLKN+BLKC and BLKNΔ151-250+BLKC) (Fig. 9D). These cells, however, showed apparently normal chromosome alignments (Fig. 9C and D). The results suggest that even in the absence of the middle region (∼1,100 amino acids) of blinkin, the addition of BLKN to BLKC can partly restore the function of blinkin.
FIG. 9.
BLKN fused to BLKC suppresses the dominant-negative effect by BLKC. (A) The blinkin mutant constructs chromosomally integrated in Flip-In TRex 293 cells are shown. Minimal Bub1- and BubR1-binding regions identified by a Y2H assay are indicated in purple and green, respectively. (B) Each cell line was treated by tet to express GFP-tagged blinkin constructs. Extracts were obtained 24 h after the addition of tet and immunoblotted by antibodies as indicated. Nonintegrated cells was used as controls. (C) Each cell line was fixed and stained by Hoechst 33342 and anti-TUB and anti-CENP-C antibodies 24 h after the tet treatment. (D) Each cell line was fixed and stained by Hoechst 33342, anti-blinkin(M), which specifically recognizes the middle region of blinkin, and anti-CENP-A antibodies 24 h after the tet treatment. (E) Time-lapse micrographs of each cell from 24 h after the addition of tet are shown. Hoechst 33342 was used for DNA staining. The number indicates the time (in minutes). Bars, 10 μm. (F) Summary of the phenotypes in four different cell lines. (G) Each cell line was cultured for 12 h after tet treatment and for an additional 18 h after the addition of nocodazole. Extracts were immunoprecipitated with anti-GFP antibodies. Input and immunoprecipitates (IP) were immunoblotted using the indicated antibodies.
To critically examine chromosome alignment and mitotic progression, a number of movies were taken. Most chromosomes aligned properly in cells that expressed BLKNΔ151-250+BLKC-GFP (Fig. 9E). The mitotic duration measured (defined as being from nuclear envelope breakdown [NEBD] to anaphase onset) was not significantly different between cells that expressed BLKN+BLKC-GFP (44.8 ± 20 min, n = 41 cells) and those that expressed BLKNΔ151-250+BLKC-GFP (42.8 ± 16.7 min, n = 40) (Fig. 9F). The N-terminal blinkin fragment that lacked the minimal Bubs-binding domains (BLKNΔ151-250) also suppressed the dominant-negative effect of BLKC in the presence of residual endogenous blinkin. The instability of endogenous blinkin was more prominent in cells that expressed BLKC than in those that expressed BLKN+BLKC or BLKNΔ151-250+BLKC (Fig. 9B). In prolonged mitosis in BLKC-expressing cells, endogenous blinkin free from the kinetochore may become unstable. Alternatively or simultaneously, BLKC might irreversibly bind to the hMis12 complex and displace endogenous blinkin more effectively than the N terminus-fused BLKC constructs so that the resulting blinkin, free from the hMis12 complex, might become unstable during mitosis.
To understand the functional difference between BLKN and BLKNΔ151-250, we examined the Bubs-binding ability of these N terminus constructs in vivo. GFP-tagged BLKC, BLKN+BLKC, and BLKNΔ151-250+BLKC were immunoprecipitated with anti-GFP antibodies from mitotic extracts of each respective cell line (Fig. 9G). Parent cells without chromosomal integration of the exogenous gene were used as a control. BLKNΔ151-250+BLKC-GFP diminished the interaction with Bubs, while BLKN+BLKC-GFP strongly coimmunoprecipitated with Bubs (Fig. 9G), and BLKC-GFP did not coimmunoprecipitate with Bubs. This result indicates that the N terminus of blinkin, BLKN (aa 1 to 728), is sufficient for the interaction with Bubs in vivo and that blinkin151-250 is essential for the proper association with Bubs, as suggested by the Y2H assay.
Minimal Bubs-binding sites are essential for chromosome alignment and segregation.
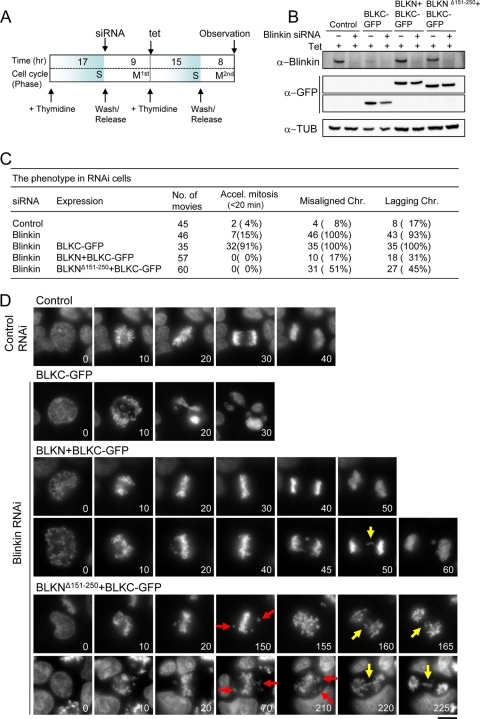
The results suggested that mitosis was supported because of the remaining endogenous blinkin in the cells that expressed BLKNΔ151-250+BLKC-GFP. To determine whether this is correct, the endogenous blinkin was depleted by using RNAi in cells that expressed BLKNΔ151-250+BLKC-GFP during mitosis in the synchronous culture (Fig. 10A). To specifically deplete endogenous blinkin and not the truncated constructs, blinkin siRNA was designed to target the middle region of blinkin that was not present in the constructs. GFP-tagged fragments (BLKC-GFP, BLKN+BLKC-GFP, and BLKNΔ151-250+BLKC-GFP) were expressed in the presence of tet (Fig. 10B, Tet, +), while the endogenous blinkin was depleted by blinkin siRNA (Fig. 10B, Blinkin siRNA, +). BLKN+BLKC-GFP- and BLKNΔ151-250+BLKC-GFP-expressing cells were well synchronized, and the extracts were analyzed just before the 2nd mitosis (Fig. 10A); in contrast, BLKC-GFP-expressing cells showed a mitotic delay during the 1st mitosis, and the protein level of endogenous blinkin was reduced before the 2nd mitosis.
FIG. 10.
Deletion of the minimal Bubs-binding domain in BLKN+BLKC fails to suppress chromosome misalignment after blinkin RNAi treatment. (A) Schematic of experimental procedures. Each cell line was synchronized using a double-thymidine block protocol. Tetracycline (tet) and siRNA were added at the indicated time points. (B) Each cell line was treated with tet to express GFP-tagged blinkin constructs. Extracts were obtained 23 h after the addition of tet and immunoblotted by antibodies as indicated. Nonintegrated cells were used as controls. (C) Summary of the phenotypes of five different RNAi cells. Chr., chromosomes. (D) Time-lapse micrographs of each cell in control RNAi or blinkin RNAi are shown. Hoechst 33342 was used for DNA staining. Misaligned and lagging chromosomes are indicated by the red and yellow arrows, respectively. The number indicates the time (in minutes). Bar, 10 μm.
A number of movies were taken to monitor mitotic timing and chromosome alignment for cells that expressed GFP-tagged proteins under the RNAi depletion of endogenous blinkin (Fig. 10C). Control RNAi cells without expression of BLKC-GFP and blinkin RNAi cells expressing BLKC-GFP showed the expected phenotypes of normal anaphase and accelerated mitosis (<20 min), respectively (Fig. 10C and D). Under these conditions, only 15% of the blinkin RNAi cells showed accelerated mitosis (4% in control cells), and the remaining cells displayed a mitotic delay, probably caused by the incomplete knockdown of blinkin protein as previously reported (17) (Fig. 10C).
To determine the functional involvement of the Bubs-binding site in blinkin, we compared the phenotypes between BLKN+BLKC- and BLKNΔ151-250+BLKC-expressing cells after blinkin RNAi treatment. In BLKN+BLKC cells, most chromosomes aligned properly (Fig. 10D, 3rd and 4th rows) and entered anaphase with normal timing (44 ± 17 min, n = 57 cells), similar to the case for control cells (43 ± 21 min, n = 45) (Fig. 10D, Control panels), though some perturbations occurred in mitosis (Fig. 10C). In sharp contrast, chromosomes in BLKNΔ151-250+BLKC cells were frequently (51%, n = 60) misaligned (Fig. 10D, red arrows) and segregated abnormally after a transient mitotic delay (114 ± 68 min). Several lagging chromosomes (Fig. 10D, yellow arrows) were observed (45%, n = 60) during anaphase in BLKNΔ151-250+BLKC cells. These results suggest that the minimal Bubs-binding site in blinkin is essential for chromosome alignment and segregation (see Discussion).
DISCUSSION
Using a Y2H assay and immunoprecipitation of human cell lines that stably expressed the tagged wild-type and mutant blinkin, we showed that different regions of human blinkin interact with five different kinetochore proteins: hMis14, Zwint-1, and Ndc80 at its C terminus and two checkpoint kinases, Bub1 and BubR1, at its N terminus. While the interaction with Ndc80 appeared to be indirect, perhaps via Zwint-1 and hMis14, the other four proteins may interact directly with blinkin, indicating that blinkin is a central player for regulating these kinetochore and checkpoint functions.
We investigated a dominant-negative effect caused by the expression of BLKC, the C-terminal fragment of blinkin. A similar negative effect was reported for the blinkin orthologue fission yeast Spc7 (15) and for Drosophila dmSpc105 (29). Overexpressed BLKC appeared to occupy hMis14 by binding and prevented endogenous blinkin from interacting with hMis14.
The negative dominance of BLKC was alleviated if the N terminus was fused to the C terminus. These results suggest that the N-terminal region is critical for the function of blinkin. Consistently, we showed that BLKN+BLKC nearly substituted for the function of blinkin, though some perturbations of mitosis occurred. It was previously shown that the large middle (M) region of blinkin weakly interacts with Bub1, as assessed by Y2H assays, and that it recruits Bub1 at kinetochores in HeLa cells (17). The M region of blinkin might function to assist interactions with Bubs and other unidentified kinetochore proteins.
Blinkin was originally discovered as a fusion partner with the mixed-lineage leukemia (MLL) gene (19). The fusion protein contains the C terminus of blinkin1793-2316 that lacks the functionally important N-terminal region and probably causes a dominant-negative effect if expressed separately. We transiently expressed GFP-tagged blinkin1793-2316 in HeLa cells and found that most cells showed chromosome misalignments, as did cells that expressed GFP-BLKC (data not shown). The negative dominance generated by the C terminus of blinkin and its relief by the fusions with the amino-terminal region might mimic the situation in generating cancer cells. For example, the fused MLL fragment in cancer cells might disrupt the strong mitotic arrest exerted by the C terminus of blinkin and result in chromosome missegregation and genomic instability, eventually producing leukemic cells. Alternatively, MLL-blinkin fusion protein may be recruited to MLL target loci, just like MLL-AF4 and -ENL fusion proteins (35), and change chromatin and transcription states by abnormally recruiting heterochromatin protein HP1 via hMis14. Since AF15q14 (blinkin) was reported to be an oncogenic agent for lung cancer (31) and mutations in Bubs are found in numerous cancer cells, blinkin might represent a useful therapeutic target. A drug that specifically targets the kinetochore localization domain of blinkin (that includes the sequence 2166PPSS or the surrounding amino acids in the C-terminal region of blinkin) (Fig. 2C) might be a useful therapeutic reagent by partially impairing the kinetochore localization of blinkin and arresting cells in mitosis.
Previous reports indicated that the N terminus of blinkin contained both unique and repetitive sequences conserved from fungi to humans (4, 8, 24, 28). However, the present mutational analysis indicates that these sequences are not involved in the interaction of blinkin with Bubs. Instead, two closely situated regions near the N terminus of blinkin may independently interact with Bub1 and BubR1. Mutations in each of the Bub interaction KI motifs [KI(D/N)XXXF(L/I)XXLK] selectively abolish the interactions of blinkin with Bub1 or BubR1. The KI motif has not yet been found in lower eukaryotes. However, the N terminus of fly dmSpc105 binds to dmBub1, as determined by a Y2H assay (29). The bulky hydrophobic amino acids in this motif may be important, and the three-dimensional (3D) structure rather than the amino acid sequence might be conserved to allow interaction with Bubs. The crystal structure of the N terminus of Bub1 and BubR1 contains similar tandem TPR repeats (1, 7), the groove of which might fit with these bulky amino acids in the KI motifs of blinkin. On the other hand, the specific recognition of Bub1 and BubR1 by blinkin may be ensured by the different sequences or structures around the KI motifs on blinkin. Mutations that cause blinkin to misrecognize Bubs may lead to Bubs dysfunction.
The results show that blinkin interacts with both Bub1 and BubR1 at the same time and functions to bring the two kinases close to each other. This delicate binding mode and ternary complex formation at the N terminus of blinkin might be important for Bubs to function properly on microtubule attachment and checkpoint signaling in a cooperative manner. The extreme N-terminal region of blinkin/hKNL1 (aa 1 to 86) binds to microtubules and PP1 phosphatase in an aurora-B-regulated fashion (20, 33). The kinetochore localization of Bubs is affected by proper microtubule attachment and aurora-B kinase activity (9, 13, 32). To understand kinetochore-based checkpoint signaling and its regulation, it is necessary to reveal how these and other binding proteins, kinases, or phosphatases affect the formation of the blinkin-Bubs ternary complex.
The present replacement experiments show that the minimal Bubs-binding site in blinkin is essential for proper chromosome alignment and segregation (Fig. 10D). Blinkin mutants lacking the Bubs-binding domain, BLKNΔ151-250+BLKC, caused an abnormal onset of anaphase with misaligned chromosomes when endogenous blinkin was depleted by RNAi, suggesting that the mitotic checkpoint was not fully functional. The mitotic delay in this mutant may derive from the residual endogenous blinkin or uncharacterized function of the N-terminal blinkin. As BLKN+BLKC promoted normal-looking mitosis with a slight perturbation in the absence of endogenous blinkin, we concluded that the Bubs-binding site is required for proper mitosis.
The interaction of Ndc80 with the hMis12 complex is somewhat enigmatic. Ndc80 is immunoprecipitated with hMis12 complex (4, 25), but the Y2H assay failed to show their interaction. A previous Y2H analysis indicated that Zwint-1 links between Ndc80/HEC1 and blinkin (17). Immunoprecipitation data indicated, however, that Zwint-1 is not sufficient to link the Ndc80 complex to blinkin in HeLa cells. Further, a Zwint-1 orthologue has not been identified for Schizosaccharomyces pombe. An evolutionarily conserved Spc7 and the complexes of Mis12 and Ndc80 of S. pombe form a stable supramolecular assembly in a near-stoichiometric fashion (Y. Shiroiwa, T. Hayashi, M. Ebe, and M. Yanagida, unpublished data). The Ndc80 complex may have direct physical contact with blinkin and/or the hMis12 complex, but the interaction might take place in only the kinetochore after the hMis12 complex has properly interacted with blinkin.
The hMis14 m2E mutant was originally isolated due to its defective interactions with heterochromatin protein 1 (HP1) in interphase and the resulting impairment of the inner centromere and kinetochore during mitosis. The same m2E mutant did not properly localize at the kinetochores (16) and did not coprecipitate with Ndc80, which suggests that hMis14 may function to properly interact with the Ndc80 complex at the kinetochore. Alternatively, the Ndc80 complex may bind to the PXVXL motif in hMis14 during mitosis. Recently, the Ndc80 complex was reported to interact in vitro with the hMis12 complex but not with the hMis12 complex containing the hMis14 m2E mutant (26). In fission yeast, the Mis12 complex also interacts with the Ndc80 complex but not with the HP1 homologues Swi6 and Chp2 (21). This binding switch of the hMis12 complex from HP1 to the Ndc80 complex may occur only in higher eukaryotes with large amounts of repetitive centromeric DNA to coordinate the formation of the inner centromere and kinetochore structure. In fact, the HP1-binding PXVXL motif on hMis14 is well conserved among mammals but not among fungi (16). While the conserved Mis12 complex serves as a basis for the assembly of blinkin/Spc105/Spc7/KNL1 and the Ndc80 complex at the kinetochore, the Mis12 complex may also have different binding partners according to the divergent centromere structure in each organism.
Acknowledgments
We are indebted to N. Nozaki for generating the BLKC, BLKM, and hNnf1 antibodies and Y. Tajima, H. Kawai, and M. Aoki for preparing the plasmids. We are grateful to I. Cheeseman for reading the manuscript and providing comments.
This study was supported by grants for Specially Promoted Research from the Ministry of Education, Sport, Culture, Science and Technology and the CREST Research Program of the Japan Science and Technology Corporation (JST), a Grant-in-Aid for Young Scientists (Start-up), and the Naito Foundation.
Footnotes
▿
Published ahead of print on 3 January 2011.
REFERENCES
- 1.Bolanos-Garcia, V. M., et al. 2009. The crystal structure of the N-terminal region of BUB1 provides insight into the mechanism of BUB1 recruitment to kinetochores. Structure 17**:**105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheeseman, I. M., J. S. Chappie, E. M. Wilson-Kubalek, and A. Desai. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127**:**983-997. [DOI] [PubMed] [Google Scholar]
- 3.Cheeseman, I. M., and A. Desai. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9**:**33-46. [DOI] [PubMed] [Google Scholar]
- 4.Cheeseman, I. M., et al. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18**:**2255-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciferri, C., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133**:**427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112**:**407-421. [DOI] [PubMed] [Google Scholar]
- 7.D'Arcy, S., O. R. Davies, T. L. Blundell, and V. M. Bolanos-Garcia. 2010. Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. J. Biol. Chem. 285**:**14764-14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai, A., et al. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17**:**2421-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ditchfield, C., et al. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161**:**267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160**:**25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goshima, G., S. Saitoh, and M. Yanagida. 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 13**:**1664-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haraguchi, T., T. Kaneda, and Y. Hiraoka. 1997. Dynamics of chromosomes and microtubules visualized by multiple-wavelength fluorescence imaging in living mammalian cells: effects of mitotic inhibitors on cell cycle progression. Genes Cells 2**:**369-380. [DOI] [PubMed] [Google Scholar]
- 13.Hauf, S., et al. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161**:**281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi, T., et al. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118**:**715-729. [DOI] [PubMed] [Google Scholar]
- 15.Kerres, A., V. Jakopec, and U. Fleig. 2007. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol. Biol. Cell 18**:**2441-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiyomitsu, T., O. Iwasaki, C. Obuse, and M. Yanagida. 2010. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J. Cell Biol. 188**:**791-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiyomitsu, T., C. Obuse, and M. Yanagida. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13**:**663-676. [DOI] [PubMed] [Google Scholar]
- 18.Kline, S. L., I. M. Cheeseman, T. Hori, T. Fukagawa, and A. Desai. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173**:**9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuefer, M. U., et al. 2003. Characterization of the MLL partner gene AF15q14 involved in t(11;15)(q23;q14). Oncogene 22**:**1418-1424. [DOI] [PubMed] [Google Scholar]
- 20.Liu, D., et al. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188**:**809-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, X., I. McLeod, S. Anderson, J. R. Yates, 3rd, and X. He. 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24**:**2919-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meraldi, P., A. D. McAinsh, E. Rheinbay, and P. K. Sorger. 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7**:**R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musacchio, A., and E. D. Salmon. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8**:**379-393. [DOI] [PubMed] [Google Scholar]
- 24.Nekrasov, V. S., M. A. Smith, S. Peak-Chew, and J. V. Kilmartin. 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14**:**4931-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obuse, C., et al. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6**:**1135-1141. [DOI] [PubMed] [Google Scholar]
- 26.Petrovic, A. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190**:**835-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Przewloka, M. R., and D. M. Glover. 2009. The kinetochore and the centromere: a working long distance relationship. Annu. Rev. Genet. 43**:**439-465. [DOI] [PubMed] [Google Scholar]
- 28.Przewloka, M. R., et al. 2007. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One 2**:**e478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schittenhelm, R. B., R. Chaleckis, and C. F. Lehner. 2009. Intrakinetochore localization and essential functional domains of Drosophila Spc105. EMBO J. 28**:**2374-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi, K., H. Yamada, and M. Yanagida. 1994. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell 5**:**1145-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takimoto, M., et al. 2002. Frequent expression of new cancer/testis gene D40/AF15q14 in lung cancers of smokers. Br. J. Cancer 86**:**1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, S. S., D. Hussein, Y. Wang, S. Elderkin, and C. J. Morrow. 2001. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114**:**4385-4395. [DOI] [PubMed] [Google Scholar]
- 33.Welburn, J. P., et al. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38**:**383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanagida, M. 2005. Basic mechanism of eukaryotic chromosome segregation. Philos Trans. R. Soc. Lond. B Biol. Sci. 360**:**609-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama, A., M. Lin, A. Naresh, I. Kitabayashi, and M. L. Cleary. 2010. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17**:**198-212. [DOI] [PMC free article] [PubMed] [Google Scholar]