The structure of human 5-lipoxygenase (original) (raw)
. Author manuscript; available in PMC: 2011 Dec 23.
Published in final edited form as: Science. 2011 Jan 14;331(6014):217–219. doi: 10.1126/science.1197203
Abstract
The synthesis of both pro-inflammatory leukotrienes and anti-inflammatory lipoxins requires the enzyme 5-lipoxygenase (5-LOX). 5-LOX activity is short-lived, apparently in part due to an intrinsic instability of the enzyme. We identified a 5-LOX-specific destabilizing sequence that is involved in orienting the carboxy-terminus which binds the catalytic iron. Herein we report the crystal structure at 2.4 Å resolution of human 5-LOX stabilized by replacement of this sequence.
Leukotrienes (LT) and lipoxins are potent mediators of the inflammatory response derived from arachidonic acid (AA). When leukocytes are activated, arachidonic acid is released from the nuclear membrane by the action of cytosolic phospholipase A2 and binds five-lipoxygenase-activating protein (FLAP). The increased Ca2+ concentration of the activated cells simultaneously promotes translocation of 5-LOX to the nuclear membrane where it acquires its substrate from FLAP (1, 2). Arachidonic acid (AA) is converted to leukotriene (LTA4) in a two-step reaction which produces the 5_S_-isomer of hydroperoxyeicosatetraenoic acid (5_S_-HPETE) as an intermediate (3, 4).
Auto-inactivation of 5-LOX activity has been described, and this loss of activity is perhaps important in limiting the synthesis of its pro- and anti-inflammatory products (5). Previous reports indicate that non-turnover based inactivation is a consequence of an O2 sensitivity linked to the oxidation state of the catalytic iron (6). However, not all LOXs display this hypersensitivity to O2. For example, 8_R_-LOX activity is stable despite a solvent exposed iron coordination sphere equivalent to that in 5-LOX (7). In similar conditions 50% of 5-LOX activity is lost in 10 hours (8). We reasoned that 5-LOX specific destabilizing features may confer susceptibility to non-turnover based inactivation. Regulatory mechanisms that facilitate transient activation include targeted degradation, phosphorylation, and allosteric control of enzyme activities. Auto-inactivation as a consequence of intrinsic protein instability may play a similar role. For example, the instability of the tumor suppressor protein p53, relative to its orthologs such as p73, has been proposed to have a functional role (9).
Based on the crystal structures of two AA-metabolizing lipoxygenases [an 8_R_-LOX from _Plexaura homomalla_ (7, 10) and a 15-LOX from rabbit reticulocyte (11, 12)], each with ~40% sequence identity to 5-LOX), we identified a 5-LOX specific lysine-rich region near the C-terminus of the enzyme that might confer instability. In the 8_R_- and 15-LOX structures, a turn centered on amino acid 655 (5-LOX numbering) leads from the C-terminal helix to the carboxyl terminal segment, allowing the terminal carboxylate to penetrate the LOX body and bind the catalytic iron (Fig 1A, 1B). In most LOXs, amino acid 655 is a highly conserved Leu, with its side chain pointing toward an invariant Arg (651). A striking 5-LOX specific feature is Lys in place of Leu at this position as part of a di- or tri- Lys peptide (Fig S1). While numerous salt links anchor the C-terminal helix to the body of the protein in the structures of the two homologues noted above, none of these salt links is conserved in the 5-LOX sequence. As a consequence of the lysine-rich sequence and the absence of helix-anchoring salt-links, the orientation of the terminal helix is less favorable and the C-terminal ligand to the active site Fe is likely to be tenuously restrained. Conservative mutations in the carboxy terminal helix have been noted to reduce enzyme expression levels and activity (13). These observations led us to replace 5-LOX KKK653-655 with the corresponding sequence from 8_R_-LOX (ENL) in an effort to stabilize the enzyme for crystallographic studies.
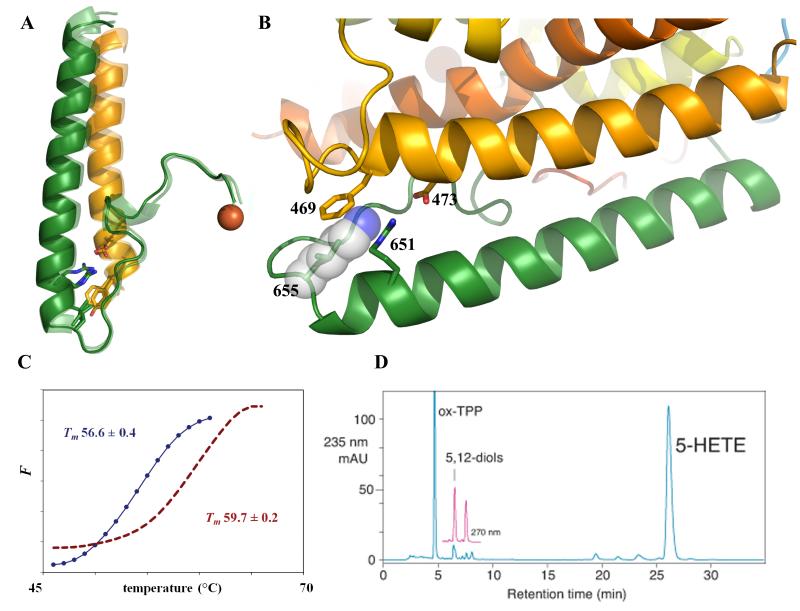
Fig 1. Stabilization of human 5-LOX.
(A) Superposition of the C-terminal regions of the structures of 15-, 8_R_-, and Stable-5-lipoxygenase. The C-terminal segment that leads to the catalytic Fe emanates from the helix which terminates at amino acid 655 (5-LOX numbering; Stable-5-LOX, pink; 8_R_-LOX green; 15-LOX, blue). Highly conserved amino acids (Leu, Phe/Tyr) and an invariant salt link (Asp-Arg) are depicted in stick rendering. (B) Detail of the turn at the end of the terminal helix. The 5-LOX specific Lys (substituted in Stable-5-LOX with Leu) is modeled at position 655 as its most common rotamer (transparent sphere rendering). As positioned, it would interfere with the invariant salt-link and cation- π interactions. All figures were generated with Pymol (29). (C) Thermal denaturation of Stable-5-LOX (red) and the parent enzyme Sol-5-LOX (blue). Fluorescence (F) is monitored as a function of temperature. Tm (with s.d.) 56.6 (±0.4) and 59.7(±0.2) °C for Sol-5-LOX and Stable-5-LOX, respectively. (D) HPLC chromatogram. Product analysis of Stable-5-LOX reveals both 5-HETE (5-HPETE reduced by the addition of triphenylphosphine, TPP) and Leukotriene A4 hydrolysis products (5,12 diols).
The mutant human enzyme (herein referred to as Stable-5-LOX) was prepared in the context of a soluble 5-LOX (Sol-5-LOX) which lacks putative membrane insertion amino acids (Δ40-44GS, W13E, F14H, W75G, L76S) as well as a pair of Cysteines (C240A, C561A) predicted to be proximal in the 5-LOX structure. Substitution of the membrane insertion loops was based on a similar approach with the Plexaura homomalla enzyme, which shares both these amino acids and Ca2+-binding residues with 5-LOX in the amino terminal membrane-binding domain (14). The substitution of KKK with ENL in this context led to a ~3°C increase in the melting temperature of the enzyme (Fig 1C). Moreover, Stable-5-LOX has a longer half-life at 37 °C (~16 hrs vs. ~7 hrs, Fig S2). Furthermore, Stable-5-LOX produces both the intermediate 5_S_-HPETE and the product leukotriene A4 (Fig 1D), as does its progenitor protein Sol-5-LOX (Fig S3). In addition, we measure a Km for AA of ~11μM (Fig S4), equivalent to that of the wild-type enzyme (15). These observations are consistent with the proposal that the KKK sequence is destabilizing, and that its substitution does not impact catalytic fidelity. The structure of Stable-5-LOX was determined to 2.4 Å resolution (Fig. 2A, S5; Table S1).
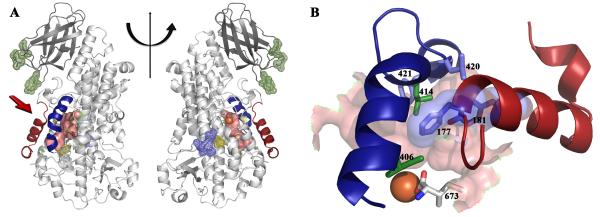
Fig 2. The structure of Stable-5-LOX.
(A) A cartoon rendering of 5-LOX. The two views differ by a 180° rotation about the vertical line. The amino terminal C2-like domain is in dark gray, and the catalytic domain in light gray. The distinctive arched helix is in blue, and helix α2 in red. The internal cavity, generated with CastP (30), is in pink and the Fe is an orange sphere. The positions of the mutated amino acids are indicated in mesh rendering: green, putative membrane insertion residues; yellow, proximal cysteines; and blue, the KKK→ENL substitution. (B). Detail of the relationship of the arched helix and helix α2 to the active site as viewed from the perspective indicated by the red arrow in panel (A). Shown in stick rendering are amino acids 406, 414, 420, 421 of the arched helix and F177 and Y181 from helix α2 (with transparent surface rendering). The latter two bulky amino acids obstruct access to the cavity. The proximity of the C-terminal Ile (673) to the corked portal is apparent.
The canonical LOX framework contains two distinct domains: the amino terminal “C2-like” domain (~120 amino acids), which in 5-LOX confers Ca2+-dependent membrane binding (16-19), and the larger catalytic domain. The latter is primarily α-helical and harbors the non-heme catalytic iron. The iron is coordinated by three conserved histidines (His- 367, 372, 550) as well as the main-chain carboxylate of the C-terminus (I673). Another structurally distinct conserved feature in this domain, previously described in detail by Minor et al (20) for soybean LOX L-1, is an arched helix that shields access to the catalytic iron. At the vertex of the Stable-5-LOX arched helix is Leu-414 (Fig 2B), an invariant amino acid that in other lipoxygenases has been proposed to control access of O2 to the substrate (21, 22) or position the substrate pentadiene for attack (7). Additional amino acids from the arched helix that help define the catalytic site are Leu-420 and Phe-421. The crystal structure of Stable-5-LOX reveals a striking variation on the classic lipoxygenase fold in helix α2 which defines one edge of the active site. In the structures of 8_R_- and 15-LOX helix α2 is 6-7 turns, while in Stable-5-LOX it is a short 3-turn helix flanked by extended loops. The shortened helix is positioned at ~45° to its counterparts in the 8_R_- and 15_S_- enzymes (Fig 3A,B). The unique orientation of helix α2 in Stable-5-LOX greatly limits access to the catalytic iron and yields a distinctive active site cavity. Specifically, the side chains of F177 and Y181 are positioned inward and close off an access channel to the catalytic iron that is observed in both the 8_R_- and 15-LOX structures (Fig. 2B, 4A). The remainder of the secondary structural elements, and their relative orientations, are maintained (Fig 3B). In addition, the structural context of the Lys-rich peptide also appears conserved as the C-terminal helices superimpose (Fig. 1A). However, it is apparent that a Lys at position 655 would interfere with invariant salt link and cation-π interactions (Fig 1B).
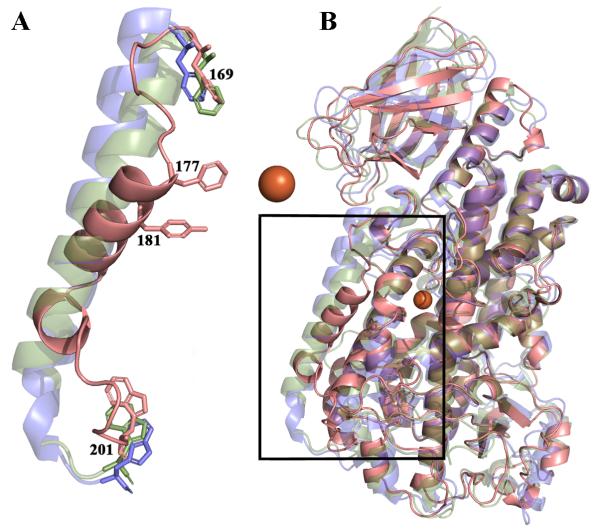
Fig. 3. (A) The positioning of helix α2 is unique in 5-LOX.
(A) A 5-LOX cartoon is rendered in pink, 15-LOX in blue and 8_R_-LOX in green. Conserved aromatic amino acids (F169,W201) that flank the region are in stick rendering. F177 and Y181 that make up the “cork” that helps define the active site are in stick. The catalytic iron is an orange sphere. (B) A full overlay of the three structures in which it is apparent that, with the exception of α2, the secondary structural elements in the enzymes are conserved. The box indicates the region amplified in (A).
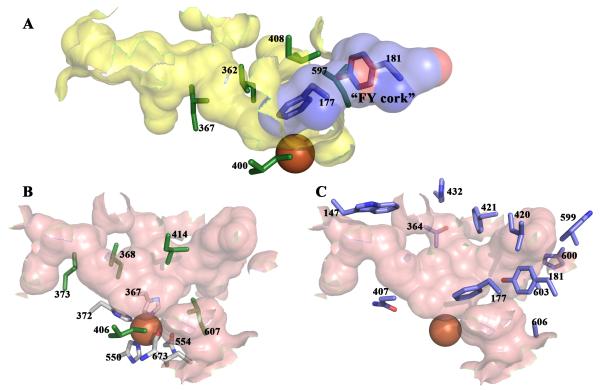
Fig 4. The 5-LOX active site.
Internal cavities calculated with CastP (30). (A) The active site cavity of 15-LOX (2P0M) is in yellow. Invariant Leu and Ile side chains are in green stick rendering. The 5-LOX “FY cork” is superposed on the 15-LOX cavity and plugs the entrance. (B) The equivalent orientation of the active site cavity of Stable-5-LOX in pink; invariant Leu and Ile side chains in green sticks. Note the similarity of the positions of these amino acids to their counterparts in 15-LOX (A). Iron coordination sphere amino acids (C, white) are in stick rendering, and the iron an orange sphere. (C) 5-LOX amino acids that contribute to the active site cavity. Entry into this cavity requires a conformational change.
In Stable-5-LOX the active site is an elongated cavity, with no clear access to bulk solvent, lined with both invariant and 5-LOX specific amino acids. Leu- 368, 373, 414, 607 and Ile-406 are conserved in all AA-metabolizing lipoxygenases (7) and form a structurally similar constellation of branched hydrophobic side chains that envelop the region where the pentadiene must be positioned for catalysis (Fig. 4A,B). Y181, A603, A606, H600 and T364 are specific to 5-LOX sequences and the small side chains of A603 and A606 appear to be required for the conformation of Y181 which, along with F177, “corks” the cavity at one end. Y181 is in van der Waals contact with A603, and the small side chains of both 603 and 606 allow both bulky aromatics (F177, Y181) to point into the cavity where they can be shielded from solvent (Fig. 4C). An additional 5-LOX specific amino acid, W599, appears to buttress the FY cork from one side. Amino acids Asn-407 and His-432 also help define the active site.
The closed cavity (volume = 663Å3) raises the question of how substrate gains access to the catalytic iron. Two possibilities can be envisioned: (1) Removal of the FY cork at one end of the cavity and/or movement of W599 that secures it, or (2) A rotamer shift of W147 at the opposite end. A rotamer shift in W147 would require only rotation of the side chain, while the former may require both side chain and main chain movements in two amino acids. This observation suggests that AA may enter 5-LOX from the opposite direction as it does in the 15_S_- or 8_R_- enzymes, which lack the FY cork. This site of entry fits well with what is known about the catalytic mechanism: H abstraction and peroxidation occur on opposite sides of the pentadiene (23). The _S_-stereochemistry of the 5-LOX product is consistent with an “inverse” orientation of AA relative to that for the 15_S_- and 8_R_- enzymes (24, 25). An opening at the W147 end would allow the AA to enter methyl end first and position the substrate for the production of the S isomer of 5-HPETE. While the above model is attractive, the structure does not rule out the alternative: that the AA enters the same portal it does in 8_R_- and 15_S_- enzymes. Carboxylate-first entry in this latter mode achieves the same binding orientation and reaction specificity.
The 2.4 Å structure of Stable-5-LOX reveals an active site which, despite a conserved constellation of five invariant amino acids, is clearly distinct from the active sites of the arachidonic acid metabolizing lipoxygenases for which structures are available. The structure provides a context for the development of 5-LOX specific inhibitors and together with the crystal structures of FLAP (26) and the downstream enzyme Leukotriene C4 Synthase (27, 28), a molecular model for early events in leukotriene biosynthesis.
Supplementary Material
SOM
One sentence summary.
Identification of a destabilizing sequence, possibly involved in function, allowed crystallization of a key enzyme of the inflammatory response.
Acknowledgments
This work was funded in part by grants from the American Heart Association (08553920E) and NSF (0818387) to MEN, and NIH (GM-15431) to ARB. Preliminary work was performed at the Center for Advanced Microstructures and Devices (Baton Rouge), funded in part by the Louisiana Governors’ Biotechnology Initiative. X-ray data were collected at Beam Line 24-ID-E of NE-CAT at the Advanced Photon Source. Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession number 3O8Y.
References
- 1.Evans JF, Ferguson AD, Mosley RT, Hutchinson JH. Trends Pharmacol Sci. 2008 Feb;29:72. doi: 10.1016/j.tips.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Dixon RA, et al. Nature. 1990 Jan 18;343:282. doi: 10.1038/343282a0. [DOI] [PubMed] [Google Scholar]
- 3.Radmark O, Samuelsson B, Lipid J. Res. 2009 Apr;50(Suppl):S40. doi: 10.1194/jlr.R800062-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu T, Radmark O, Samuelsson B. Proc Natl Acad Sci U S A. 1984 Feb;81:689. doi: 10.1073/pnas.81.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy RC, Gijon MA. Biochem J. 2007 Aug 1;405:379. doi: 10.1042/BJ20070289. [DOI] [PubMed] [Google Scholar]
- 6.Percival MD, Denis D, Riendeau D, Gresser MJ. Eur J Biochem. 1992 Nov 15;210:109. doi: 10.1111/j.1432-1033.1992.tb17397.x. [DOI] [PubMed] [Google Scholar]
- 7.Neau DB, et al. Biochemistry. 2009 Aug 25;48:7906. doi: 10.1021/bi900084m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang YY, Hamberg M, Radmark O, Samuelsson B. Anal Biochem. 1994 Jul;220:28. doi: 10.1006/abio.1994.1294. [DOI] [PubMed] [Google Scholar]
- 9.Canadillas JM, et al. Proc Natl Acad Sci U S A. 2006 Feb 14;103:2109. doi: 10.1073/pnas.0510941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldham ML, Brash AR, Newcomer ME. J Biol Chem. 2005 Nov 25;280:39545. doi: 10.1074/jbc.M506675200. [DOI] [PubMed] [Google Scholar]
- 11.Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner MF. Nat Struct Biol. 1997;4:1003. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 12.Choi J, Chon JK, Kim S, Shin W. Proteins. 2008 Feb 15;70:1023. doi: 10.1002/prot.21590. [DOI] [PubMed] [Google Scholar]
- 13.Kuhn H, Anton M, Gerth C, Habenicht A. Arterioscler Thromb Vasc Biol. 2003 Jun 1;23:1072. doi: 10.1161/01.ATV.0000074167.01184.48. [DOI] [PubMed] [Google Scholar]
- 14.Neau DB, Gilbert NC, Bartlett SG, Dassey A, Newcomer ME. Acta Crystallographica Section F. 2007;63:972. doi: 10.1107/S1744309107050993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aharony D, Stein RL. J Biol Chem. 1986 Sep 5;261:11512. [PubMed] [Google Scholar]
- 16.Chen XS, Zhang YY, Funk CD. J Biol Chem. 1998 Nov 20;273:31237. doi: 10.1074/jbc.273.47.31237. [DOI] [PubMed] [Google Scholar]
- 17.Chen XS, Funk CD. J Biol Chem. 2001 Jan 5;276:811. doi: 10.1074/jbc.M008203200. [DOI] [PubMed] [Google Scholar]
- 18.Hammarberg T, Reddy KV, Persson B, Radmark O. Adv Exp Med Biol. 2002;507:117. doi: 10.1007/978-1-4615-0193-0_19. [DOI] [PubMed] [Google Scholar]
- 19.Kulkarni S, Das S, Funk CD, Murray D, Cho W. J Biol Chem. 2002 Apr 12;277:13167. doi: 10.1074/jbc.M112393200. [DOI] [PubMed] [Google Scholar]
- 20.Minor W, et al. Biochemistry. 1996;35:10687. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- 21.Knapp MJ, Klinman JP. Biochemistry. 2003 Oct 7;42:11466. doi: 10.1021/bi0300884. [DOI] [PubMed] [Google Scholar]
- 22.Knapp MJ, Seebeck FP, Klinman JP. J Am Chem Soc. 2001 Mar 28;123:2931. doi: 10.1021/ja003855k. [DOI] [PubMed] [Google Scholar]
- 23.Schneider C, Pratt DA, Porter NA, Brash AR. Chem Biol. 2007 May;14:473. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walther M, Ivanov I, Myagkova G, Kuhn H. Chem Biol. 2001 Aug;8:779. doi: 10.1016/s1074-5521(01)00050-3. [DOI] [PubMed] [Google Scholar]
- 25.Coffa G, Brash AR. Proc Natl Acad Sci U S A. 2004 Oct 20; doi: 10.1073/pnas.0406727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson AD, et al. Science. 2007 Jul 27;317:510. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 27.Ago H, et al. Nature. 2007 Aug 2;448:609. doi: 10.1038/nature05936. [DOI] [PubMed] [Google Scholar]
- 28.Molina D. Martinez, et al. Nature. 2007 Aug 2;448:613. [Google Scholar]
- 29.DeLano WL. 2002 http://www.pymol.org/
- 30.Dundas J, et al. Nucleic Acids Res. 2006 Jul 1;34:W116. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SOM