BrainAligner: 3D Registration Atlases of Drosophila Brains (original) (raw)
. Author manuscript; available in PMC: 2011 Dec 1.
Published in final edited form as: Nat Methods. 2011 May 1;8(6):493–500. doi: 10.1038/nmeth.1602
Abstract
Analyzing Drosophila neural expression patterns in thousands of 3D image stacks of individual brains requires registering them into a canonical framework based on a fiducial reference of neuropil morphology. Given a target brain labeled with predefined landmarks, the BrainAligner program automatically finds the corresponding landmarks in a subject brain and maps it to the coordinate system of the target brain via a deformable warp. Using a neuropil marker (the antibody nc82) as a reference of the brain morphology and a target brain that is itself a statistical average of 295 brains, we achieved a registration accuracy of 2µm on average, permitting assessment of stereotypy, potential connectivity, and functional mapping of the adult fruitfly brain. We used BrainAligner to generate an image pattern atlas of 2,954 registered brains containing 470 different expression patterns that cover all the major compartments of the fly brain.
Introduction
An adult Drosophila brain has about 100,000 neurons with cell bodies at the outer surface and neurites extending into the interior to form the synaptic neuropil. Specific types of neurons can be labeled using antibody detection1 or genetic methods such as the UAS-GAL4 system2, where each GAL4 line drives expression of a fluorescent reporter in a different subpopulation of neurons. Computationally registering, or aligning, images of fruit fly brains in three-dimensions (3D) is useful in many ways. First, automated 3D alignment of multiple identically labeled brains allows quantitative assessment of stereotypy: how much the expression pattern or the shape of identified neurons varies between individuals. Second, aligning brains that have different antibody or GAL4 patterns reveals areas of overlapping or distinctive expression that might be selected for genetic intersectional strategies3. Third, comparison of aligned neural expression patterns suggests potential neuronal circuit connectivity. Fourth, aligning images of a large collection of GAL4 lines gives an estimate of how extensively they cover different brain areas. Finally, for behavioral screens that disrupt neural activity in parts of a brain using GAL4 collections, accurate alignment of images is a prerequisite for detecting anatomical features in brains that correlate with behavior phenotypes.
Earlier 3D image registration approaches4,5,6 use surface- or landmark-based alignment modules of the commercial 3D visualization software AMIRA (Visage Imaging, Inc.) to align sample specimens to a template. The major disadvantages of these approaches are the huge amount of time taken for a user to manually segment the surfaces or to define the landmarks in each subject brain, as well as the potential for human error.
The earliest and most relevant parallel line of research for automated alignment is for two-dimensional (2D) or 3D biomedical images such as CT and MR human brain scans7,8,9, and for 2D mouse brain in situ hybridization images as part of the Allen Brain Atlas project10. Previous efforts to automatically register images of the fruit fly nervous system based on image features includes work on adult brains11,12, on the adult ventral nerve cord and larval nervous system13. We conducted comparison tests (Supplementary Note) on several widely used methods for registration14,15,16,11,12,13 and all produced unsatisfactory alignments at a rate that make them unsuitable for use in a pipeline that involves thousands of high-resolution 3D laser scanning microscope (LSM) images of Drosophila brains.
In this study we developed an automatic registration program, BrainAligner, for Drosophila brains and used it to align large 3D LSM images of thousands of brains with different neuronal expression patterns. Our algorithm combines several existing approaches into a new strategy based on reliably detecting landmarks in images. BrainAligner is hundreds of times faster than several competitive methods and automatically assesses alignment accuracy with a quality score. We validated alignment accuracy using biological ground truth represented by co-expression of patterns in the same brain. We have used BrainAligner to assemble a preliminary 3D Drosophila brain atlas, for which we assessed the stereotypy of neurite tract patterns throughout a Drosophila brain.
Results
BrainAligner
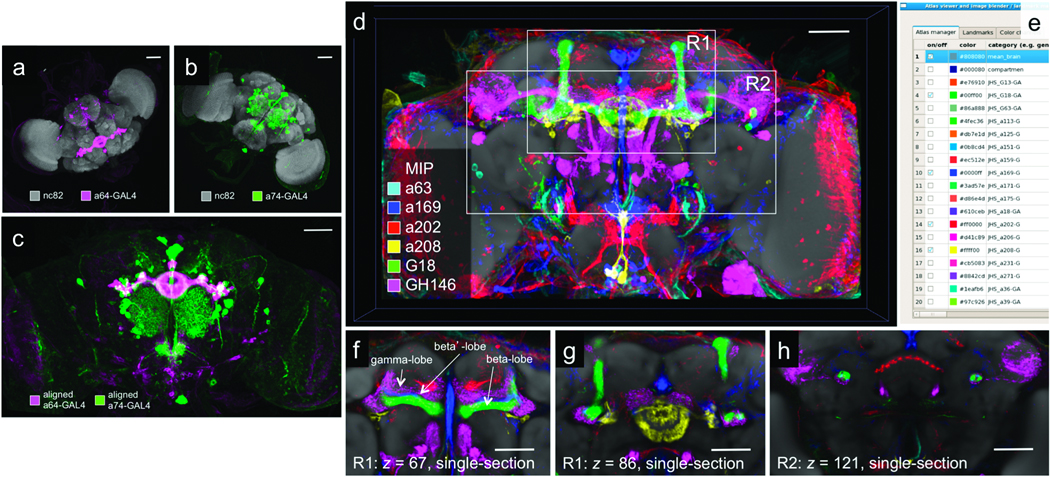
BrainAligner registers 3D images of adult Drosophila brain into a common coordinate system (Fig. 1). Brains that express GFP in various neural subsets were dissected and labeled with an antibody to GFP (colors in Fig. 1a–b); this is the pattern channel. Brains were also labeled with nc82, an antibody that detects a ubiquitous presynaptic component and marks the entire synaptic neuropil17 (gray in Fig. 1a–b); this is the reference channel. The brains to be registered have different orientations, sizes, and morphological deformations that are either biological or introduced in sample preparation. For each subject brain, BrainAligner maps the reference channel to a standardized target brain image using a nonlinear geometrical warp. Using the same transformation, the pattern channel from the subject image is then warped onto the target. Multiple subject images are aligned to a common target so that their patterns can be compared in the same coordinate space (Fig. 1c and Supplementary Video 1). In this way, we have mapped a large collection of GAL4 patterns into a common framework to identify intersecting expression patterns in various anatomical structures (Fig. 1d–h and Supplementary Video 2).
Figure 1.
BrainAligner registers images of neurons from different brains onto a common coordinate system. (a–b) Maximum intensity projections of confocal images of a64-GAL4 and a74-GAL4 brains. Neurons are visualized by membrane-targeted GFP and brain morphology is visualized by staining with the antibody nc82. (c) Aligned and overlaid neuronal patterns of (a) and (b). (d) Alignment of many GAL4 expression patterns. Patterns of interest can be selected and displayed in the common coordinate system. R1 and R2, regions of interest. (e) V3D-AtlasViewer software for viewing the 3D pattern atlas. (f–h) Zoomed-in single-section views of R1 and R2 in (d). Scale bars in all sub-figures: 50 µm.
BrainAligner registers subject to target using a global 3D affine transformation followed by a nonlinear local 3D alignment. For large-scale applications, brains may have different orientations, brightness, sizes, evenness of staining, morphological damage, and other types of image noise, which requires our algorithm to be robust. Thus we optimized only the degrees of freedom that are necessary.
In global alignment, we sequentially optimized the displacement, scaling and rotation parameters of an affine transform from subject to target to maximize the correlation of voxel intensities between the two images (Fig. 2a and Methods). We visually examined the transformed brains after the global alignment, and found no transformation errors in over 99% of our samples. The failure cases typically corresponded to poorly dissected brains that were either damaged structurally or for which excess tissues were present.
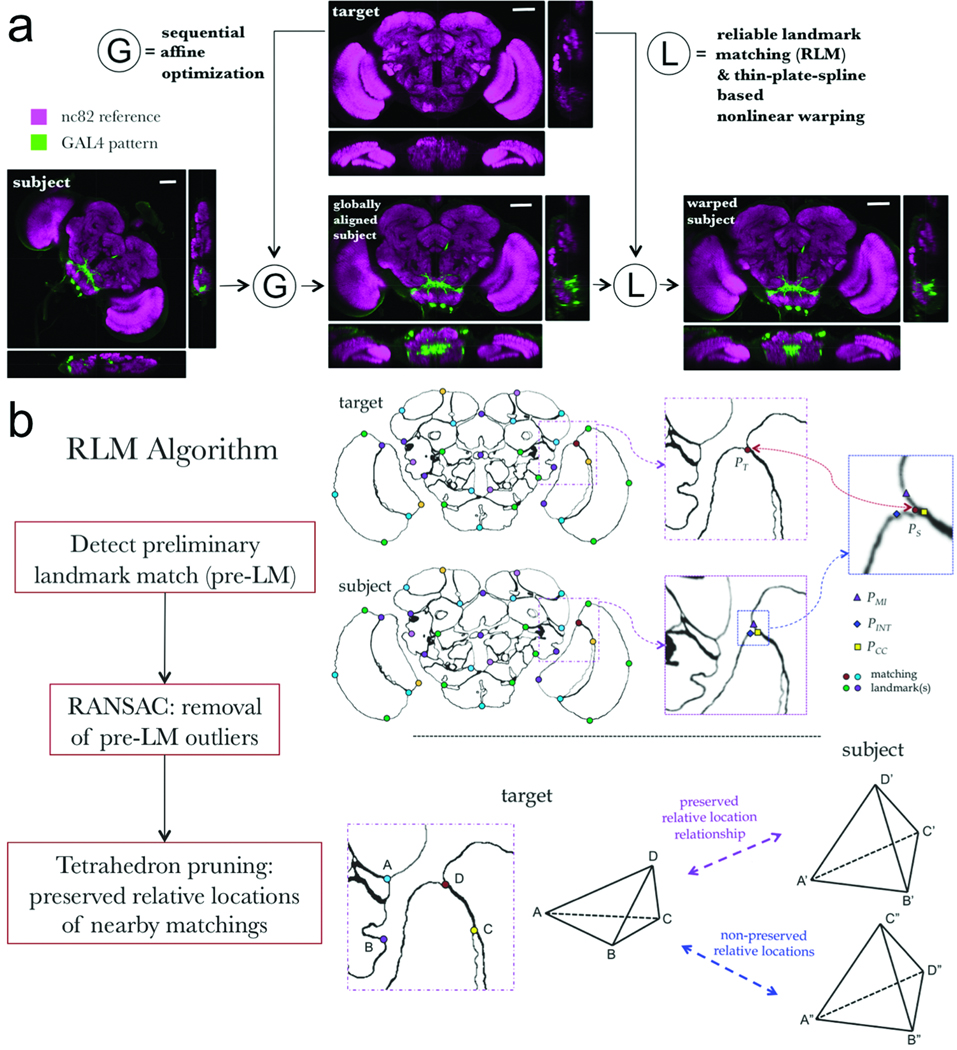
Figure 2.
Schematic illustration of the BrainAligner algorithm. (a) BrainAligner performs a global alignment (G) followed by nonlinear local alignments (L) using landmarks. Scale bars: 50 µm. (b) The Reliable Landmark Matching (RLM) algorithm for detecting corresponding feature points in subject and target images. Dots of the same color indicate the matching landmarks; PT, a target brain landmark position; PS, a subject brain landmark; PMI, PINT, PCC, the best matching positions based on mutual information (MI), voxel intensity (INT), and correlation coefficient (CC) of local image patches. In the tetrahedron-pruning step, the landmarks in a subject image that clearly violate the relative position relationships of the target are discarded.
In the local alignment step, we designed a reliable landmark matching (RLM) algorithm (Fig. 2b) to detect corresponding 3D feature points, each of which is called a landmark, in every target-subject pair. For the target brain we manually defined 172 landmarks that correspond to the points of high curvature (“corners” or edge points) of brain compartments as indicated by abrupt image contrast changes in the neuropil labeling. For each target landmark, RLM first searches for its matching landmark in the subject image using two or more independent matching criteria such as maximizing: (a) mutual information18,11, (b) inverse intensity difference, (c) correlation, and (d) similarity of invariant image moments15, within a small region around the landmark in the target and its potential match in the subject. A match confirmed by a consensus of these criteria is superior to a match based on only a single criterion. Therefore, when a consensus of the best matching locations of these criteria are close to each other (<5 voxels apart), RLM reports a preliminary landmark match (pre-LM), which is the site within the 3D bounding box of these best matching locations that gives the maximal product of the individual matching scores. These pre-LMs may violate the smoothness constraint, which states that in toto all matching-landmark pairs should be close to a single global affine transform, and locally, that relative location relationships should be preserved. Therefore, RLM uses a random sample consensus (RANSAC) algorithm19 to remove the outliers from the set of pre-LMs with respect to the global affine transform that produces the fewest outliers. Next, RLM optionally checks the remaining set of pre-LM pairs and detects those violating the relative location relationship in every corresponding tetrahedron formed by three additional neighboring matching points. Pre-LM pairs that clearly create a spatial twist with respect to nearby neighbors are removed. The landmarks that remain are usually highly faithful matching locations, and are called reliable landmarks.
We used the reliable landmarks to generate a thin-plate-spline warping field20 and thus mapped the reference channel of a subject image to the target. We then applied the same warping field derived from the reference channel to the pattern channel. We also optimized BrainAligner’s running speed. For instance, to generate the warping field we used hierarchical interpolation (Methods) instead of using all image voxels directly, improving the speed 50-fold without visible loss of alignment quality. Typically BrainAligner needs only 40 minutes on a current single CPU to align two images with 1024×1024×256 voxels.
One advantage of RLM is that the percentage, Qi, of the target landmarks that are reliably matched can be used to score how many image features are preserved in the automatic registration. The larger Qi, the better the respective alignment. We visually inspected the aligned brains and ranked alignment quality using a score Qv (range = 0~10, the larger the better). The automatic score Qi and the manual score Qv correlated significantly on 805 randomly selected alignments in the central brain, left optic lobe and right optic lobe (Supplementary Fig. 1), suggesting that Qi is a good indicator of alignment quality. Empirically, when Qi > 0.5, the respective alignment was good; when Qi > 0.75, the alignment was excellent. Low Qi scores typically correspond to poor nc82 staining and brains damaged during sample preparation.
While BrainAligner can be used to align subject brains to any target brain, we prefer to use an optimized “average” target brain obtained as follows. We first selected one image of a real brain, TR (Supplementary Fig. 2), as the target for an initial alignment for 295 brains that aligned to TR with Qi > 0.75. We then computed the mean-brain, TA, for the respective local alignments (Supplementary Fig. 2). Although TA was smoother than TR, it preserved detailed information, reflected in the strong correlation between TA and TR (Supplementary Fig. 2). We used TA as a new, and statistically more meaningful, target image for BrainAligner. Compared to the results for TR, this led to 38% more brains aligning with a Qi score larger than 0.7, and 14% more aligning with a Qi score higher than 0.5, on a data set consisting of 496 brains (Supplementary Fig. 3).
Assessment of BrainAligner accuracy and biological variance
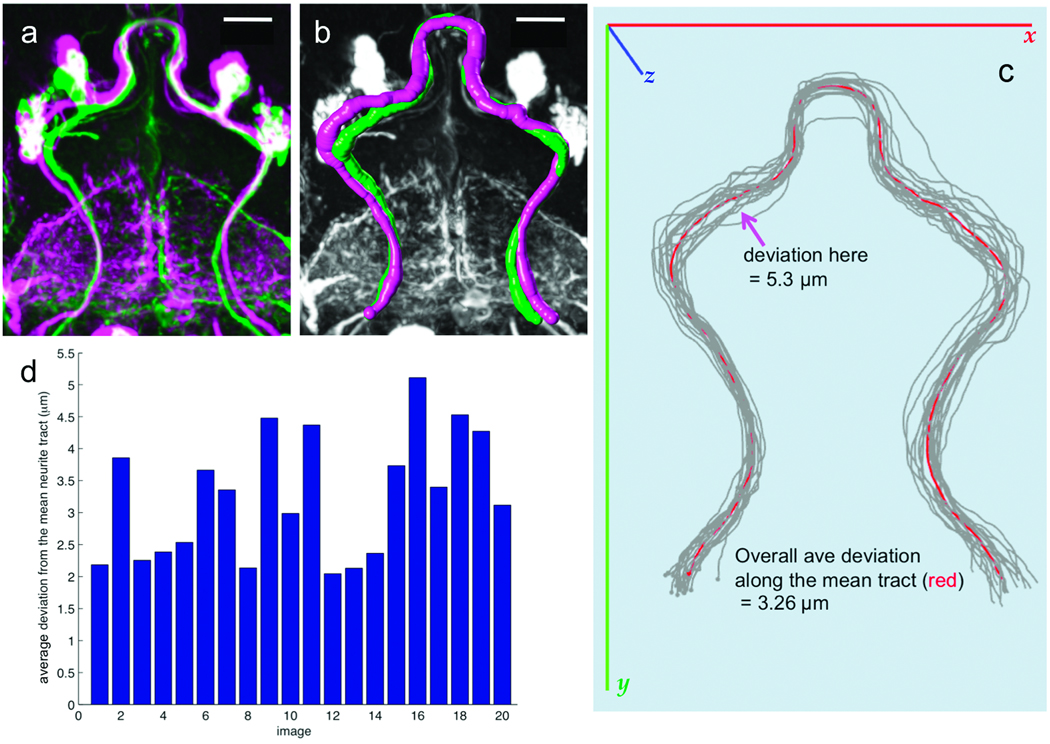
The variation between individual aligned brains of the same genotype is a combination of biological difference, variation introduced during sample preparation or imaging, and alignment error. In a previous study11, the variance of axon position was estimated to be approximately 2.5 ~ 4.3µm in the inner antennal cerebral tract (iACT) and at its neurite bifurcation point. We addressed a similar question by aligning 20 samples of a278-GAL4; UAS-mCD8-GFP to the common target TA. The large neurite bundles in aligned samples (Fig. 3a) were traced in 3D (Fig. 3b) using V3D-Neuron21,22. A mean tract model, Rm, of all these tracts was computed (Fig. 3c, and Supplementary Video 2). The neurite tracts were compared to Rm, at 243 evenly spaced locations. The variability of tract position was 3.26 µm (about 5.6 voxels in our images) with a range of 2.1 to 5.1 µm (Fig. 3d).
Figure 3.
Stereotypy of neuronal morphology and reproducibility of GAL4 expression patterns. (a) Two aligned and overlaid examples (magenta and green) of the a278-GAL4 expression pattern, from different brains. Scale bar: 20 µm. (b) 3D reconstruction of the major neurite tracts in (a). Magenta and green, surface representations of the reconstructed tracts. Gray, GAL4 pattern. Scale bar: 20 µm. (c) 3D reconstructed neurite tracts (gray) from 20 aligned a278-GAL4 images, along with their mean tract model (red). (d) Average deviation of the mean tract model from each reconstructed tract.
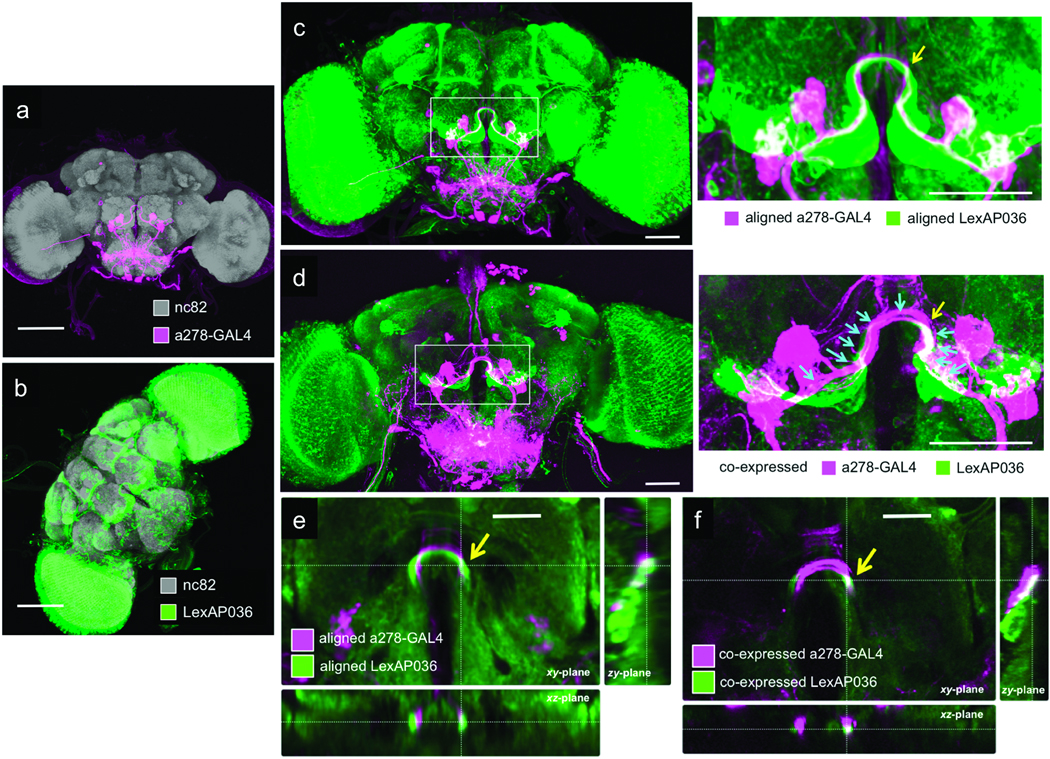
With ~3µm variance, BrainAligner produces reliable results. We further differentiated biological variability from aligner variance. The existence of two binary expression systems, GAL4 and LexA23, permits rigorous comparison of a computational prediction of overlap with a biological test of co-expression. The LexA line (LexAP036) showed potential overlap with the a278-GAL4 line used above in the Ω-shaped antennal lobe commissure (ALC) (Fig. 4a and 4b) when registered with BrainAligner (Fig. 4c and 4e). We then co-expressed distinct reporter constructs using the LexA and GAL4 systems simultaneously in the same fly and showed that there is indeed overlapping expression in the ALC (Fig. 4d and 4f). We estimated the precision of BrainAligner’s registration using the absolute value of the difference of the biological spatial distance of co-localized patterns and their respective spatial distance measured from the computationally aligned patterns. The average distance measured at 11 different spatial locations (Fig. 4d) along ALC of the aligned patterns and physically overlapping patterns was 1.8±1.1µm. Therefore the estimated registration precision was 0.8 to 2.9 µm. We also saw agreement between the aligned and co-expressed images of other GAL4-LexA pairs with expression patterns in the optic tubercle (Supplementary Fig. 4, Supplementary Videos 4a–b).
Figure 4.
Expression pattern overlap by computational and biological methods. (a) Maximum intensity projection of a278-GAL4; UAS-mCD8-GFP. Scale bar: 100 µm. (b) Maximum intensity projection of LexAP036; lexop-CD2-GFP. Scale bar: 100 µm. (c) Aligned image of GAL4 and LexA expression patterns in (a) and (b), with a zoomed-in view to the right. Scale bar: 50 µm. (d) Co-expression of the GAL4 and LexA patterns, with a zoomed-in view to the right. Scale bar: 50 µm. Arrows indicate the 11 locations where colocalization of the two patterns was measured; the yellow arrow indicates a region of substantial overlap. (e–f) Cross-sectional views of single slices of the aligned (e) and co-expressed (f) samples at a position corresponding to the yellow arrow in (c) and (d). Scale bars: 25 µm.
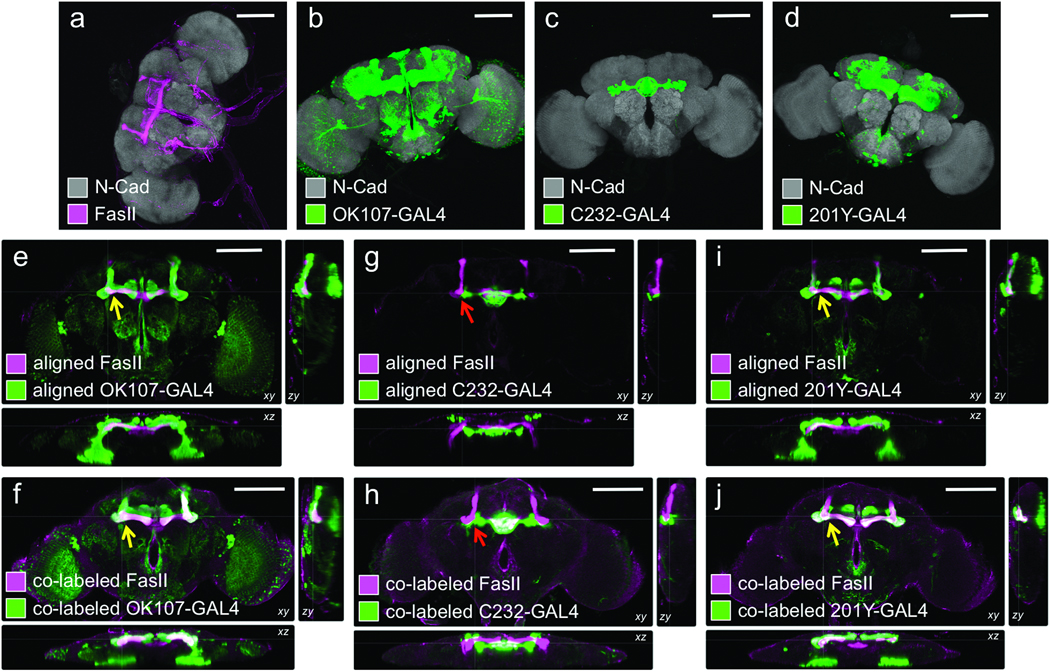
We performed an independent test of BrainAligner’s accuracy by comparing the computational alignment and co-labeling for FasII antibody staining, which labels the mushroom bodies, and various GAL4 lines that express in or near the mushroom bodies. BrainAligner accurately predicted the overlap of FasII antibody staining with the 201Y and OK107-GAL4 patterns, while C232-GAL4, which expresses in the central complex, does not co-localize with FasII (Figure 5).
Figure 5.
Comparison of computational alignment of separate brains with co-expression within the same brain. For all images, grey shows N-cadherin (N-Cad) labeling, which serves as the reference signal for alignment to the nc82-labeled target. Magenta, FasII antibody staining; green, GAL4 expression pattern (anti-GFP stain). (a) Wild-type w1118 adult brain. (b–d) Expression patterns of the indicated lines shown as maximum intensity projections of 20X confocal image stacks. (e, g, i) Cross-sectional views of computational alignments of FasII expression from (a) with GAL4 patterns from (b–d). (f, h, j) Matched cross-sectional views of brains expressing the GAL4 lines and labeled with both anti-GFP and anti-FasII to show biological co-localization. OK107 and 201Y expression patterns overlap with FasII (yellow arrows), but C232 expresses in adjacent but non-overlapping brain regions (red arrow). Scale bars, 100 µm.
Finally, the Flp-Out technique24 allows the expression of GFP reporters in a random subset of neurons within a given GAL4 expression pattern. Therefore, the computational alignment of Flp-Out subsets (“clones”) should correlate well with the expression pattern of the parent GAL4 lines. We aligned the CG8916-GAL4 expression pattern (Supplementary Fig. 5a) and Flp-Out clones (Supplementary Fig. 5b and 5c) of this GAL4 line. Nested expression patterns were visible in the superior clamp (SCL), posterior ventrolateral protocerebrum (PVLP), anterior VLP (AVLP), superior lateral protocerebrum (SLP), and SOG (Supplementary Fig. 5d and 5e, Supplementary Video 5).
Building a 3D image atlas of Drosophila brain
We automatically registered 2,954 brain images from 470 enhancer trap GAL4 lines (unpublished data) to our optimized target brain. We selected a well-aligned representative image of each GAL4 pattern (with a Qi score > 0.5) and arranged them as a 3D image pattern atlas (Fig. 1d, Supplementary Video 2). To effectively browse, search, and compare the expression patterns in these brains, we developed V3D-AtlasViewer software (Fig. 1e) based upon our fast 3D image visualization and analysis system V3D21. V3D-AtlasViewer organizes the collection of registered GAL4 patterns using a spreadsheet (Fig. 1e), within which a user can select and display any subset of patterns on top of a standard brain for visualization. This 3D image atlas reveals interesting anatomical patterns. For example, visualizing different sections in six GAL4 patterns demonstrates the previously reported subdivision of the mushroom body horizontal lobe into gamma-lobe (red/purple), beta’-lobe (blue) and beta-lobe (green)25 (Fig. 1e, Supplementary Videos 6a and 6b).
With this atlas, we were able for the first time to analyze the distribution of GAL4 patterns in different brain regions in a common coordinate system. The 470 GAL4 lines covered all known brain compartments (Supplementary Fig. 6a), with the SOG, superior lateral protocerebrum (SLP), prow (PRW), mushroom bodies (MB), and antennal lobes (AL) the five most represented compartments in this GAL4 collection. A relatively small number of GAL4 lines expressed in superior posterior slope (SPS), inferior posterior slope (IPS), inferior bridge (IB), and gorget (GOR). For the central complex of a Drosophila brain, less than 20% of GAL4 lines expressed in the fan-shape body (FB), ellipsoid body (EB), and noduli (NO). It may not be surprising that a large fraction of lines express in the SOG, since this neuropil region represents 7.43% of the brain by volume (Supplementary Fig. 6c). Therefore we also produced the density map of the neuronal pattern distribution within each compartment, normalizing the distribution by volume (Supplementary Fig. 6b). The central complex was over-represented in our GAL4 collection while the SOG, normalized for its volume, was actually under represented.
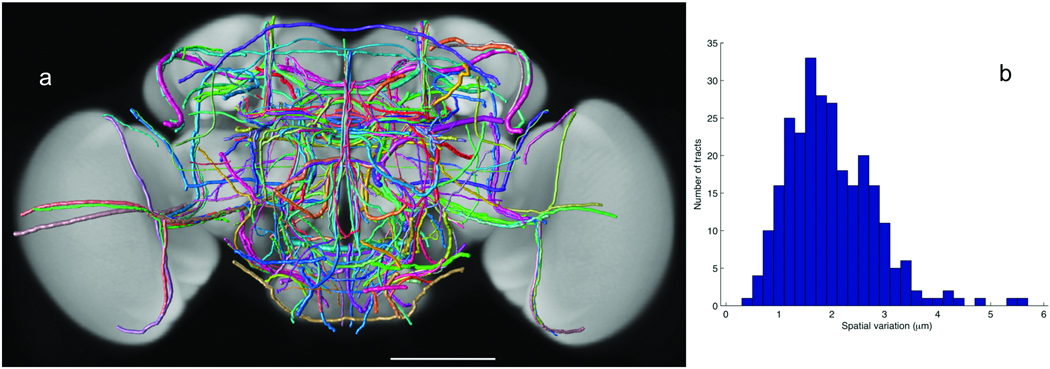
We examined the stereotypy of 269 neurite tracts that project throughout all brain compartments. We reconstructed each tract from at least two aligned brains of each GAL4 line. The spatial variations were computed and used as the width of each tract in visualization (Fig. 6a and Supplementary Video 7). The average variation was 1.98±0.83 µm (Fig. 6b), consistent with our previous independent test of 111 tracts21. This range of variation is within the upper bound of biological stereotypy of the neurite tracts themselves and noise introduced in sample preparation, imaging, and image analysis including registration and tracing. The tracing error was close to 021. Compared to the typical size of an adult fly brain (590 µm × 340 µm × 120 µm), this small variation indicates strong stereotypy of the neurite tracts.
Figure 6.
A 3D atlas of neurite tracts reconstructed from aligned GAL4 patterns. (a) 269 stereotyped neurite tracts and their distribution in the brain. The width of each tract equals the respective spatial deviation. The tracts are color-coded randomly for better visualization. Scale bar: 100 µm. (b) Distribution of the spatial deviation of the neurite tracts.
Discussion
BrainAligner is a new landmark-detection-based algorithm and software package for the large-scale automatic alignment of confocal images of Drosophila brains. We have used it, in combination with an optimized virtual target brain, consistent tissue preparation and imaging, and a library of GAL4 lines, to generate a pilot 3D atlas of neural expression patterns for Drosophila. We have also applied BrainAligner to our on-going FlyLight project that will produce an even higher resolution 3D digital map of the Drosophila brain. BrainAligner has robustly registered over 17,000 brain images of thousands of GAL4 lines within a few days, without any manual intervention during the alignment. The alignability of new samples, determined by their Qi scores, serves as an important quality control check. We are developing further methods to expand and query this resource, but it is already in use for anatomical and behavioral investigation of neural circuit principles.
Expression patterns generated by recombinase-based methods to label neurons of a common developmental lineage (MARCM26) and images in which single neurons are labeled can be aligned with our GAL4 reference atlas to identify lines that have GAL4 expression in those cells, allowing investigation of their behavioral roles. Examination of different GAL4 expression patterns for proximity or overlap suggests which areas might be functionally connected. The Drosophila brain is subdivided into large regions based on divisions in the synaptic neuropil caused by fiber tracts, glial sheaths, and cell bodies, but these anatomical regions may be further subdivided by gene expression patterns revealed by the GAL4 lines. When the GAL4 lines are aligned to a template brain upon which anatomical regions have been labeled, we can annotate the expression patterns using the VANO software27 in a faster and more uniform way. Alignment permits imaged-based searching, a significant improvement over keyword searching based on anatomical labels. Accurate alignment of images will also make it easier to correlate anatomy with behavioral consequences. Integration of aligned neuronal patterns with other genetic and physiological screening tools may be used to study different neuron types.
We have optimized BrainAligner to run on large datasets of GAL4 lines expressed in the adult fly brain and ventral nerve cord, but these are not the only type of data that can be aligned. Antibody expression patterns, in situs, and protein-trap patterns28,29 are also suitable; if the same reference antibody is included, images from different sources can be aligned using BrainAligner. Although BrainAligner was developed using the nc82 pre-synaptic neuropil marker, we have also successfully aligned brains where the reference channel was generated by staining with rat anti-N-Cadherin antibody (see Figure 5). Other reference antibodies that label a more restricted area of the brain, such as anti-FasII, may also work with the algorithm. It is also possible to align any pair of brains directly, rather than aligning both to a common template.
BrainAligner can be used in many situations where the image data has different properties than the data presented in this study. The optic lobes of an adult Drosophila brain shift in relation to the central brain and distort alignments. We developed an automated method to segregate the optic lobes from the central brain30, which was then registered using BrainAligner. For the larval nervous system and the adult ventral nerve cord of Drosophila, we detected and aligned the principal skeletons of these images13, followed by BrainAligner registration. BrainAligner automatically detects the corresponding landmarks, but it permits using manually added landmarks to improve critical alignments or to optimize alignments in a particular brain region. Indeed the brains to be aligned may also be imaged using different magnification scales. Higher resolution images may have only a part of the brain in the field of view, complicating registration. In such a case, the user can manually supply as few as four to five markers using V3D software21 to generate a globally aligned brain, which can then be automatically aligned using BrainAligner.
Despite a number of successfully used image registration methods in other scenarios such as building the Allen mouse brain atlas10, we have not found another automated image registration method that performs as well as BrainAligner on our large scale applications. Indeed, the key algorithm in BrainAligner, the RLM method, can be viewed as an optimized combination of several existing methods. It compares the results produced using different criteria and only uses results that agree with each other. BrainAligner is not limited to Drosophila and could be applied to other image data such as mouse brains.
Methods
Immunohistochemistry and confocal imaging
Males from enhancer-GAL4 lines (unpublished collection, JHS and B. Ganetzky) were crossed to virgin UAS-mCD8-GFP (Bloomington #5137 26) that produces a membrane-targeted fluorescent protein in the neurons. Adult brains were dissected in phosphate-buffered saline (PBS), fixed overnight in 2% paraformaldehyde, washed extensively in PBS-0.5% Triton, and then incubated overnight at 4°C rotating in primary antibodies (nc82 1:50 Developmental Studies Hybridoma Bank17 and rabbit anti-GFP 1:500 Molecular Probes/Invitrogen A11122). After washing all day at room temperature, brains were incubated overnight at 4°C rotating in secondary antibodies: goat anti-mouse-Alexa568 and goat anti-rabbit-Alexa488 1:500 Molecular Probes/Invitrogen A11034 and A11031. After another day of washing, brains were cleared and mounted in the glycerol-based Vectashield (on glass slides with 2 clear re-enforcement rings as spacers (Avery 05721). Samples were imaged on Zeiss Pascal Confocal with 0.84 µm z-steps using a 20X air immersion lens. Sequential scanning was used to ensure that there was no bleed-through between the reference- and pattern-channels. The raw images collected had 1024×1024×N voxels (the number of z-sections, N, typically was around 160), 8bits (voxel size = 0.58µm × 0.58µm × 0.84 µm), and two color-channels. The gain was increased as the imaging depth increased to maintain optimal use of detector range: the pattern channel intensity was maintained between over- and under-saturation. This resulted in a gain ramp of roughly 10% from lens-proximal to lens-distal surface of sample.
The enhancer-LexA lines (AMS and JHS, unpublished collection) were crossed to LexOp-CD2-GFP (in attP2 generated by AMS) and stained with nc82 and anti-GFP as above. For the double-label experiments, combination stocks of LexOp-CD2-GFP, UAS-mCD8-RFP; enhancer-GAL4 32,33 were built and crossed to the LexA lines. These were stained with rabbit anti-GFP 1:500 and rat anti-CD8 1:400 Invitrogen. The secondary antibodies were Alexa488 anti-rabbit and Alexa568 anti-rat (1:500). For the Flp-Out clones, GAL4 lines were crossed to hs-Flp; UAS-FRT-CD2-FRT-mCD8-GFP stocks31 and heat-shocked at the end of embryonic development or in adulthood.
Other fly stocks: C232-GAL434, 201Y-GAL435, and OK107-GAL436 were obtained from the Bloomington Stock Center. Other antibodies: FasII37 (1D4) and N-Cadherin38 (DNEX#8), both from the Developmental Studies Hybridoma Bank (developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242) and both were used 1:50.
BrainAligner implementation
To maximize the robustness of the automatic alignment and avoid being entrapped in local minima, in BrainAligner we used sequential global affine alignment in three steps. First we aligned the center of mass of a subject image to that of the target image. Then we rescaled a subject image proportionally so that its principal axis (obtained via principal component analysis) had the same length with that of the target image. Finally, we rotated a subject image around its center of mass, and thus detected the angle for which the target image and the rotated subject image had the greatest overlap. Since normally we did not have shearing in the 3D images, we did not optimize it for the affine transform. The rescaling step might also be skipped as brains imaged under the same microscopic setting had the similar size.
For the local nonlinear alignment, we computed the features based on adaptively determined image patches. The radius of an image patch was calculated using the formula 48×S/512, where S is the largest image dimension in 3D. To reduce the computational complexity, we searched matching landmarks hierarchically, first at a coarse level (grid spacing = 16 voxels) and then at a fine level (grid spacing = 1 voxel) around the best matching location (within a 13×13×7 window) detected at the coarse level. The mutual information was calculated on discretized image voxel intensity, by binning the grayscale intensity into an evenly spaced 16 intensity levels. For the RANSAC step, we constrained that all matching landmark pairs would satisfy a global affine transformation. Thus we computed the Euclidian distances of all initial matching landmark pairs after such a transformation, and removed the matching pairs that had more than 2 times the standard deviation of the distance distribution.
We designed a fast way to compute the thin-plate-spline (TPS)20 based displacement field, which was used to warp images. We computed the displacement field using TPS for a sub-grid (with 4×4×4 downsampling) of an entire image, followed by tri-linear interpolation for all remaining voxels to approximate the entire TPS transform. This method resulted in very similar displacement field compared to a direct implementation of TPS, but is about 50 times faster.
Data analyses
For the co-localization analysis using co-expressed GAL4 and LexA patterns, we measured distances between a series of pairs of high-curvature locations along the respective co-expressed GAL4 and LexA patterns in the ALC tract. We treated these distances as the ground truth of characteristic features that should be matched in computationally aligned brains. Then in the aligned brains, we visually detected these matching locations and produced the respective distance measurements. The error of registration was defined as the absolute value of the difference between the corresponding distances.
The correlation analysis for the Flp-Out data was performed around each of the co-localized subset clone patterns and the parent pattern. We first used V3D-Neuron21 to trace the co-localized neurite tracts, which were used to define the “foreground” image region of interest (ROI) for the correlation analysis. Suppose a foreground ROI had K voxels, then we randomly sampled another K voxels from the remaining brain area as the negative control for calculating the correlation coefficient for this co-localized subset clone pattern and the parent pattern.
In the analysis of GAL4 pattern distribution, for an aligned brain image, we calculated the mean value, m, and standard deviation, σ, of the entire brain area. We defined a brain compartment as having neuronal pattern(s) if (1) it had any absolutely visible voxels (typically intensity > 50 for an 8-bit image), and (2) its voxel intensities were outstanding compared to the average expression signal in the entire brain area (i.e. intensity > m + 3×σ). The names of brain compartments we used are consistent with the on-going effort of an international fruit fly brain nomenclature group.
Data and software
The BrainAligner and V3D-AtlasViewer software are available as Supplementary Software 1. BrainAligner, the optimized target brain, as well as additional information about BrainAligner, can also be downloaded from http://penglab.janelia.org/proj/brainaligner. The V3D-AtlasViewer program is a module of V3D21, which can also be freely downloaded from the web site http://penglab.janelia.org/proj/v3d.
Supplementary Material
1
2
3
Acknowledgements
The BrainAligner algorithm and the associated software packages were designed and developed by Hanchuan Peng, with support from Fuhui Long, Lei Qu, and Eugene Myers. The enhancer trap GAL4 collection used for initial development of the BrainAligner was generated by JHS and Barry Ganetzky (University of Wisconsin, Madison). The LexA enhancer trap collection was generated by AMS, JHS, and the Janelia FlyCore. We thank Gerry Rubin, Barret Pfeiffer, and Karen Hibbard for the UAS-mCD8-RFP, lexAop-CD2-GFP stock and LexAP vector. We thank Benny Lam for visually scoring the quality of aligned brains, Yan Zhuang for tracing and proofreading neurite tracts, Yang Yu and Jinzhu Yang for manual landmarking. We acknowledge Chris Zugates, and the FlyLight project team for discussion of aligner optimization. We thank Guorong Wu (University of North Carolina, Chapel Hill) for help in testing a registration method, and Dinggang Shen (University of North Carolina, Chapel Hill) for discussion when we initially developed BrainAligner. We thank Gerry Rubin for commenting on this manuscript.
Footnotes
Author Contributions
H.P. designed and implemented BrainAligner, did the experiments, and performed the analyses. H.P. and J.H.S. designed the biological experiments. F.L. and E.W.M. helped design the algorithm. P.C. prepared the samples and acquired confocal images. L.Q. helped implement the RANSAC algorithm and some comparison experiments. A.J. produced the brain compartment label field. A.W.S. helped generate LexA lines. H.P., E.W.M. and J.H.S. wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Buchner E, Bader R, Buchner S, Cox J, Emson PC, Flory E, Heizmann CW, Hemm S, Hofbauer A, Oertel WH. Cell-specific immuno-probes for the brain of normal and mutant Drosophila melanogaster. I. Wildtype visual system. Cell and Tissue Research. 1988;253:357–370. doi: 10.1007/BF00222292. [DOI] [PubMed] [Google Scholar]
- 2.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 3.Luan H, White BH. Combinatorial methods for refined neuronal gene targeting. Current opinion in neurobiology. 2007;17:572–580. doi: 10.1016/j.conb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Broughton SJ, Kitamoto T, Greenspan RJ. Excitatory and Inhibitory Switches for Courtship in the Brain of Drosophila melanogaster. Curr Biol. 2004;14:538–547. doi: 10.1016/j.cub.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 5.Jenett A, Schindelin JE, Heisenberg M. The Virtual Insect Brain protocol: creating and comparing standardized neuroanatomy. BMC Bioinformatics. 2006;7:544. doi: 10.1186/1471-2105-7-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boerner J, Duch C. Average shape standard atlas for the adult Drosophila ventral nerve cord. The Journal of Comparative Neurology. 2010;518:2437–2455. doi: 10.1002/cne.22346. [DOI] [PubMed] [Google Scholar]
- 7.Maintz JBA, Viergever MA. A survey of medical image registration. Medical Image Analysis. 1998;2:1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- 8.Zitova B, Flusser J. Image registration methods: a survey. Image and Vision Computing. 2003;21:977–1000. [Google Scholar]
- 9.Fischer B, Dawant B, Lorenz C, editors. Biomedical Image Registration; Proceedings of the 4th International Workshop, WBIR; Lübeck, Germany. Springer; 2010. Jul-13. 2010. [Google Scholar]
- 10.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 11.Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang AS, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Current Biology. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 13.Qu L, Peng H. A principal skeleton algorithm for standardizing confocal images of fruit fly nervous systems. Bioinformatics. 2010;26:1091–1097. doi: 10.1093/bioinformatics/btq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thirion JP. Image matching as a diffusion process: an analogy with Maxwell's demons. Medical Image Analysis. 1998;2:243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- 15.Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Transactions on Medical Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 16.Vercauteren T, Pennec X, Perchant A, Ayache N. Symmetric log-domain diffeomorphic registration: a demons-based approach. Lecture Notes in Computer Science. 2008;5241:754–761. doi: 10.1007/978-3-540-85988-8_90. [DOI] [PubMed] [Google Scholar]
- 17.Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. "Multimodality image registration by maximization of mutual information,". IEEE Transactions on Medical Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 19.Fischler MA, Bolles RC. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Communications of the ACM. 1981;24:381–395. [Google Scholar]
- 20.Bookstein FL. Principal warps: thin-plate spline and the decomposition. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1989;11:567–585. [Google Scholar]
- 21.Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nature Biotechnology. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H, Ruan Z, Atasoy D, Sternson S. Automatic reconstruction of 3D neuron structures using a graph-augmented deformable model. Bioinformatics. 2010;26:i38–i46. doi: 10.1093/bioinformatics/btq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nature Neuroscience. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 24.Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- 25.Crittenden JR, Skoulakis E, Han K, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learning and Memory. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 27.Peng H, Long F, Myers EW. VANO: a volume-object image annotation system. Bioinformatics. 2009;25:695–697. doi: 10.1093/bioinformatics/btp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelso RJ, Buszczak M, Quinones AT, Castiblanco C, Mazzalupo S, Cooley L. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Research. 2004;32:D418–D420. doi: 10.1093/nar/gkh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knowles-Barley S, Longair M, Douglas AJ. BrainTrap: a database of 3D protein expression patterns in the Drosophila brain. Database. 2010 doi: 10.1093/database/baq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam SC, Ruan Z, Zhao T, Long F, Jenett A, Simpson J, Myers EW, Peng H. Segmentation of center brains and optic lobes in 3D confocal images of adult fruit fly brains. Methods. 2010;50:63–69. doi: 10.1016/j.ymeth.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 32.Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Dell KMC, Armstrong JD, Yang MY, Kaiser K. Functional dissection of the Drosophila mushroom bodies by selective feminization of genetically defined subcompartments. Neuron. 1995;15:55–61. doi: 10.1016/0896-6273(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 35.Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- 36.Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- 37.Grenningloh G, Rehm EJ, Goodman CS. Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell. 1991;67:45–57. doi: 10.1016/0092-8674(91)90571-f. [DOI] [PubMed] [Google Scholar]
- 38.Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3