Targeted Genome Editing Across Species Using ZFNs and TALENs (original) (raw)
. Author manuscript; available in PMC: 2012 Nov 5.
Published in final edited form as: Science. 2011 Jun 23;333(6040):307. doi: 10.1126/science.1207773
Although entire genome sequences are available for numerous species, lack of reverse genetic tools has hindered cross-species comparisons of gene function. We demonstrate a widely applicable solution: targeted genome editing using site-specific nucleases. These engineered nucleases have fusions between the DNA cleavage domain of FokI and a custom-designed DNA binding domain: C2H2 zinc-finger motifs for zinc finger nucleases (ZFNs) (1) or truncated transcription activator-like effector (TALE) domains for efficient TALE nucleases (TALENs) (2). ZFNs and TALENs induce double-strand breaks at desired loci that can be repaired by error-prone non-homologous end-joining to yield small insertions and deletions (indels) at the break sites. We developed efficient procedures for targeted, heritable disruption of genes and cis-acting regulatory elements in the model nematode Caenorhabditis elegans and applied them to C. briggsae, a species diverged by 15 to 30 million years. Precisely targeted genome modifications had not been achieved in nematodes.
As proof of principle, we used ZFNs to disrupt a single-copy transgene expressing green fluorescent protein (GFP) in the C. elegans germline (Fig. 1A and table S1) (3). We overcame prior obstacles to attaining heritable, ZFN-induced mutations in worms (4) by injecting gonads with ZFN-encoding mRNAs carrying 5′ and 3′ untranslated regions favorable for germline translation (fig. S1). Progeny lacking green fluorescence carried indels at the target site, indicating heritable, targeted gene disruption (Fig. 1A and fig. S2A).
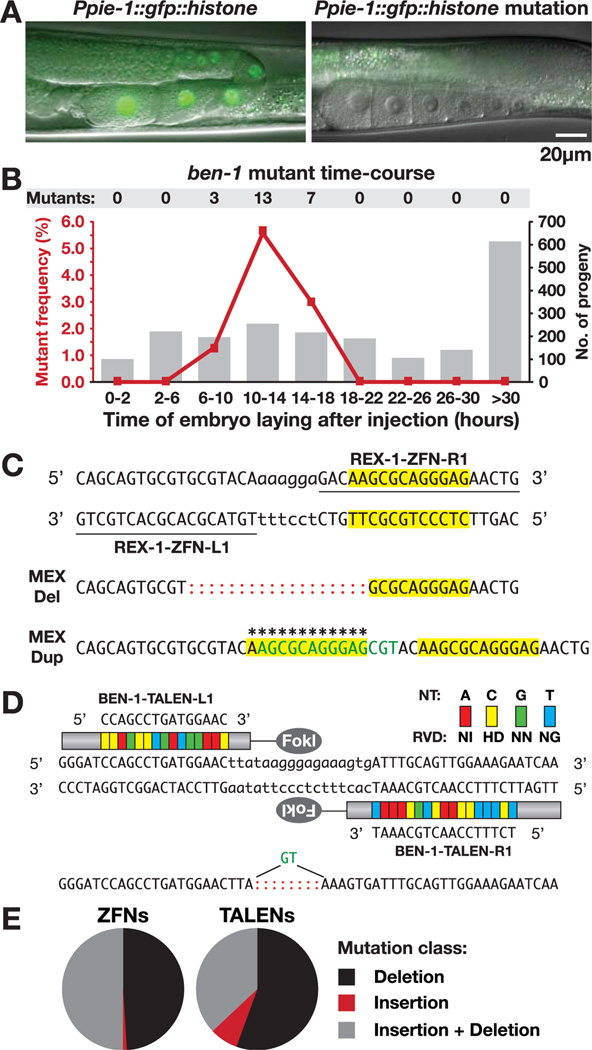
Fig. 1.
Heritable, targeted nematode mutations induced by ZFNs and TALENs. (A) Hermaphrodite gonads with an intact (left) or ZFN-disrupted (right) GFP::histone transgene. Residual signal (right) is gut autofluorescence. (B) Frequency of ben-1 mutants (left y axis, red line) from time periods after ZFN mRNA injection (x axis) relative to total progeny per period (right y axis, histogram). (C) ZFN-induced mutations in cis regulatory element rex-1 cause partial deletion and full duplication (asterisks) of MEX motif (yellow). Represented are ZFN recognition sequences (underlined), FokI cleavage region (lowercase nucleotides), insertions (green), and deletions (red colons). (D) TALEN recognition sequences in ben-1 and TALEN-induced mutation. Each RVD (colored blocks) recognizes a nucleotide (NT) in the target site. (E) Proportion of each mutation class induced by ZFNs (_n_=102) and TALENs (n = 27).
We next defined the time period after injection that yielded the highest proportion of mutant progeny (Fig. 1B and fig. S3, A to C).We targeted the endogenous gene ben-1 with ZFNs, because ben-1/+ mutants have an easily scorable phenotype: mobility on the paralysis-inducing drug benomyl (table S1).We found 5.1% of progeny from the peak 4-hour window to be mutants compared with 1.1% of total progeny. One in three injected animals produced mutants. A single animal could yield many mutant progeny, all with independent mutations (fig. S3C and table S2).
In ben-1 mutants, we found no mutations at the top 39 off-target ZFN sites predicted by empirical specificity data (table S3, A and B). Moreover, sequence analysis of the entire ben-1(y462) mutant genome revealed only two indels: the ZFN-induced ben-1 mutation and a deletion unlikely to have been caused by ZFN activity. The latter arose at a site unrelated to the ZFN site (fig. S4) and at a frequency consistent with spontaneous C. elegans indels (3).
To target mutagenesis of endogenous cis-regulatory elements, we devised methods to identify mutants by molecular genotyping, that is, without reliance on phenotype (fig. S3D) (3).We targeted rex-1 on the X chromosome. rex-1 recruits the dosage compensation complex (DCC) to reduce gene expression (5). Pivotal to DCC recruitment by rex sites is a 12–base pair (bp) motif called MEX that is enriched on X. We identified 32 rex-1 mutations, including a partial deletion and a full duplication of the MEX motif (Fig. 1C and fig. S2B). Thus, ZFN-driven mutagenesis is sufficiently robust to obtain mutations without a priori knowledge of phenotype and is suitably precise to target small regulatory motifs.
Genome-editing protocols developed for ZFNs proved to be robustly portable to TALENs. DNA binding domains of TALEs have tandem repeat units (each 33 to 35 amino acids) in which the nucleotide binding preference is determined by two adjacent amino acids called the repeat variable diresidue (RVD) (6). By using previously described rules for altering DNA binding specificity of TALEs (6), we designed a pair of TALENs to target the same ben-1 region as the ZFNs.
TALENs induced many ben-1 mutations (Fig. 1D and fig. S2E) (3). Heterozygous mutants composed 3.5% of progeny in the peak window of mutagenesis defined for ZFNs. The spectrum of mutations was similar for ZFNs and TALENs (Fig. 1E and fig. S2, A to E).
To assess the nuclease delivery and mutant screening procedures across species, we applied them, without modification, to C. briggsae. By using ZFNs, we targeted the C. briggsae ortholog of the C. elegans gene sdc-2, which directs the DCC to assemble onto X and induces hermaphrodite sexual differentiation (5). Five independent sdc-2 mutations were obtained (1.5% of progeny), thus establishing the efficacy of nuclease technology in C. briggsae (fig. S2C) (3). Our studies demonstrate a broadly useful strategy that brings targeted mutagenesis to nonmodel organisms and facilitates dissection of diverse biological processes.
Supplementary Material
wood som.pdf
Acknowledgments
A.J.W. is a Sir Henry Wellcome Post-Doctoral Fellow. NIH (GM30702) and HHMI funded these studies.
Footnotes
References and Notes
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Nat. Rev. Genet. 2010;11:636. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Miller JC, et al. Nat. Biotechnol. 2011;29:143. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 3.Materials and methods are available as supporting material on Science Online.
- 4.Morton J, Davis MW, Jorgensen EM, Carroll D. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16370. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer BJ. Curr. Opin. Genet. Dev. 2010;20:179. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanove AJ, Schornack S, Lahaye T. Curr. Opin. Plant Biol. 2010;13:394. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
wood som.pdf