Estrogen Receptor (ESR1) mRNA Expression and Benefit From Tamoxifen in the Treatment and Prevention of Estrogen Receptor–Positive Breast Cancer (original) (raw)
Abstract
Purpose
Several mechanisms have been proposed to explain tamoxifen resistance of estrogen receptor (ER) –positive tumors, but a clinically useful explanation for such resistance has not been described. Because the ER is the treatment target for tamoxifen, a linear association between ER expression levels and the degree of benefit from tamoxifen might be expected. However, such an association has never been demonstrated with conventional clinical ER assays, and the ER is currently used clinically as a dichotomous marker. We used gene expression profiling and ER protein assays to help elucidate molecular mechanism(s) responsible for tamoxifen resistance in breast tumors.
Patients and Methods
We performed gene expression profiling of paraffin-embedded tumors from National Surgical Adjuvant Breast and Bowel Project (NSABP) trials that tested the worth of tamoxifen as an adjuvant systemic therapy (B-14) and as a preventive agent (P-1). This was a retrospective subset analysis based on available materials.
Results
In B-14, ESR1 was the strongest linear predictor of tamoxifen benefit among 16 genes examined, including PGR and ERBB2. On the basis of these data, we hypothesized that, in the P-1 trial, a lower level of ESR1 mRNA in the tamoxifen arm was the main difference between the two study arms. Only ESR1 was downregulated by more than two-fold in ER-positive cancer events in the tamoxifen arm (P < .001). Tamoxifen did not prevent ER-positive tumors with low levels of ESR1 expression.
Conclusion
These data suggest that low-level expression of ESR1 is a determinant of tamoxifen resistance in ER-positive breast cancer. Strategies should be developed to identify, treat, and prevent such tumors.
INTRODUCTION
The antiestrogen tamoxifen is a commonly used treatment for patients with estrogen-receptor (ER) –positive breast cancer. As adjuvant therapy in patients with ER-positive early breast cancer, tamoxifen improves overall survival1and reduces risk for development of hormone-dependent breast cancer in women at increased risk for developing breast cancer.2 Unfortunately, some patients who receive adjuvant tamoxifen eventually experience relapse and die as a result of the disease1; in the National Surgical Adjuvant Breast and Bowel Project (NSABP) prevention trial (P-1), 30% of ER-positive tumors were not prevented by tamoxifen.2
The mechanisms of de novo and acquired resistance to tamoxifen in ER-positive breast cancer are not clear and have been the subject of studies by many investigators.3 From a biologic viewpoint, the amount of ER should be predictive of the degree of benefit from tamoxifen, which targets the receptor. However, there has been no clear demonstration of this relationship.4 Instead, only a threshold effect has been demonstrated, in that patients diagnosed with ER-negative breast cancer (defined by < 10 fmol/g protein by ligand binding assay [LBA]) did not gain significant benefit from adjuvant tamoxifen.5–7 Such observations have led to hypotheses that mutations of the ER gene (ESR1) or changes in molecules other than ER are responsible for tamoxifen resistance.3 However, studies exploring these possibilities have not led to the development of clinically useful predictors of tamoxifen resistance.3
Here, we provide evidence from retrospective gene expression analyses of available tumor blocks collected from two pivotal trials conducted by the NSABP that a determinant of tamoxifen resistance in both adjuvant treatment and prevention settings is a low level of ESR1 mRNA. ESR1 expression level is the strongest linear predictor of benefit from tamoxifen among 16 genes from the 21-gene recurrence score assay8 using tumor samples from NSABP trial B-14,9 which tested the worth of adjuvant tamoxifen in the treatment of ER-positive, node-negative breast cancer. In the P-1 prevention trial2 tamoxifen failed to prevent 30% of ER-positive breast cancer. We hypothesized that the ER-positive breast cancer that developed in women on the tamoxifen arm, which by definition is tamoxifen resistant, would have lower levels of ESR1 mRNA than would those from women in the placebo arm. Data from microarray gene expression analyses of cancer events from P-1 supported this hypothesis.
PATIENTS AND METHODS
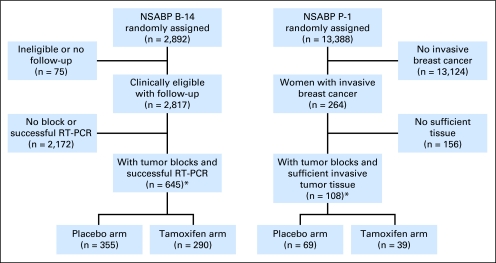
Human investigations were performed after approval by a local human investigations committee and were in accordance with an assurance filed with and approved by the Department of Health and Human Services. This is a retrospective subset analysis that is based on available materials. A CONSORT diagram for the B-14 and P-1 trials is shown in Figure 1.
Fig 1.
CONSORT diagram. NSABP, National Surgical Adjuvant Breast and Bowel Project; RT-PCR, reverse transcriptase polymerase chain reaction. (*) Included in analysis.
Patients
Paraffin blocks containing sufficient invasive breast cancer for RNA extraction were available from 645 of the 2,817 randomly assigned patients in the NSABP B-14 study (n = 355 from the placebo arm and n = 290 from the tamoxifen arm).10 ER and progesterone receptor (PR) proteins were measured by ligand binding at the time of enrollment. Ten fmol/mg protein was the ligand binding cutoff point for ER positivity. The proportion of patients who did not have distant recurrence at 10 years after surgery was 75.4% (95% CI, 70.7% to 80.1%) for the placebo arm, and it was 83.6% (95% CI, 79.2% to 88.1%) for the tamoxifen arm of the study subset.
Of 13,388 women who participated in the P-1 trial, 264 experienced invasive breast cancer events (n = 175 in the placebo arm and n = 89 in the tamoxifen arm) before the trial results were reported and treatment was unblinded.2 Paraffin blocks with sufficient invasive tumor tissue were available from 108 (n = 69 from the placebo arm and n = 39 from the tamoxifen arm). Central ER immunohistochemistry (IHC) identified 84 of these as ER positive (n = 57 from the placebo arm and n = 27 from the tamoxifen arm).
Histopathology and Other Markers
From hematoxylin and eosin–stained whole-tissue sections, histologic subtype and tumor grade (modified Bloom-Richardson grading criteria) were assessed centrally (by F.L.B. for B-14 and by O.L.B. for P-1). ER IHC was performed with the PharmDx kit (Dako, Carpinteria, CA), and Allred scores were determined. Amplification of HER2 and MYC were examined by fluorescence in situ hybridization with commercial probes (Vysis, Downers Grove, IL).
Gene Expression Profiling for B-14
Gene expression profiling for B-14 specimens was performed by Genomic Health and was blinded to clinical outcome data by using the previously described Onco_type_DX (Genomic Health, Redwood City, CA) assay.8 In brief, the expression levels of 16 cancer-related genes comprising the recurrence score were measured in triplicate with TaqMan real-time quantitative reverse transcription polymerase chain reaction (RT-PCR) and were normalized relative to five reference genes. Normalized expression levels ranged from 0 to 15 units, for which each 1-unit increase reflected a two-fold increase in mRNA. The recurrence score was calculated on a scale from 0 to 100 and was derived from the reference-normalized expression measurements for the 16 cancer-related genes.
Gene Expression Profiling for P-1
Microarray gene expression profiling of blocks from P-1 was achieved by using an in-house–developed protocol (Data Supplement). Hybridization intensity data were compiled by using the Partek Genomics Suite (Partek, St Louis, MO). After quantile normalization, genes with a mean intensity less than 500 were filtered out, and 7,734 probes with informative data remained. Data were log_2_ transformed for statistical analyses. Raw microarray data files and anonymized clinical data have been deposited to Gene Expression Omnibus (GSE 12,665).
Statistical Analyses
In the B-14 retrospective study, the primary end point was distant recurrence–free interval. Contralateral disease, other second primary cancers, and deaths before distant recurrence were considered censoring events. Ipsilateral breast recurrence, local chest wall recurrence, and regional recurrences were not considered either as events or as censoring events. Cox proportional hazards models were utilized to determine if clinical or gene expression variables were predictive of tamoxifen response. This was accomplished by using the likelihood ratio test to evaluate the statistical significance of the interaction term in a model that included parameters for treatment, the individual variable, and the interaction between treatment and the individual variable. A P value less than .05 was considered statistically significant. Penalized Cox proportional hazards models were used to test whether log-hazard was a linear function of predictors or not.11
Statistical analysis for microarray data was performed by using the Partek Genomics Suite. For P-1, a t test was performed to identify differentially expressed genes between two phenotypes. Two-sided P values were reported.
RESULTS
Assessment to Determine Predictive Markers of Tamoxifen Benefit in NSABP B-14
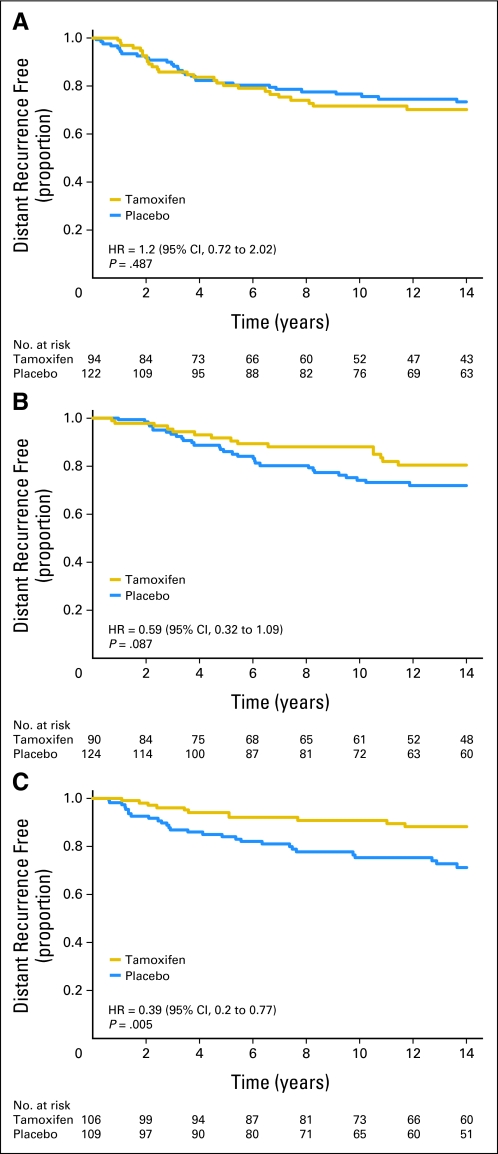
The B-14 study subset was similar to all eligible randomly assigned patients in the B-14 (Data Supplement). The results from the assessment to determine markers of tamoxifen benefit in B-14 are listed in Table 1. Among clinical variables, ER by the LBA (< 50 fmol/mg v ≥ 50 fmol/mg) and patient age (< 50 years v ≥ 50 years) showed a significant interaction with tamoxifen treatment (interaction P = .024 and 0.023, respectively). The interaction P value for ER by LBA for the parent B-14 study cohort approached, but did not achieve, statistical significance (P = .075). By gene expression, the quantitative assessment of two genes, ESR1 (P < .001) and SCUBE2 (P = .004), and the ER group (P = .008) showed a highly significant interaction with tamoxifen treatment, and higher gene expression was associated with greater degree of benefit from tamoxifen. Even after adjusting for the interaction between treatment and age, ESR1 (P = .005) and SCUBE1 (P = .009) still had significant interactions with treatment. Notably, PGR or HER2 were not predictive. The 21-gene recurrence score showed a trend for interaction (P = .06) in which high-risk patients derived little benefit from tamoxifen. Kaplan-Meier estimates of placebo- and tamoxifen-treated patients according to tertiles of ESR1 mRNA expression showed increased benefit of tamoxifen treatment with increasing levels of ESR1 expression (Fig 2).
Table 1.
Hazard Ratios for Tamoxifen Treatment According to Clinical Variables and the Gene Expression Variables for Distant Recurrence-Free Interval in NSABP Clinical Trial B-14
| Variable Interacting With Treatment | Evaluable Patients (n = 645) | All Patients (N = 2,817) | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | Interaction P | Hazard Ratio | 95% CI | Interaction P | |
| Clinical variable | ||||||
| Age ≥ 50 years* | 0.440 | 0.217 to 0.892 | .023 | 0.892 | 0.631 to 1.261 | .518 |
| Tumor size > 2 cm† | 1.179 | 0.600 to 2.316 | .633 | 1.257 | 0.9 to 1.754 | .179 |
| Quantitative ER ≥ 50 | 0.457 | 0.232 to 0.899 | .024 | 0.74 | 0.53 to 1.032 | .075 |
| Quantitative PR ≥ 50 | 0.831 | 0.425 to 1.625 | .588 | 0.975 | 0.7 to 1.36 | .885 |
| Grade | ||||||
| Poor‡ | 1.854 | 0.639 to 5.380 | .123 | — | — | |
| Moderate‡ | 1.074 | 0.392 to 2.947 | — | — | ||
| Gene expression variable§ | ||||||
| Recurrence score‖ | 1.967 | 0.978 to 3.958 | .06 | — | — | |
| ER gene group | 0.757 | 0.619 to 0.926 | .008 | — | — | |
| ESR1 | 0.744 | 0.630 to 0.878 | < .001 | — | — | |
| SCUBE2 | 0.806 | 0.695 to 0.934 | .004 | — | — | |
| PGR | 0.965 | 0.832 to 1.120 | .643 | — | — | |
| BCL2 | 0.770 | 0.579 to 1.023 | .074 | — | — | |
| Proliferation gene group | 1.075 | 0.747 to 1.546 | .698 | — | — | |
| MKI67 | 1.224 | 0.884 to 1.696 | .224 | — | — | |
| SURV | 1.053 | 0.813 to 1.365 | .693 | — | — | |
| STK15 | 0.912 | 0.622 to 1.336 | .636 | — | — | |
| CCNB1 | 0.857 | 0.587 to 1.253 | .428 | — | — | |
| MYBL2 | 1.138 | 0.890 to 1.455 | .303 | — | — | |
| Invasion gene group | 0.927 | 0.621 to 1.386 | .712 | — | — | |
| MMP11 | 0.886 | 0.693 to 1.132 | .334 | — | — | |
| CTSL2 | 1.102 | 0.823 to 1.477 | .515 | — | — | |
| HER2 gene group | 1.046 | 0.777 to 1.408 | .768 | — | — | |
| HER2 | 0.942 | 0.677 to 1.310 | .721 | — | — | |
| GRB7 | 1.052 | 0.788 to 1.404 | .732 | — | — | |
| Nongrouped genes | ||||||
| GSTM1 | 0.901 | 0.722 to 1.125 | .359 | — | — | |
| BAG1 | 0.786 | 0.511 to 1.208 | .272 | — | — | |
| CD68 | 0.939 | 0.585 to 1.506 | .791 | — | — |
Fig 2.
Quantitative estrogen receptor expression by reverse transcriptase polymerase chain reaction and distant recurrence at 10 years. Each Kaplan-Meier plot represents tamoxifen and placebo arms of patients diagnosed with tumors that express (A) low, (B) middle, and (C) high tertile levels of ESR1 mRNA. HR, hazard ratio.
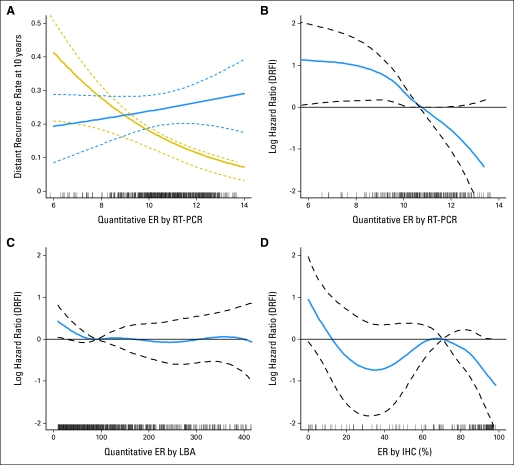
The reason for the different strength of interaction observed between two measures of ER (LBA v RT-PCR) was investigated by examining the linearity of the relationship between distant recurrence and the quantitative level of ER as continuous measures by these assays. The relative risk reduction by tamoxifen (the differential between two curves in Fig 3A) increases with increasing ESR1 mRNA expression. A formal statistical test for nonlinearity of the relationship between the log-hazard of tamoxifen-treated patients and ESR1 mRNA expression was not significant (P = .457), which confirmed the linear nature of the association (Fig 3B). On the basis of data from the 2,817 randomly assigned patients on B-14, ER protein by LBA was neither a significant prognostic factor for tamoxifen-treated patients (P = .1) nor a significant predictor of treatment effect (P = .14; Fig 3C).
Fig 3.
RNA and protein measurement of estrogen receptor (ER) and clinical outcome. (A) Rate of distant recurrence at 10 years as a function of quantitative ER by reverse transcriptase polymerase chain reaction (RT-PCR) in the placebo (blue line) and tamoxifen (gold line) groups. Solid lines are estimates, and dashed lines are 95% confidence bands. (B) Log-hazard ratio against the mean versus ESR1 mRNA on the basis of 290 patients randomly assigned to tamoxifen. Dashed lines are 95% confidence bands. (C) Log-hazard ratio against the mean versus ER by ligand binding assay (LBA) on the basis of 1,345 patients randomly assigned to tamoxifen, with ER by LBA less than 450 fmol/mg. (D) Log-hazard ratio against the mean versus ER by immunohistochemistry (IHC) on the basis of 177 patients randomly assigned to tamoxifen. DRFI, distant recurrence–free interval.
In a subset of 177 of the 290 tamoxifen-treated patients, the ER protein expression level of tumors was measured by using quantitative image analysis after staining with a US Food and Drug Administration– approved ER immunostaining kit (PharmDx; Dako). ER by IHC was strongly associated with distant recurrence (P = .004); its relationship with distant recurrence–free interval tended to be volatile, though the test for nonlinearity was not significant (P = .129; Fig 3D). When data were combined from 115 tamoxifen-registered patients on B-14, the nonlinearity was significant (P = .008; Data Supplement). These data suggest that a low level of ESR1 mRNA expression is an important determinant of tamoxifen resistance in ER-positive breast cancer.
Microarray Gene Expression Profiling of P-1 Breast Cancer Events
It would be of great interest to confirm in another data set the observation that low ESR1 mRNA is associated with tamoxifen resistance. However, to our knowledge, no other clinical trial cohort of randomly assigned patients with an annotated tissue bank available exists to directly confirm these findings. Therefore, we examined the whole genome expression profiles of all available tumor blocks with sufficient material from the NSABP P-1 trial that tested the worth of tamoxifen in prevention of breast cancer in women at high risk.
In the P-1 trial, 70% of the expected ER-positive tumors were prevented by tamoxifen. The 30% that developed on tamoxifen were functionally resistant to tamoxifen.2 Analysis of data in all available blocks from P-1 also showed a decrease in the percentage of ER-positive tumors that arose in the tamoxifen arm compared with the placebo arm, albeit this decrease was somewhat smaller than that seen in the entire cohort (Table 2). In the placebo arm, 57 tumors (82.6%) were ER positive, and 12 were ER negative (17.4%); in the tamoxifen arm, 27 (69.2%) were ER positive, and 12 (30.8%) were ER negative. As seen in B-14, ER-positive breast cancers from the tamoxifen arm of P-1 (which by definition were tamoxifen-resistant) had lower levels of ESR1 mRNA.
Table 2.
Distribution of Invasive Cancer Cases From the Breast Cancer Prevention Trial, NSABP P-1, by Treatment Group and Tumor Characteristics
| Tumor Characteristic | All Cases | ER Positive Only | ||||||
|---|---|---|---|---|---|---|---|---|
| Placebo | Tamoxifen | Placebo | Tamoxifen | |||||
| No. | % | No. | % | No. | % | No. | % | |
| ER IHC | ||||||||
| Negative | 12 | 17.4 | 12 | 30.8 | ||||
| Positive | 57 | 82.6 | 27 | 69.2 | ||||
| ER IHC Allred score | ||||||||
| 0 | 9 | 13.0 | 11 | 28.2 | ||||
| 1 | 1 | 1.4 | 0 | 0.0 | ||||
| 2 | 2 | 2.9 | 1 | 2.6 | ||||
| 3 | 2 | 2.9 | 2 | 5.1 | 2 | 3.5 | 2 | 7.4 |
| 4 | 3 | 4.3 | 5 | 12.8 | 3 | 5.3 | 5 | 18.5 |
| 5 | 1 | 1.4 | 6 | 15.4 | 1 | 1.8 | 6 | 22.2 |
| 6 | 16 | 23.2 | 5 | 12.8 | 16 | 28.1 | 5 | 18.5 |
| 7 | 12 | 17.4 | 6 | 15.4 | 12 | 21.1 | 6 | 22.2 |
| 8 | 23 | 33.3 | 3 | 7.7 | 23 | 40.4 | 3 | 11.1 |
| HER2 status | ||||||||
| Negative | 58 | 84.1 | 34 | 87.2 | 51 | 89.5 | 24 | 88.9 |
| Positive | 11 | 15.9 | 5 | 12.8 | 6 | 10.5 | 3 | 11.1 |
| Histologic grade | ||||||||
| 1 | 18 | 26.1 | 9 | 23.1 | 17 | 29.8 | 7 | 25.9 |
| 2 | 30 | 43.5 | 17 | 43.6 | 27 | 47.4 | 13 | 48.1 |
| 3 | 21 | 30.4 | 13 | 33.3 | 13 | 22.8 | 7 | 25.9 |
| Histologic type | ||||||||
| Ductal | 64 | 92.8 | 33 | 84.6 | 52 | 91.2 | 22 | 81.5 |
| Lobular | 5 | 7.2 | 6 | 15.4 | 5 | 8.8 | 5 | 18.5 |
| cMYC amplification* | ||||||||
| Negative | 66 | 95.6 | 38 | 97.4 | 54 | 94.7 | 26 | 96.3 |
| Positive | 2 | 2.9 | 1 | 2.6 | 2 | 3.5 | 1 | 3.7 |
| Total | 69 | 100.0 | 39 | 100.0 | 57 | 100.0 | 27 | 100.0 |
We first tested this by examining ER IHC results with the semi-quantitative score as described by Harvey et al.12 A comparison by treatment group of the distribution of this variable is listed in Table 2. ER protein expression was lower in ER-positive tumors in the tamoxifen arm than in the placebo arm (P < .001; Fig 4B). We also performed global gene expression profiling, because B-14 demonstrated that ESR1 mRNA is a better quantitative measure of ER level than IHC or LBA, and we wanted to determine if other genes might contribute to tamoxifen resistance.
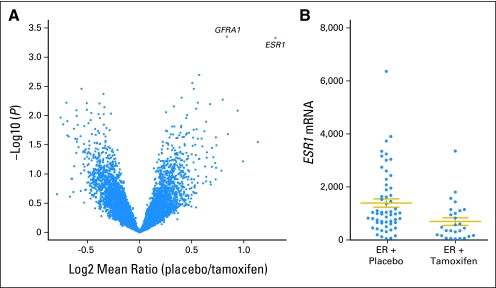
Fig 4.
Results of microarray gene expression analysis of National Surgical Adjuvant Breast and Bowel Project (NSABP) Breast Cancer Prevention Trial (P-1) estrogen receptor (ER) –positive cancer events. (A) Volcano plot of the results of t test for differences of expression levels of all filtered genes (N = 7,743) from Agilent array (Agilent Technologies, Santa Clara, CA) between ER-positive invasive cancer events that occurred initially in the tamoxifen versus the placebo arm from NSABP P-1 (N = 84). The x axis represents log (fold difference between two phenotypes); the y axis represents log (P value). Each spot represents a gene. Locations of the ER gene probe (ie, ESR1) and of GRFA1 are noted. (B) Scattergram of normalized ESR1 mRNA levels of ER-positive invasive breast cancer events from the NSABP P-1 according to treatment assignment (N = 84). Note the absence of tumors with high levels of ESR1 mRNA in the tamoxifen arm. Horizontal bars represent mean with standard errors. P value for the difference was less than .001. Data are plotted in a natural scale to better demonstrate the differences in higher expression level data range.
When differences between the ER-positive tumors that developed in the placebo arm were compared with those in the tamoxifen arm by microarray, the only differentially expressed gene with a fold difference greater than a two was ESR1, which was significantly underexpressed in the tumors from the tamoxifen arm (Data Supplement). The mean log_2_ intensity of ESR1 was 9.89 for the placebo arm, and it was 8.59 for the tamoxifen arm, with a 1.30 log_2_-fold (or 2.47-fold) difference. This is best demonstrated with a volcano plot (Fig 4A), with log(fold difference) in the _x_-axis and log(P value) from the t test in the _y_-axis for all genes examined. ESR1 and GFRA1 are distinctly separated from other genes in this plot (Fig 4A). The GFRA1 gene was differentially expressed by 1.79-fold13 (Data Supplement). Because the volcano plot does not provide insight into the distribution of occurrences for a single gene within each cohort, a scattergram of the ESR1 mRNA expression level of individual occurrences categorized by treatment arm was constructed. Visual inspection of the plot shows that tamoxifen was less effective in preventing tumors with low expression of ESR1 mRNA (Fig 4B).
There are other genes that were differentially expressed in the ER-positive tumors that occurred in the tamoxifen arm, but the degree of difference was not as marked as it was for ESR1 or GFRA1. It is possible that some of these are false-positive findings. These genes are listed in the Data Supplement.
DISCUSSION
This study represents a retrospective subset analysis of available tissues from two prominent and different clinical trials: NSABP B-14, a treatment trial to test the benefit of adjuvant tamoxifen, and NSABP P-1, a prevention trial to test the ability of tamoxifen in preventing breast cancers. Data from the B-14 trial showed that both ER protein and ESR1 RNA levels had a highly significant interaction with tamoxifen and that the maximum benefit from tamoxifen was achieved in the highest tertile of ESR1 mRNA expression levels.
Data from the P-1 trial validated this relationship between ER and tamoxifen. ER protein and ESR1 RNA levels and whole-transcriptome expression analysis of P-1 tumors confirmed the observation that low levels of ER were associated with tamoxifen resistance. ER protein expression was lower in ER-positive tumors that arose in the tamoxifen arm than those in the placebo arm (P < .001). ESR1 expression was underexpressed by 2.5-fold in tumors that arose in the tamoxifen arm compared with tumors that arose in the placebo arm (P = .0005). Expression profiling of the entire human genome (more than 44,000 probes on Agilent arrays) identified ESR1 as the most significantly differentially expressed gene between the placebo and tamoxifen treated tumors. These two different trials with both protein and RNA analyses both indicated that low ER levels were associated with the tamoxifen resistance.
The limitations of subset analysis have been detailed elsewhere and are always a concern in interpretation of data. However, the confirmation of B-14 results by P-1 results supports the conclusion that low levels of ER are at least in part responsible for tamoxifen resistance in breast cancer.
ER is used as a dichotomous variable in clinical decision making. This practice has been appropriate, because the examination of B-14 data never demonstrated a statistically significant interaction between ER protein levels (measured by LBA) and the degree of benefit from tamoxifen,4 and because the current generation of IHC assays is essentially bimodal in distribution as a result of saturation of the assay at relatively low levels of ER expression.14 However, examination of the NSABP B-14 trial with a quantitative assay method using Onco_type_DX demonstrated a clear linear relationship between ESR1 mRNA and the degree of benefit from tamoxifen; patients with lower levels of ESR1 mRNA did not gain significant benefit from adjuvant tamoxifen, even though their tumors were classified as ER positive by the dichotomous clinical definition.9
This finding has important implications, because it suggests that lower expression levels of ESR1 mRNA may be one of the mechanisms responsible for tamoxifen resistance. Cancer events from the P-1 trial provided a unique cohort in which to test this hypothesis developed from B-14, because ER-positive tumors arising in the tamoxifen arm are, by definition, tamoxifen resistant. Microarray gene expression profiling was used to examine differentially expressed genes between the ER-positive tumors that arose in the tamoxifen arm compared with the placebo arm, and an a priori hypothesis stated that ESR1 would be the major differentially expressed gene. The result is in agreement with that hypothesis. The only gene that was significantly differentially expressed between the two cohorts by more than two-fold was ESR1 (2.47-fold). Another gene, GFRA1, which is associated with the ER-positive phenotype, was also distinctly differentially expressed, although only by 1.79 fold.13 Tamoxifen was more effective in preventing ER-positive tumors with higher levels of ESR1 mRNA but was not effective in preventing those with lower levels. The absence of other significant differentially expressed genes suggests that low ESR1 mRNA levels are associated with tamoxifen resistance in many of these tumors, but other mechanisms are possible.
Because tamoxifen primarily prevented tumors with high ESR1 mRNA, an interpretation could be made that breast cancer chemoprevention with tamoxifen is clinically meaningless, because it only prevents tumors with good prognoses. However, ESR1 itself was not prognostic in untreated patients (Data Supplement), and there was only modest correlation between ESR1 and the 21-gene recurrence score in such a way that even high ESR1 tumors can have a high recurrence scores (Data Supplement).15 Therefore, one could have a tumor high in ESR1 and still have a poor prognosis. Furthermore, prevention of even good-prognosis breast cancer is a desirable and meaningful outcome.
Because tamoxifen is less effective in preventing the occurrence and relapse of ER-positive tumors with low levels of ESR1 expression, it is important to develop strategies to treat and prevent such tumors. A report by the Trans-ATAC (ie, Arimidex, tamoxifen, alone or in combination trial) investigators16 provided an important perspective on this question. In that study, benefit from anastrazole was independent of the quantile levels of ER measured by IHC or ESR1 mRNA. Therefore, it is possible that aromatase inhibitors could be better preventive agents than tamoxifen by inhibiting the occurrence of even low-ER tumors.
HER2 has been implicated as an instigator of tamoxifen resistance in some studies.17–19 However, in our two study cohorts, HER2 gene amplification or expression levels did not directly predict tamoxifen resistance; as reported previously,20 there were generally fewer instances of high ESR1 expression among the _HER2_-amplified tumors in B-14 (data not shown).
In summary, these data suggest that the low-level expression of ESR1 is associated with tamoxifen resistance in ER-positive breast cancer. Strategies should be developed to identify, treat, and prevent such tumors.
Supplementary Material
Data Supplements
Footnotes
Supported by Public Health Service Grants No. U10-CA-37377, U10-CA-69974, U10-CA-12027, U10-CA-69651, and U24-CA-114732 from the National Cancer Institute, Department of Health and Human Services; in part by a grant with the Pennsylvania Department of Health; and by Genomic Health.
The Pennsylvania Department of Health specifically disclaims responsibility for any analysis, interpretations, or conclusions.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00003906.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Frederick L Baehner, Genomic Health (C); Joffre Baker, Genomic Health (C); Maureen T. Cronin, Genomic Health (C); Drew Watson, Genomic Health (C); Steven Shak, Genomic Health (C) Consultant or Advisory Role: Victor G Vogel, AstraZeneca (U); D. Lawrence Wickerham, Eli Lilly (U) Stock Ownership: Frederick L Baehner, Genomic Health; Joffre Baker, Genomic Health; Maureen T Cronin, Genomic Health; Drew Watson, Genomic Health; Steven Shak, Genomic Health Honoraria: Charles E Geyer Jr, Genomic Health; D. Lawrence Wickerham, AstraZeneca Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Chungyeul Kim, Gong Tang, Katherine L. Pogue-Geile, Joffre Baker, Drew Watson, Steven Shak, Norman Wolmark, Soonmyung Paik
Financial support: Steven Shak, Soonmyung Paik
Administrative support: Joseph P. Costantino, Steven Shak, Worta McCaskill-Stevens, D. Lawrence Wickerham, Soonmyung Paik
Provision of study materials or patients: Victor G. Vogel, D. Lawrence Wickerham, Soonmyung Paik
Collection and assembly of data: Chungyeul Kim, Gong Tang, Katherine L. Pogue-Geile, Joseph P. Costantino, Frederick L. Baehner, Maureen T. Cronin, Drew Watson, Steven Shak, Olga L. Bohn, Debora Fumagalli, Yusuke Taniyama, Ahwon Lee, Megan L. Reilly, Victor G. Vogel, Worta McCaskill-Stevens, D. Lawrence Wickerham, Soonmyung Paik
Data analysis and interpretation: Gong Tang, Katherine L. Pogue-Geile, Maureen T. Cronin, Steven Shak, Olga L. Bohn, Debora Fumagalli, Leslie G. Ford, Charles E. Geyer Jr, Soonmyung Paik
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Early Breast Cancer Trialists' Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomized trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 3.Clarke R, Liu MC, Bouker KB, et al. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 4.Bryant J, Fisher B, Gunduz N, et al. S-phase fraction combined with other patient and tumor characteristics for the prognosis of node-negative, estrogen-receptor-positive breast cancer. Breast Cancer Res Treat. 1998;51:239–253. doi: 10.1023/a:1006184428857. [DOI] [PubMed] [Google Scholar]
- 5.Fisher B, Redmond C, Brown A, et al. Influence of tumor estrogen and progesterone receptor levels on the response to tamoxifen and chemotherapy in primary breast cancer. J Clin Oncol. 1983;1:227–241. doi: 10.1200/JCO.1983.1.4.227. [DOI] [PubMed] [Google Scholar]
- 6.McGuire WL. Estrogen receptors in human breast cancer. J Clin Invest. 1973;52:73–77. doi: 10.1172/JCI107175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan LR, Jr, Schein PS, Woolley PV, et al. Therapeutic use of tamoxifen in advanced breast cancer: Correlation with biochemical parameters. Cancer Treat Rep. 1976;60:1437–1443. [PubMed] [Google Scholar]
- 8.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 9.Fisher B, Costantino J, Redmond C, et al. A randomized clinical trial evaluating tamoxifen in the treatment of patients with node-negative breast cancer who have estrogen-receptor-positive tumors. N Engl J Med. 1989;320:479–484. doi: 10.1056/NEJM198902233200802. [DOI] [PubMed] [Google Scholar]
- 10.Fisher B, Jeong JH, Bryant J, et al. Treatment of lymph-node-negative, oestrogen-receptor-positive breast cancer: Long-term findings from National Surgical Adjuvant Breast and Bowel Project randomized clinical trials. Lancet. 2004;364:858–868. doi: 10.1016/S0140-6736(04)16981-X. [DOI] [PubMed] [Google Scholar]
- 11.Gray RJ. Spline-based tests in survival analysis. Biometrics. 1994;50:640–652. [PubMed] [Google Scholar]
- 12.Harvey JM, Clark GM, Osborne CK, et al. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 13.Esseghir S, Todd SK, Hunt T, et al. A role for glial cell derived neurotrophic factor induced expression by inflammatory cytokines and RET/GFR alpha 1 receptor up-regulation in breast cancer. Cancer Res. 2007;67:11732–11741. doi: 10.1158/0008-5472.CAN-07-2343. [DOI] [PubMed] [Google Scholar]
- 14.Collins LC, Botero ML, Schnitt SJ. Bimodal frequency distribution of estrogen receptor immunohistochemical staining results in breast cancer: An analysis of 825 cases. Am J Clin Pathol. 2005;123:16–20. doi: 10.1309/hcf035n9wk40etj0. [DOI] [PubMed] [Google Scholar]
- 15.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 16.Dowsett M, Allred C, Knox J, et al. Relationship between quantitative estrogen and progesterone receptor expression and human epidermal growth factor receptor 2 (HER-2) status with recurrence in the Arimidex, tamoxifen, alone or in combination trial. J Clin Oncol. 2008;26:1059–1065. doi: 10.1200/JCO.2007.12.9437. [DOI] [PubMed] [Google Scholar]
- 17.De Placido S, De Laurentiis M, Carlomagno C, et al. Twenty-year results of the Naples GUN randomized trial: Predictive factors of adjuvant tamoxifen efficacy in early breast cancer. Clin Cancer Res. 2003;9:1039–1046. [PubMed] [Google Scholar]
- 18.Dowsett M, Houghton J, Iden C, et al. Benefit from adjuvant tamoxifen therapy in primary breast cancer patients according oestrogen receptor, progesterone receptor, EGF receptor, and HER2 status. Ann Oncol. 2006;17:818–826. doi: 10.1093/annonc/mdl016. [DOI] [PubMed] [Google Scholar]
- 19.Ellis MJ, Coop A, Singh B, et al. Letrozole is more effective neoadjuvant endocrine therapy than tamoxifen for ErbB-1-, estrogen receptor-positive primary breast cancer: Evidence from a phase III randomized trial. J Clin Oncol. 2001;19:3808–3816. doi: 10.1200/JCO.2001.19.18.3808. [DOI] [PubMed] [Google Scholar]
- 20.Konecny G, Pauletti G, Pegram M, et al. Quantitative association between HER-2/neu and steroid receptors in hormone receptor-positive primary breast cancer. J Natl Cancer Inst. 2003;95:142–153. doi: 10.1093/jnci/95.2.142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Supplements