Association between BRCA2 but not BRCA1 Mutations and Beneficial Survival, Chemotherapy Sensitivity, and Gene Mutator Phenotype in Patients with Ovarian Cancer (original) (raw)
. Author manuscript; available in PMC: 2014 Sep 9.
Published in final edited form as: JAMA. 2011 Oct 12;306(14):1557–1565. doi: 10.1001/jama.2011.1456
Abstract
Context
Attempts to determine the clinical significance of BRCA1/2 mutations in ovarian cancer (OvCa) have produced conflicting results.
Objective
To determine the relationships between BRCA1/2 deficiency (i.e., mutation and promoter hypermethylation) and overall survival (OS), progression-free survival (PFS), chemotherapy response, and whole exome mutation rate in OvCa.
Design, Setting, and Patients
Observational study of multidimensional genomics and clinical data on 316 high-grade serous OvCa cases that were made public between 2009 and 2010 via The Cancer Genome Atlas project.
Main Outcome Measures
OS and PFS rates (primary outcomes) and chemotherapy response (secondary outcome).
Results
BRCA2 mutations (29 cases) were associated with significantly better OS (adjusted hazard ratio [HR], 0.33; 95% CI, 0.16–0.69, _P_=0.003; 5-year OS: 61% for BRCA2 mutated vs. 25% for BRCA wild-type [wt] cases) and PFS (adjusted HR, 0.40; 95% CI, 0.22–0.74, _P_=0.004; 3-year PFS: 44% for BRCA2 mutated vs. 16% for BRCA wt cases), whereas neither BRCA1 mutations (37 cases) nor BRCA1 methylation (33 cases) were associated with prognosis. Moreover, BRCA2 mutations were associated with a significantly higher primary chemotherapy sensitivity rate (100% for BRCA2 mutated vs. 82% [_P_=0.02] and 80% [_P_=0.05] for BRCA wt and BRCA1 mutated cases, respectively) and longer platinum-free duration (median platinum-free duration: 18.0 months for BRCA2 mutated vs. 11.7 [_P_=0.02] and 12.5 [_P_=0.04] months for BRCA wt and BRCA1 mutated cases, respectively). Further investigation revealed that BRCA2 mutated, but not BRCA1 mutated cases, exhibited a “mutator phenotype” by containing significantly more mutations than BRCA wt cases across the whole exome (median mutation number per sample: 84 for BRCA2 mutated vs. 52 for BRCA wt cases, false-discovery rate <0.1).
Conclusions
BRCA2 mutation, but not BRCA1 deficiency, is associated with improved survival, chemotherapy response, and genome instability compared with BRCA wild-type.
Keywords: BRCA1, BRCA2, mutations, survival, platinum-based drug response
Increased surveillance of BRCA1/2 germ-line mutation carriers is a generally accepted strategy for detecting ovarian cancer early. Women with BRCA1 mutations have a 39%–54% cumulative lifetime risk of developing OvCa, and women with BRCA2 mutations have a 11%–23% risk1–3.
Both BRCA1 and BRCA2 tumor suppressor genes are involved in DNA repair via homologous recombination. Cells with alterations in homologous recombination pathway genes are unable to repair DNA double-strand breaks by homologous recombination, which is mostly error-free. This can result in genomic instability and a predisposition to malignant transformation4, 5. On the other hand, because homologous recombination pathway deficiencies can also impair tumor cells’ ability to repair DNA-cross links introduced by chemotherapy agents such as cisplatin, it has been hypothesized that BRCA-deficient patients will likely have higher survival rates because of an improved response to platinum-based chemotherapy15. However, conflicting data exist regarding the outcome of BRCA-deficient patients after OvCa develops. Some researchers have found that OvCa patients with BRCA1/2 germ-line mutations have a more favorable clinical course6–12, whereas others have shown the opposite13, 14. Second, whether the effect of BRCA1/2 mutations on patient outcome is directly attributable to their influence on platinum-based chemotherapy response has not been well elucidated. Most studies that have investigated the clinical features of BRCA1/2 mutation carriers lack detailed chemotherapy information6–9, 11, 12, apart from occasional studies reporting improved responses to platinum-based therapy in small cohorts15.
Using multidimensional genomic and clinical data on 316 high-grade serous OvCa patients in The Cancer Genome Atlas (TCGA) project, we evaluated the association between BRCA1/2 deficiencies in OvCa and patient overall and progression-free survival (OS and PFS) rates, chemotherapy response, and whole-exome mutation rates.
PATIENTS AND METHODS
Patients
We searched the TCGA database of 316 high-grade serous OvCa patients on September 1, 2010. Detailed patient information, including age at diagnosis, tumor stage and grade, and surgical outcome, is listed in Table 1. All OvCa specimens were surgically resected before systemic treatment and were selected to have >70% tumor cell nuclei and <20% necrosis. Ninety-five percent of tumors were stage III or IV, and all were high grade. According to the TCGA database, 86% of patients were non-Ashkenazi Jewish whites; 7% were Ashkenazi Jews, 3% were African-Americans, and 3% were Asian (Table 1). All patients received a platinum agent, and 94% received a taxane. Specimens in the TCGA network were obtained from patients with appropriate consent from the relevant institutional review board. All TCGA database information remained de-identified.
Table 1.
Age and tumor characteristics of patients with different BRCA1/2 status
| Characteristic | All cases | BRCAwild-type | _BRCA1_mutation | _BRCA2_mutation | _BRCA1_methylation | _P_valuec |
|---|---|---|---|---|---|---|
| No. of casesa | 316 | 219 | 35 | 27 | 33 | |
| Age (years) | ||||||
| Mean (SD) | 60.6 (0.7) | 61.8 (0.8) | 55.9 (1.9) | 60.9 (2.4) | 57.3 (1.6) | .01 |
| Range | 27–88 | 35–88 | 41–76 | 27–79 | 40–77 | |
| Ethnic background (%) | ||||||
| Non-Ashkenazi Jewish | 266 (86) | 190 (89) | 24 (71) | 21 (78) | 29 (91) | |
| white | ||||||
| Ashkenazi Jewish | 20 (7) | 9 (4) | 7 (21) | 3 (11) | 1 (3) | |
| African-American | 10 (3) | 7 (3) | 1 (3) | 2 (7) | 0 (0) | .02 |
| Asian | 10 (3) | 6 (3) | 2 (6) | 0 (0) | 2 (6) | |
| Other | 2 (1) | 1 (1) | 0 (0) | 1 (4) | 0 (0) | |
| Missing, no. | 8 | 6 | 1 | 0 | 1 | |
| Tumor stage (%) | ||||||
| II | 14 (4) | 9 (4) | 2 (6) | 1 (4) | 2 (6) | |
| III | 248 (79) | 169 (77) | 27 (77) | 24 (92) | 26 (79) | .65 |
| IV | 53 (17) | 41 (19) | 6 (17) | 1 (4) | 5 (15) | |
| Missing, no. | 1 | 0 | 0 | 1 | 0 | |
| Tumor grade (%) | ||||||
| 2 | 28 (9) | 20 (9) | 2 (6) | 2 (8) | 3 (9) | |
| 3 | 281 (91) | 193 (91) | 32 (94) | 24 (92) | 30 (91) | .92 |
| Missing, no. | 7 | 6 | 0 | 1 | 0 | |
| Residual tumor (%) | ||||||
| 0 cmb | 58 (21) | 37 (19) | 7 (23) | 5 (21) | 8 (27) | |
| ≤1 cm | 150 (54) | 103 (54) | 15 (5) | 14 (58) | 17 (57) | |
| 1–2 cm | 14 (5) | 11 (6) | 2 (7) | 1 (4) | 0 (0) | .95 |
| >2 cm | 56 (20) | 41 (21) | 6 (20) | 4 (17) | 5 (17) | |
| Missing, no. | 38 | 27 | 5 | 3 | 3 |
OS duration was defined as the interval from the date of initial surgical resection to the date of death or last contact (censored). The PFS duration was defined as the interval from the date of initial surgical resection to the date of progression/recurrence or last contact (censored). The drug (platinum)-free interval was defined as the interval from the end of adjuvant platinum-based treatment to the date of progression/recurrence, or last contact (censored).
Analysis of Chemotherapy Response Data
Two aspects of chemotherapy response were investigated: primary response to platinum-treatment and platinum-free duration after treatment. The patient was designated as primary sensitive if she had experienced a complete or partial response to adjuvant chemotherapy as noted in the TCGA data16 and as primary resistant if she had stable or progressive disease. On the basis of this criterion, 225 cases were primary sensitive and 36 were primary resistant. Fifty-five cases with no information on primary response to adjuvant therapy were excluded (eFigure 1). In 225 primary sensitive cases, we used the drug (platinum)-free duration to scale chemotherapy response; the shorter the platinum-free duration, the more resistance the patient had. Given that suboptimal debulking can contribute to rapid disease progression17, 33 patients with >2 cm residual tumor were excluded from the platinum-free duration survival analysis (eFigure 1).
Analysis of Whole-Exome Mutation Data
In total, 19,359 mutations across 316 OvCa samples were downloaded from the TCGA Data Portal (http://tcga-data.nci.nih.gov/tcga/findArchives.htm). The sequencing and quality control procedures were recently described18. In brief, whole-exome capture (~180,000 exons from ~18,500 genes) and sequencing was performed on 316 OvCa samples and matched (normal) controls. Among them, 236 sample pairs were performed on the Illumina GAIIx platform and 80 sample pairs on the ABI SOLiD 3 platform.
We used an enrichment score (ES)19 to determine whether cases with BRCA1 or BRCA2 mutations were enriched among hypermutated cases with high mutation rates across the whole exome. First, all 316 OvCa cases were decreasingly ordered on the basis of their total mutation numbers. For each patient group (i.e., _BRCA1_-or BRCA2_-mutated), we calculated the ES, which is a normalized Kolmogorov-Smirnov statistic. Considering the samples S1,‥,S_i,‥, S_N_=316, which are ordered on the basis of total mutations, and a patient group P that contains G members, we defined
if S_i_ was not a member of P and
if S_i_ was a member of P. We then computed a running sum across all N samples. The ES was defined as
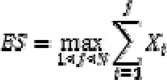 |
(3) |
|---|
Intuitively, the ES calculated by going down the decreasingly ordered sample list. If a sample was included in the target group (i.e., _BRCA1_- or _BRCA2_-mutated), we increased the running-sum statistic and otherwise decreased the statistic. ES reached a higher positive score when samples in the target group were consistently ranked at the top of the sample list. The maximum ES was obtained when the N samples in the target group were ranked the top N most mutated samples among all 316 OvCa cases. The ES was measured for each _BRCA1_- and _BRCA2_-mutated patient group. To determine whether any given patient group was significantly associated with hypermutation, we permuted the BRCA mutation status 106 times, which generated a background ES distribution to calculate the false-discovery rate (FDR).
Analysis of Methylation and Expression Data
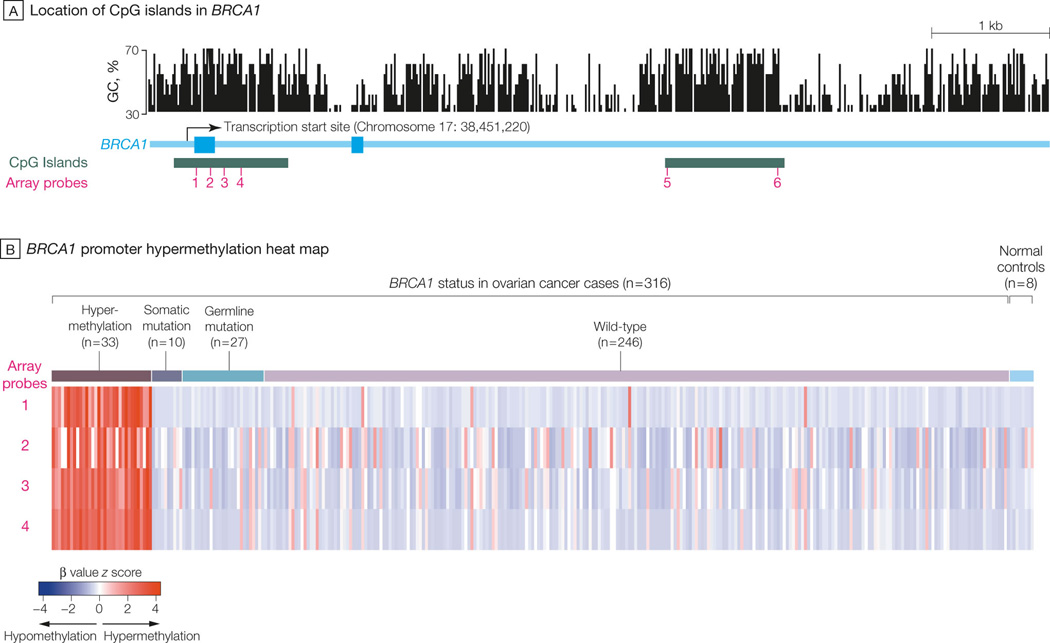
Level 3 Illumina Infinium DNA methylation and Agilent 244K gene expression data of 316 TCGA ovarian samples and eight normal fallopian tube samples were downloaded on September 1, 2010, from the Open-Access tiers of TCGA Data Portal (http://tcga-data.nci.nih.gov/tcga/findArchives.htm). The Illumina Infinium HumanMethylation27 arrays interrogate 27,578 CpG sites located in proximity to the transcription start sites of 14,475 consensus coding sequences in the NCBI Database (Genome Build 36).
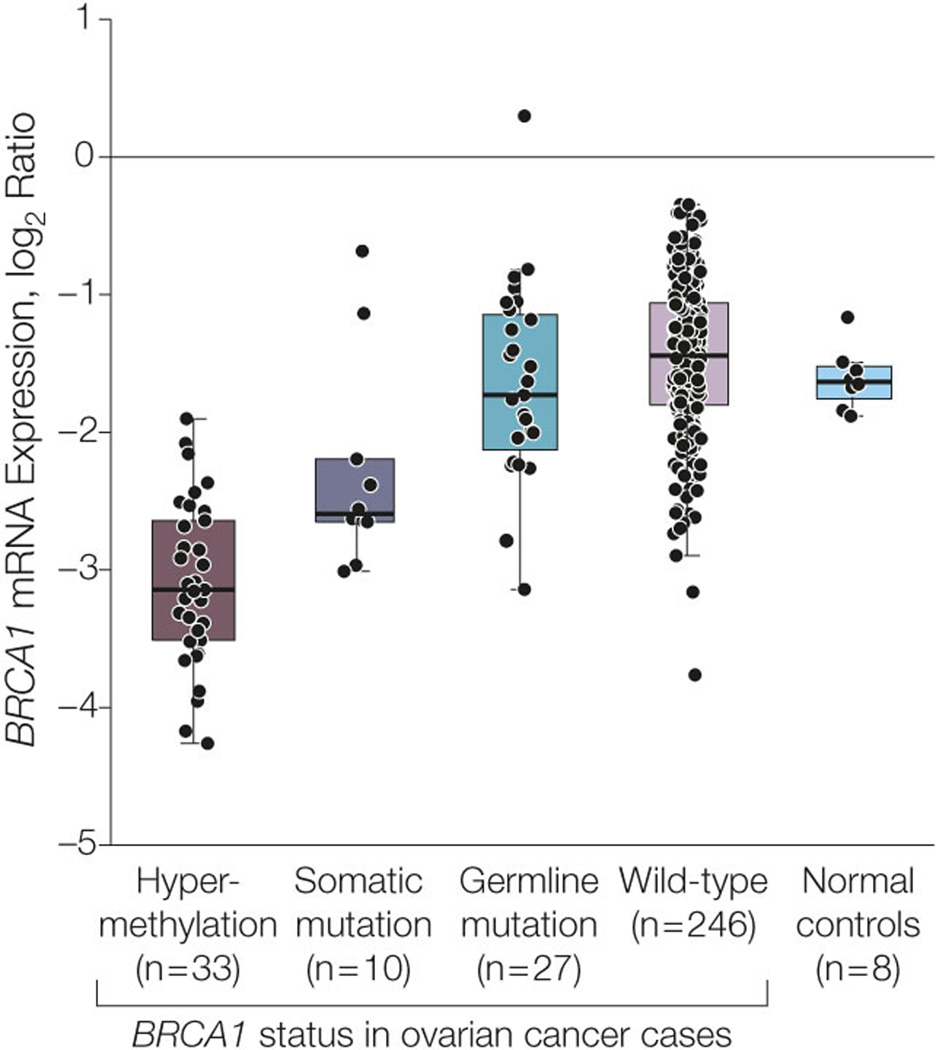
We calculated the Spearman’s rank correlation between DNA methylation and gene expression for nine different probes located in three CpG islands near the BRCA1 region and found statistically significant inverse correlations (Benjamini-Hochberg adjusted FDR <0.0001) for four probes (cg19531713, cg19088651, cg08993267, and cg04658354) located in the CpG island near the transcription start site (TSS) (see Figure 5A). We then used _K_-means consensus clustering (_K_=2) on the DNA methylation (beta values) of four probes across 316 samples to separate the epigenetically silenced and non-epigenetically silenced groups of samples. For the five probes located in the other two CpG islands far away from the TSS, no significant inverse correlation with BRCA1 expression was observed. No inverse correlation between probes near the BRCA2 region and BRCA2 mRNA expression was observed.
Figure 5. Hypermethylation of BRCA1 promoter in 316 OvCa cases.
A. Locations of BRCA1 CpG islands and methylation microarray probes. The GC percentage (top panel), CpG island (blue), and six probes (red bars) annotated to BRCA1 in an Illumina Infinium DNA methylation microarray were visualized using the University of California Santa Cruz (UCSC) genome browser. The GC percentage track shows the percentage of G (guanine) and C (cytosine) bases in 5-base windows. Three other probes (see Methods) annotated to BRCA1 are not shown here because they were too far away from the BRCA1 transcription start site (TSS). B. Heatmap shows the DNA methylation of BRCA1 promoter across 316 cases. Four probes located in the promoter CpG island of BRCA1 are arranged in columns. Three hundred sixteen OvCa and eight normal samples are arranged in rows. Red: hypermethylation; blue: hypomethylation. The color bar to the right of the heatmap indicates BRCA1 status: red: _BRCA1_-hypermethylated OvCa; green: BRCA1 somatically mutated OvCa; yellow: BRCA1 germ-line-mutated OvCa; blue: wt BRCA1 OvCa (excluding the hypermethylated cases); black: normal tissue. C. BRCA1 mRNA expression in different status groups. OvCa cases are grouped by BRCA1 deficiency type, as shown along the right side of Figure 5B. Each data point represents the BRCA1 mRNA expression level (x axis) in the tumor of the corresponding group. Red data points: BRCA1 hypermethylated OvCa; green data points: BRCA1 somatically mutated OvCa; yellow data points: BRCA1 germ-line mutated OvCa; blue data points: wt BRCA1 OvCa; black data points: normal tissue. The box plots indicate the minimum, lower quartile, median, upper quartile, and maximum for each group.
Statistical Analysis
Standard statistical tests were used to analyze the clinical and genomics data, including the chi-square test, Fisher’s exact test, Kruskal-Wallis test, Wilcoxon rank-sum test, log-rank test, and Cox proportional hazard analysis. Significance was defined as P < 0.05. Benjamini-Hochberg multiple testing correction20 was used to estimate the false-estimated rate, when multiple testing correction applied. Analyses were primarily performed using R 2.10.0 and SPSS v.18 (SPSS, Chicago, IL).
Results
BRCA1 and BRCA2 Mutations in OvCa
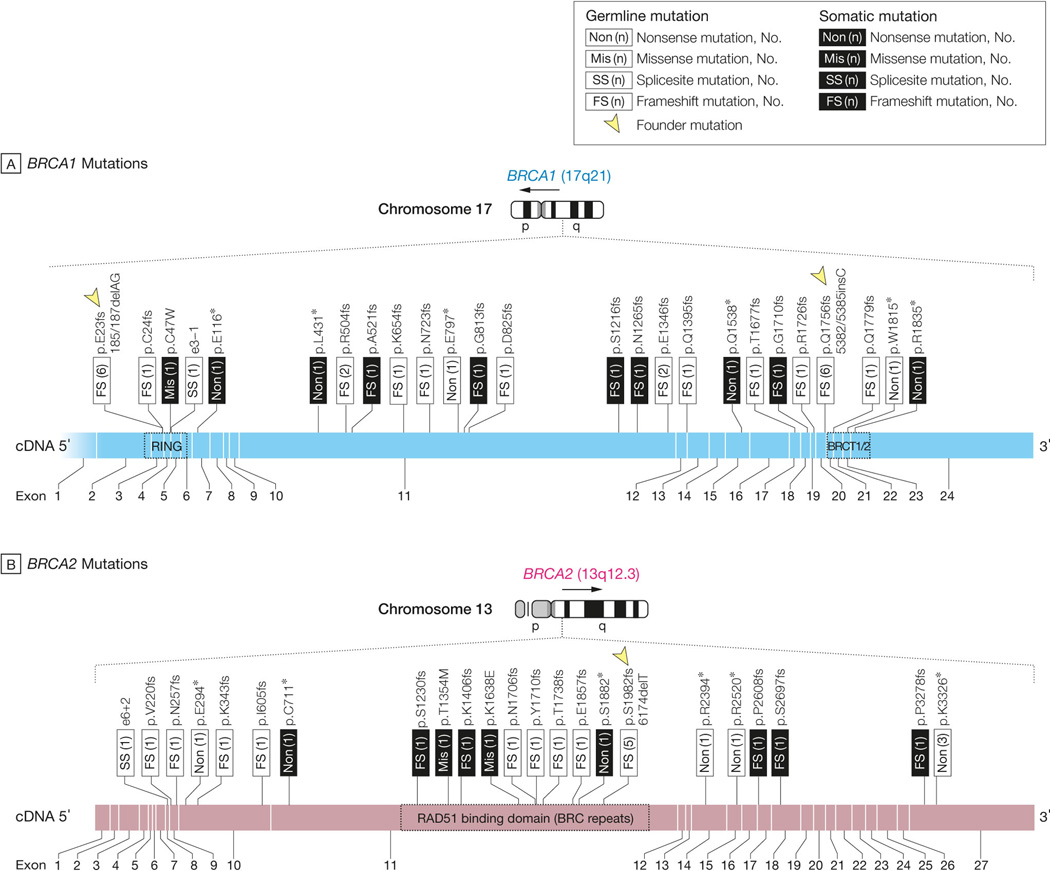
BRCA1 and BRCA2 were non-synonymously mutated in 37 (11.7%) and 29 (9.2%) of 316 cases, respectively. Two cases had both BRCA1 and BRCA2 mutations and were excluded from analyses comparing BRCA1 and BRCA2 mutated groups. All but one BRCA1 mutation were null mutations (frame shift or nonsense mutations). Among the 37 BRCA1 mutated cases, 27 were germ-line mutations and 10 were somatic mutations. Twelve of the observed BRCA1 germ-line mutations corresponded to the well-known “founder” mutations, 185/187delAG (E23fs) in the RING-type zinc finger domain and 5382/5385insC (Q1756fs), both of which have been extensively studied in Ashkenazi Jewish populations21 (Figure 1A). Among the 29 BRCA2 mutated cases, 20 were germ-line mutations and nine were somatic mutations. Only five of the observed BRCA2 germ-line mutations corresponded to the well-known 6174delT (S1982fs) founder mutation22 (Figure 1B).
Figure 1. BRCA1/2 mutations and their association with age at diagnosis in 316 OvCa cases.
A, B. Germ-line and somatic mutations in BRCA1 and BRCA2, respectively. Mutations are mapped to the corresponding exons of BRCA1 and BRCA2. Red: germ-line mutation; green: somatic mutation. Twelve BRCA1 germ-line mutations corresponded to the well-known founder mutations 185/187delAG and 5382/5385insC (indicated by arrowheads in A); five BRCA2 germ-line mutations corresponded to the well-known founder mutation 6174delT (indicated by the arrowhead in B). C. Scatter plot indicates the distribution of age at diagnosis of patients with different BRCA statuses. The P values are calculated by Wilcoxon rank-sum test. Abbreviations: wt, wild-type; mt, mutation.
Patients with both types of mutations did not differ significantly from each other with respect to tumor stage, grade, or histologic type (Table 1), but patients with BRCA1 mutations were younger at diagnosis (mean age, 55.9 years) than were those with wt BRCA (mean age, 61.8 years; _P_=0.006, Wilcoxon rank test) or BRCA2 mutation (mean age, 60.9 years; _P_=0.03, Wilcoxon rank test, Figure 1C). No differences in OS and PFS duration were observed between germ-line and somatic mutations; therefore, these mutation types were pooled for downstream analysis.
_BRCA2_-, but not _BRCA1_-, Mutated Cases Show Improved Survival in OS and PFS Duration
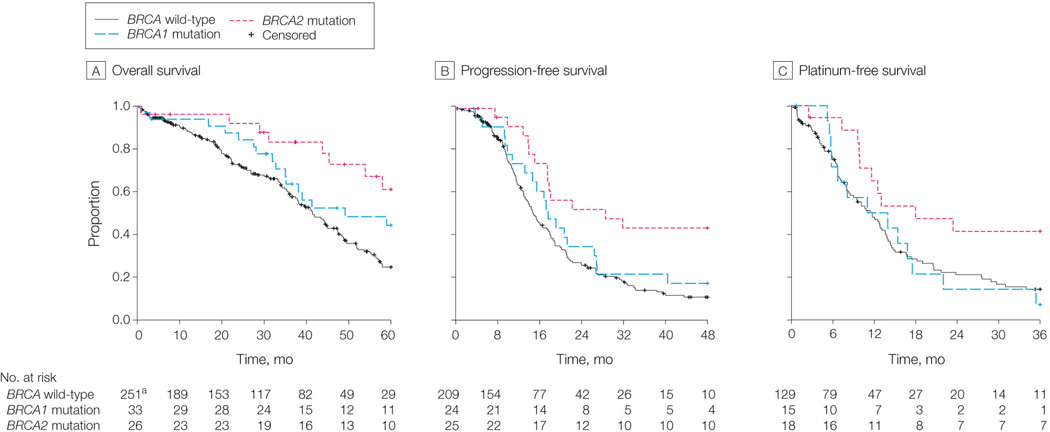
The 5-year survival rate of BRCA2 mutation carriers was 61% (95% CI [confidence interval] 43%-87%), which is significantly better than that of wt BRCA cases, with 5-year survival rates of 25% in the unadjusted (log-rank _P_=0.002, Figure 2A) and adjusted (_P_=0.003, HR [hazard ratio]=0.33, 95% CI 0.16–0.69) models (Table 2). In contrast, BRCA1 mutation carriers had non-significant difference in survival compared with wt BRCA cases in the unadjusted model (_P_=0.09, log-rank test, Figure 2A). BRCA1 mutation carriers’ non-significantly longer survival durations vanished when we adjusted for age (_P_=0.35, HR=0.76, 95% CI=0.43–1.35, Table 2), suggesting that their survival duration was attributable to younger age at diagnosis. BRCA2 mutation carriers had significantly longer PFS durations than did wt BRCA carriers (_P_=0.004, HR=0.40, 95% CI=0.22–0.74, Table 2); no difference was found for BRCA1 mutation carriers (Figure 2B and Table 2).
Figure 2. Association of BRCA1/2 mutations with survival and chemotherapy responses.
A, B. Kaplan-Meier survival curves for OS and PFS durations of _BRCA1_-mutated, _BRCA2_- mutated, and wt BRCA OvCa cases. The percent probability of survival is plotted versus time since diagnosis in months. C. Bar chart indicates the distribution of primary chemotherapy sensitivity in _BRCA1_-, _BRCA2_-mutated, and wt BRCA OvCa cases. The P values are calculated by two-tailed Fisher’s exact test. D. Kaplan-Meier survival curves for platinum-free duration of BRCA1, BRCA2, and wt BRCA OvCa cases. The percent probability of survival is plotted versus time since the end of adjuvant therapy.
Table 2.
Multivariable models for overall survival and progression-free survival in women with ovarian cancer
| Overall survival | ||||
|---|---|---|---|---|
| 3-year OS rate (%) (95% CI) | 5-year OS rate (%) (95% CI) | Hazard ratio (95% CI) | P valuec | |
| BRCA statusa | ||||
| BRCA wild-type | 58 (51–66) | 25 (19–34) | 1 [reference] | |
| BRCA1 mutation | 64 (49–84) | 44 (29–67) | 0.76 (0.43–1.35) | .35 |
| BRCA2 mutation | 83 (69–100) | 61 (43–87) | 0.33 (0.16–0.69) | .003 |
| BRCA1 methylation | 68 (51–89) | 24 (11–55) | 1.06 (0.62–1.81) | .83 |
| Tumor stage | ||||
| II | 92 (79–100) | 54 (28–100) | 1 [reference] | |
| III and IV | 60 (54–67) | 30 (24–37) | 4.12 (1.01–16.74) | .05 |
| Residual tumor | ||||
| 0 cmd | 73 (61–88) | 49 (34–72) | 1 [reference] | |
| ≤1 cm | 59 (50–68) | 21 (14–31) | 1.82 (1.09–3.03) | .02 |
| 1–2 cm | 61 (39–95) | 31 (13–77) | 1.42 (0.61–3.28) | .42 |
| >2 cm | 47 (34–64) | 27 (16–47) | 1.93 (1.07–3.50) | .03 |
| Age increase of 1.0 | - | - | 1.03 (1.01–1.05) | <.001 |
| year | ||||
| Progression-free survival | ||||
| 3-year PFS rate (%) (95% CI) | 5-year PFS rate (%) (95% CI) | Hazard ratio (95% CI) | P valuec | |
| BRCA statusa | ||||
| BRCA wild-type | 16 (11–23) | 10 (6–18) | 1 [reference] | |
| BRCA1 mutation | 22 (10–47) | 13 (5–38) | 0.81 (0.48–1.38) | .44 |
| BRCA2 mutation | 44 (27–69) | 39 (23–65) | 0.40 (0.22–0.74) | .004 |
| BRCA1 methylation | 5 (1–32) | NAb | 1.33 (0.82–2.15) | .24 |
| Tumor stage | ||||
| II | 56 (33–95) | 37 (14–98) | 1 [reference] | |
| III and IV | 16 (12–23) | 12 (8–18) | 3.31 (1.21–9.06) | .02 |
| Residual tumor | ||||
| 0 cmd | 36 (23–55) | 22 (11–45) | 1 [reference] | |
| ≤1 cm | 11 (6–19) | 9 (5–17) | 1.92 (1.25–2.95) | .003 |
| 1–2 cm | 15 (2–89) | 15 (2–89) | 2.29 (0.93–5.64) | .07 |
| >2 cm | 9 (3–25) | 4 (1–25) | 1.72 (1.03–2.87) | .04 |
| Age increase of 1.0 | - | - | 1.00 (0.98–1.01) | .88 |
| year |
A direct comparison between BRCA1 and BRCA2 mutation carriers revealed significant difference in PFS between BRCA1 and BRCA2 mutation carriers: 44% (95% CI: 27%-69%, Table 2) of BRCA2 mutated cases remained progression free 3 years after surgical resection compared with only 22% (95% CI: 10%–47%, Table 2) of BRCA1 mutated cases (_P_=0.05, log-rank test, Figure 2B).
_BRCA1_-, _BRCA2_-Mutated, and wt BRCA Patients Experience Different Responses to Chemotherapy
Among all 316 patients treated with platinum-based adjuvant chemotherapy, 261 experienced primary responses. We determined the association of BRCA1/2 mutations with chemotherapy response by investigating both primary chemotherapy response and platinum-free duration. Patients who experienced complete or partial responses to adjuvant chemotherapy were defined as primary sensitive, whereas patients with stable or progressive disease during therapy were defined as primary resistant (eFigure 1). We identified 225 sensitive and 36 resistant cases on the basis of this criterion. Among _BRCA2_-mutated cases, 100% (25 of 25) were primary sensitive compared with 85% (175 of 205) of wt BRCA cases _P_=0.05, chi-square test, Figure 2C). Interestingly, only 80% (24 of 30) of _BRCA1_-mutated cases were primary sensitive to platinum-based therapy (_P_=0.02 compared with BRCA2 mutated cases, chi-square test) (Figure 2C).
We next determined the association between BRCA1/2 mutations and platinum-free duration. As shown in Figure 2D, _BRCA2_-mutated cases had significantly longer platinum-free duration than those with BRCA1 mutations (_P_=0.04, log-rank test, median platinum-free duration: 18.0 months for BRCA2-mutated vs. 12.5 months for _BRCA1_-mutated cases) and wt BRCA cases (_P_=0.02, log-rank test, median platinum-free duration: 11.7 months). Again, we found no difference between BRCA1 mutation and wt BRCA cases in platinum-free survival duration. In summary, BRCA2 mutations were associated with significantly improved primary chemotherapy response and longer platinum-free durations than were _BRCA1_-mutated and wt BRCA OvCa patients, whereas BRCA1 mutations had no statistically significant association with primary chemotherapy sensitivity or platinum-free survival compared with wt BRCA cases.
_BRCA2_-Mutated Cases Exhibit a Mutator Phenotype in OvCa
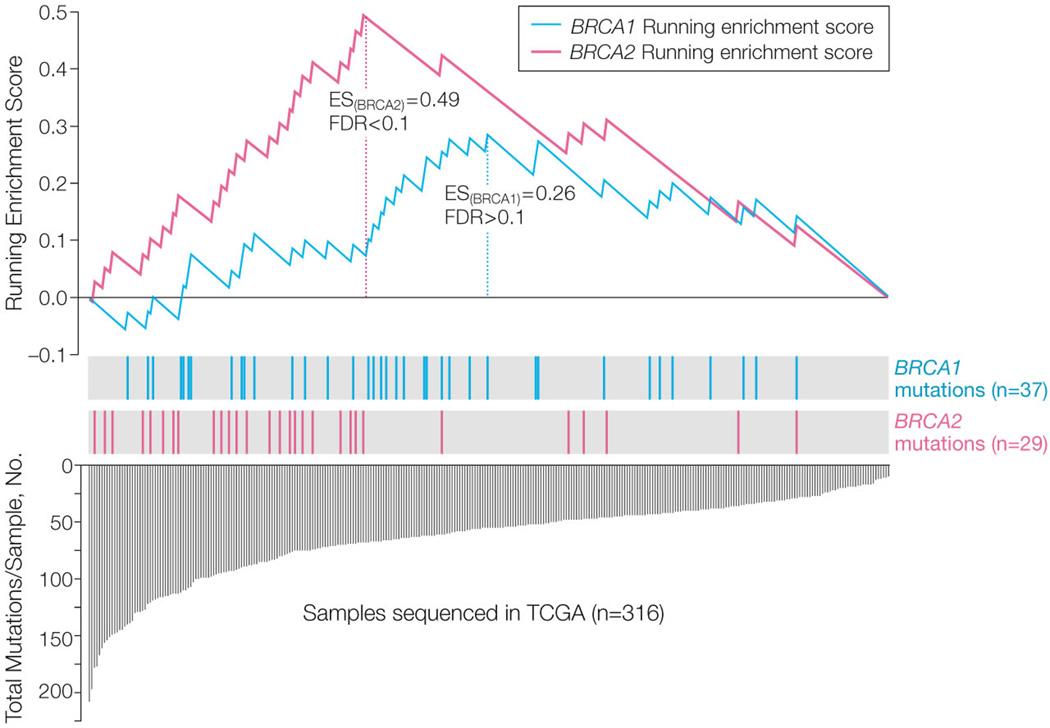
Using whole-exome deep-sequencing data on 316 TCGA cases, we further examined the association between BRCA1 and BRCA2 mutations with the mutation rate in the OvCa exome. The ES score, detailed in the Methods, was chosen to describe the degree of enrichment of hypermutated OvCa cases in _BRCA1_- and _BRCA2_-mutated patient groups. As we expected, _BRCA2_-mutated cases were highly enriched with hypermutated samples (ES=0.49, FDR <0.1, median mutation number per sample: 84 for _BRCA2_ mutated vs 52 for BRCA wt cases, Figure 3). However, we observed no enrichment of hyper-mutated samples in _BRCA1_-mutated cases (ES=0.26, FDR >0.1, Figure 3). We then identified 61 genes that were differentially mutated between BRCA2 and wt BRCA cases (P<0.005, FDR <0.2, Figure 4). A number of these genes are involved in response to DNA damage (e.g., TP63, BLM, and BCL3) (eTable 1). We could not identify differentially mutated genes between BRCA1 and wt BRCA cases using the same criteria.
Figure 3. Association of BRCA1/2 mutations with genome instability.
Enrichment Score (ES) test (detailed in Methods) of hypermutated sample enrichment in _BRCA1_- and _BRCA2_-mutated cases. The bottom portion of the plot shows the total numbers of non-synonymous mutations of 316 decreasingly ranked OvCa cases. The height of each blue discrete line indicates the number of non-synonymous mutations in each OvCa case. The middle portion of the plot shows where the samples with BRCA1 (green bar) or BRCA2 (red bar) mutations appear in the ranked list of samples in the bottom portion. The top portion of the plot shows the running ES for the _BRCA1_- or _BRCA2_-mutated cases.
Figure 4. Heatmap indicates the mutation landscape of _BRCA1_-, _BRCA2_-mutated, and wt BRCA cases.
Sixty-one differentially mutated genes between _BRCA2_-mutated and wt BRCA cases are arranged in rows; samples are in columns. Yellow: mutation. Blue: non-mutation. Each sample is also labeled as described in the top color bar for BRCA mutation status (green: BRCA1 mutation; red: BRCA2 mutation; blue: wt BRCA), tumor grade, clinical stage, and primary chemotherapy response status.
BRCA1 Hypermethylation Did Not Improve OvCa Prognosis
Using the procedures described in the Methods, we identified 33 of 316 samples (10.5%) with BRCA1 inactivation via promoter hypermethylation (Figure 5A, B). No promoter hypermethylation of BRCA2 was observed across 316 TCGA samples. The BRCA1 hypermethylated cases were mutually exclusive with _BRCA1_-mutated cases (_P_=0.02, Fisher’s exact test, Figure 5B). BRCA1 mRNA levels were significantly lower in hypermethylated BRCA1 cases than in wt BRCA1 cases and normal tissues (P <0.001, Wilcoxon rank-sum test, >2-fold change for both comparisons; Figure 5C), indicating that promoter hypermethylation indeed silenced BRCA1 expression. Similar to _BRCA1_-mutated patients, _BRCA1_-hypermethylated patients were significantly younger than wt BRCA patients (_P_=0.03, mean age at diagnosis: 57.3 years for BRCA1 hypermethylated vs. 61.8 years for BRCA wt cases, Table 1). _BRCA1_-hypermethylated cases exhibited no significant differences in OS or PFS duration compared with BRCA wt cases (Table 2) but had significantly shorter durations than those with BRCA2 mutations (median OS: 86.8 months for _BRCA2_-mutated vs. 41.5 months for _BRCA1_-hypermethylated cases, _P_=0.01, log-rank test; median PFS: 28.6 months for _BRCA2_-mutated vs. 14.8 months for _BRCA1_-hypermethylated cases, _P_=0.002, log-rank test; eTable 2). This observation indicates that BRCA1 inactivation, whether by genomic or epigenomic mechanisms, is not associated with improved OvCa patient outcome.
Comment
In this study, an analysis of 316 high-grade serous OvCa cases revealed that only BRCA2 mutations were an independent predictor of OvCa survival, whereas BRCA1 mutations were not significantly associated with beneficial OS. In a further analysis, we found no difference in PFS between _BRCA1_-mutated cases and wt BRCA cases, whereas _BRCA2_-mutated patients had significantly longer PFS durations than did _BRCA1_-mutated and wt BRCA patients. Furthermore, using DNA methylation data from the same 316 OvCa cases, we identified 33 BRCA1 promoter-hypermethylated cases. Similar to the _BRCA1_-mutated cases, _BRCA1_-hypermethylated cases had similar survival rate to that of wt BRCA cases but significantly shorter survival relative to BRCA2 mutated cases.
Previous studies have mostly combined BRCA1/2 mutations to assess potential associations with OvCa survival, and some have observed improved outcomes in patients with BRCA1/2 mutations. These include four reports on the association between three Ashkenazi Jewish founder BRCA1/2 mutations and survival of Jewish women with OvCa7, 9, 10, 12. Most of these studies are limited by a small sample size and a focus on germ-line founder mutations and thus do not have enough statistical power to allow adequate differentiation between _BRCA1_’s and _BRCA2_’s effects on survival. Our study provides a more representative spectrum of BRCA mutations than have previous studies, given that only 7% of patients in the present study were of Ashkenazi Jewish background and 27% of our _BRCA_-positive patients had Jewish founder mutations.
Our analyses of chemotherapy response confirmed our observations regarding survival by demonstrating that all _BRCA2_-mutated cases had significantly higher chemotherapy sensitivity rates and longer platinum-free durations than did _BRCA1_-mutated and wt BRCA cases. In accordance with our observations for prognosis and chemotherapy response, _BRCA2_-mutated cases, but not _BRCA1_-mutated cases, exhibited a “mutator phenotype” that contained significantly more mutations, as determined from whole-exome mutation data. These findings suggest that the different associations between survival and BRCA1 and BRCA2 deficiencies likely result from patients’ distinct responses to platinum-based treatment, which may be caused by the differing nature of the dysfunction of these two genes.
Differences between BRCA1 and BRCA2 mutations have been suggested by the results of previous studies. Clinically, although germ-line mutations in BRCA1 and BRCA2 result in a higher risk for breast and ovarian cancer, carriers of these genes have different risk factors1–3. Unlike BRCA1 mutations, which are almost exclusively associated with female breast and ovarian cancer, BRCA2 families also have an increased risk for male breast cancer and pancreatic and prostate cancers23, 24.
Functionally, both BRCA1 and BRCA2 have been reported to play key roles in DNA damage repair, but they appear to have distinct but complementary functions4, 5. _BRCA2_’s primary function appears to be regulation of RAD51 protein, which is required for double-strand break repair by homologous recombination25. It has been established by several research groups that _BRCA2_-mutated cells are recombination deficient and undergo a significantly reduced homology-directed repair of DNA double-strand breaks26–28. This explains our observation of a “mutator” phenotype among _BRCA2_-mutated cases, and improved chemotherapeutic responses. In contrast, BRCA1 plays a more versatile role in tumor suppression through its ability to participate in DNA damage response29–32, checkpoint control33, mitotic spindle assembly34, sister-chromatid decatenation35, and centrosome duplication36, 37. The failure of one of these functions could predispose _BRCA1_-mutated cells to tumorigenesis but not necessarily render the developed cancer cell sensitive to DNA-crosslink agents such as cisplatin, as we observed in the present study.
Our observations provide evidence that BRCA1 and BRCA2 mutations are differentially associated with patient survival compared with wt BRCA and that this difference may be a result of distinct response to platinum-based treatment and different associations with genome instability. However, there are potential limitations in our study. Although the patient cohort (316 cases) represents the most comprehensive data composition (both genomic and clinical) ever assembled, it is still relatively small, and our findings should be further validated. In addition, the associations between BRCA2 mutation and chemotherapy sensitivity and higher exome mutation rate do not necessarily imply that BRCA2 mutations affect chemotherapy sensitivity and genome instability. To fully understand and exploit these results, functional studies are required.
Nevertheless, the discovery that BRCA1 and 2 deficiencies are associated with differential effects on patient survival and chemotherapy response in OvCa may have an immediate effect on clinical prediction and trial design and sheds new light on the function of these two genes.
Supplementary Material
Supplementary material
Acknowledgments
Funding/Support: This study was supported by a grant from the National Institutes of Health (U24CA143835) to Drs. Shmulevich and Zhang, a grant from the Blanton-Davis Ovarian Cancer Research Program to Dr. Zhang, and an Ovarian Cancer SPORE grant (P50 CA083639) to Dr. Sood. Dr Yang is an Odyssey Fellow at MD Anderson Cancer Center. Dr. Khan was partially supported by an ASLA-Fulbright Research Grant for a Junior Scholar.
Role of the Sponsor: The sponsor had no role in the design or conduct of the study; the collection, analysis, and interpretation of data; or the preparation, review, or approval of the manuscript.
Additional Contributions: We thank Ms. Ann Sutton, a scientific editor in the Department of Scientific Publications of MD Anderson Cancer Center, for editing this manuscript. She received no specific compensation for editing this paper.
Footnotes
The manuscript has not been presented elsewhere. No potential conflicts of interest exist.
Author Contributions: Drs. Zhang and Yang had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Drs. Yang, Khan, and Zhang. Acquisition of data: Drs. Yang and Khan. Analysis and interpretation of data: Drs. Yang, Khan, Sun, Hess, Shmulevich, Sood, and Zhang. Drafting of the manuscript: Drs. Yang and Zhang; Critical revision of the manuscript for important intellectual content: Drs. Yang, Khan, Sood, and Zhang; Statistical analysis: Drs. Yang, Khan, and Hess; Obtained funding: Drs. Shmulevich and Zhang; Administrative, technical, or material support: Drs. Shmulevich and Zhang; Study supervision: Drs. Zhang, Sood, and Shmulevich.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Drs. Hess, Shmulevich, Sood, and Zhang are funded by the NIH and various foundations and institutional funds.
Publisher's Disclaimer: Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
Contributor Information
Da Yang, Department of Pathology, The University of Texas MD Anderson Cancer Center, Houston, Texas.
Sofia Khan, Department of Pathology, The University of Texas MD Anderson Cancer Center, Houston, Texas.
Yan Sun, Department of Pathology, The University of Texas MD Anderson Cancer Center, Houston, Texas; Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China (Dr. Sun); and Institute for Systems Biology, Seattle, Washington.
Kenneth Hess, Department of Biostatistics, The University of Texas MD Anderson Cancer Center.
Ilya Shmulevich, Institute for Systems Biology, Seattle, Washington.
Anil K. Sood, Department of Gynecologic Oncology and Reproductive Medicine, Department of Cancer Biology, and Center for RNAi and Non-Coding RNA, The University of Texas MD Anderson Cancer Center, Houston, Texas.
Wei Zhang, Department of Pathology, The University of Texas MD Anderson Cancer Center, Houston, Texas.
References
- 1.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003 Oct 24;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 2.Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003 May;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brose MS, Rebbeck TR, Calzone KA, Stopfer JE, Nathanson KL, Weber BL. Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J Natl Cancer Inst. 2002 Sep 18;94(18):1365–1372. doi: 10.1093/jnci/94.18.1365. [DOI] [PubMed] [Google Scholar]
- 4.Tutt A, Ashworth A. The relationship between the roles of BRCA genes in DNA repair and cancer predisposition. Trends Mol Med. 2002 Dec;8(12):571–576. doi: 10.1016/s1471-4914(02)02434-6. [DOI] [PubMed] [Google Scholar]
- 5.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002 Jan 25;108(2):171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 6.Aida H, Takakuwa K, Nagata H, et al. Clinical features of ovarian cancer in Japanese women with germ-line mutations of BRCA1. Clin Cancer Res. 1998 Jan;4(1):235–240. [PubMed] [Google Scholar]
- 7.Ben David Y, Chetrit A, Hirsh-Yechezkel G, et al. Effect of BRCA mutations on the length of survival in epithelial ovarian tumors. J Clin Oncol. 2002 Jan 15;20(2):463–466. doi: 10.1200/JCO.2002.20.2.463. [DOI] [PubMed] [Google Scholar]
- 8.Rubin SC, Benjamin I, Behbakht K, et al. Clinical and pathological features of ovarian cancer in women with germ-line mutations of BRCA1. N Engl J Med. 1996 Nov 7;335(19):1413–1416. doi: 10.1056/NEJM199611073351901. [DOI] [PubMed] [Google Scholar]
- 9.Boyd J, Sonoda Y, Federici MG, et al. Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA. 2000 May 3;283(17):2260–2265. doi: 10.1001/jama.283.17.2260. [DOI] [PubMed] [Google Scholar]
- 10.Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer. 2003 May 1;97(9):2187–2195. doi: 10.1002/cncr.11310. [DOI] [PubMed] [Google Scholar]
- 11.Majdak EJ, Debniak J, Milczek T, et al. Prognostic impact of BRCA1 pathogenic and BRCA1/BRCA2 unclassified variant mutations in patients with ovarian carcinoma. Cancer. 2005 Sep 1;104(5):1004–1012. doi: 10.1002/cncr.21276. [DOI] [PubMed] [Google Scholar]
- 12.Chetrit A, Hirsh-Yechezkel G, Ben-David Y, Lubin F, Friedman E, Sadetzki S. Effect of BRCA1/2 mutations on long-term survival of patients with invasive ovarian cancer: the national Israeli study of ovarian cancer. J Clin Oncol. 2008 Jan 1;26(1):20–25. doi: 10.1200/JCO.2007.11.6905. [DOI] [PubMed] [Google Scholar]
- 13.Johannsson OT, Ranstam J, Borg A, Olsson H. Survival of BRCA1 breast and ovarian cancer patients: a population-based study from southern Sweden. J Clin Oncol. 1998 Feb;16(2):397–404. doi: 10.1200/JCO.1998.16.2.397. [DOI] [PubMed] [Google Scholar]
- 14.Pharoah PD, Easton DF, Stockton DL, Gayther S, Ponder BA. Survival in familial, BRCA1-associated, and BRCA2-associated epithelial ovarian cancer. United Kingdom Coordinating Committee for Cancer Research (UKCCCR) Familial Ovarian Cancer Study Group. Cancer Res. 1999 Feb 15;59(4):868–871. [PubMed] [Google Scholar]
- 15.Tan DS, Rothermundt C, Thomas K, et al. "BRCAness" syndrome in ovarian cancer: a case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J Clin Oncol. 2008 Dec 1;26(34):5530–5536. doi: 10.1200/JCO.2008.16.1703. [DOI] [PubMed] [Google Scholar]
- 16.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011 Jun 30;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi DS, Eisenhauer EL, Lang J, et al. What is the optimal goal of primary cytoreductive surgery for bulky stage IIIC epithelial ovarian carcinoma (EOC)? Gynecol Oncol. 2006 Nov;103(2):559–564. doi: 10.1016/j.ygyno.2006.03.051. [DOI] [PubMed] [Google Scholar]
- 18.TCGA. Integrated Genomic Analyses of Ovarian Carcinoma. Nature. doi: 10.1038/nature10166. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003 Jul;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Royal Statistical Society Series B. 1995;57(1):289. [Google Scholar]
- 21.Berchuck A, Heron KA, Carney ME, et al. Frequency of germline and somatic BRCA1 mutations in ovarian cancer. Clin Cancer Res. 1998 Oct;4(10):2433–2437. [PubMed] [Google Scholar]
- 22.Foster KA, Harrington P, Kerr J, et al. Somatic and germline mutations of the BRCA2 gene in sporadic ovarian cancer. Cancer Res. 1996 Aug 15;56(16):3622–3625. [PubMed] [Google Scholar]
- 23.Cancer risks in BRCA2 mutation carriers. The Breast Cancer Linkage Consortium. J Natl Cancer Inst. 1999 Aug 4;91(15):1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 24.Thompson D, Easton DF. Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002 Sep 18;94(18):1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 25.Davies AA, Masson JY, McIlwraith MJ, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001 Feb;7(2):273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 26.Patel KJ, Yu VP, Lee H, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998 Feb;1(3):347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 27.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001 Feb;7(2):263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 28.Xia F, Taghian DG, DeFrank JS, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007 May 25;316(5828):1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 30.Kim H, Huang J, Chen J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol. 2007 Aug;14(8):710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol. 2007 Aug;14(8):716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 32.Wang B, Matsuoka S, Ballif BA, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007 May 25;316(5828):1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat Genet. 2002 Mar;30(3):285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- 34.Joukov V, Groen AC, Prokhorova T, et al. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006 Nov 3;127(3):539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 35.Lou Z, Minter-Dykhouse K, Chen J. BRCA1 participates in DNA decatenation. Nat Struct Mol Biol. 2005 Jul;12(7):589–593. doi: 10.1038/nsmb953. [DOI] [PubMed] [Google Scholar]
- 36.Sankaran S, Crone DE, Palazzo RE, Parvin JD. BRCA1 regulates gamma-tubulin binding to centrosomes. Cancer Biol Ther. 2007 Dec;6(12):1853–1857. doi: 10.4161/cbt.6.12.5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starita LM, Machida Y, Sankaran S, et al. BRCA1-dependent ubiquitination of gamma tubulin regulates centrosome number. Mol Cell Biol. 2004 Oct;24(19):8457–8466. doi: 10.1128/MCB.24.19.8457-8466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material