Spinal cord glioneuronal tumor with neuropil-like islands with 1p/19q deletion in an adult with low-grade cerebral oligodendroglioma (original) (raw)
. Author manuscript; available in PMC: 2020 Aug 25.
Published in final edited form as: J Neurooncol. 2011 Nov 15;107(2):421–426. doi: 10.1007/s11060-011-0760-9
Abstract
Glioneuronal tumor with neuropil-like islands (GTNI) is considered a rare variant of astrocytoma, characterized by discrete aggregates of cells expressing neuronal markers that punctuate a GFAP-positive glial background. Of the 24 published GTNI cases, only two occurred in adult spinal cords; none occurred concurrent with another CNS tumor; and none of those tested exhibited the 1p/19q deletion typical of oligodendroglioma. A 48-year-old man without significant past medical history was diagnosed with a WHO grade II oligodendroglioma by stereotactic biopsy of a lesion discovered after the patient suffered a generalized tonic-clonic seizure. By FISH analysis, this tumor exhibited the 1p/19q deletion present in up to 80% of oligodendrogliomas. The patient received 14 monthly cycles of temozolomide, and his cerebral tumor had a minor response. When the patient subsequently reported progressive paresthesias of his lower extremities, an MRI revealed an enhancing, cystic tumor of the thoracic spinal cord that was diagnosed as GTNI by histological analysis. By FISH analysis, this lesion exhibited the same 1p/19q deletion present in the concurrent cerebral oligodendroglioma. This case of a spinal cord GTNI with 1p/19q deletions constitutes the third report of a spinal cord GTNI in an adult patient; the first report of a GTNI in an individual with a separate CNS neoplasm; and the first report of a GTNI with 1p/19q deletions. This case establishes a potential genetic kinship between GTNI and oligodendroglioma that warrants further investigation.
Keywords: Glioneuronal tumor with neuropil-like islands (GTNI), 1p/19q deletion, Oligodendroglioma, Astrocytoma
Introduction
Glioneuronal tumor with neuropil-like islands (GTNI) is considered a rare variant of infiltrating astrocytoma, usually WHO grade II or III [1]. This tumor was recently added to the 2007 WHO Classification of Tumors of the Central Nervous System as a subtype of astrocytoma. Histologically, these lesions demonstrate distinctive oval islands of neuropil-like tissue that stains for synaptophysin. Oligodendrocyte-like cells, which may express the neuronal markers NeuN and Hu, exist within or around these islands; mature-appearing neurons may also be present [2]. These islands typically punctuate a background of GFAP-positive fibrillary, gemistocytic, or protoplasmic elements characteristic of astrocytoma [1]. Since this tumor’s initial description [3], approximately 25 cases of GTNI have been published (Table 1). With the exception of two spinal cord lesions, one in a 44-year-old woman [4] and one in a 48-year-old man [5], all tumors in adult patients have been intracranial [2, 3, 6–12]. In contrast, all reported pediatric GTNIs have originated in the spinal cord [13–15]. Heretofore, no published cases of GTNI have exhibited whole-arm 1p/19q deletions, as occur in many oligodendrogliomas [8], though two pediatric cases have demonstrated isolated 1p deletions [13, 15]. In this context, we report the case of a spinal cord GTNI with 1p/19q deletions in an adult patient with a prior diagnosis of low-grade, 1p/19q-deleted cerebral oligodendroglioma.
Table 1.
Summary of all published GTNI cases
| Case | Age (years)/gender | GTNI location | 1p/19q Status | Reference |
|---|---|---|---|---|
| 1 | 40/F | CC, L/R frontal lobes | Unknown | Teo et al. [3] |
| 2 | 31/M | R temporal lobe | Unknown | Teo et al. [3] |
| 3 | 37/M | R temporal lobe | Unknown | Teo et al. [3] |
| 4 | 25/F | L frontal lobe | Unknown | Teo et al. [3] |
| 5 | 44/F | Cervical/thoracic SC | Unknown | Harris et al. [4] |
| 6 | 23/F | L frontal/temporal lobes | Unknown | Prayson et al. [2] |
| 7 | 43/M | L parietal lobe | Intact | Keyvani et al. [6] |
| 8 | 42/M | R frontal lobe | Unknown | Bisson et al. [7] |
| 9 | 16/F | Cervical/thoracic SC | 1p whole-arm deletion | Rickert et al. [13] |
| 10 | 38/M | Frontal lobe | Intact | Barbashina et al. [8] |
| 11 | 56/M | Frontal lobe | Intact | Barbashina et al. [8] |
| 12 | 40/M | Frontal lobe | Intact | Barbashina et al. [8] |
| 13 | 40/M | Parietal lobe | Intact | Barbashina et al. [8] |
| 14 | 57/M | Frontal lobe | Intact | Barbashina et al. [8] |
| 15 | 47/M | Parietal lobe | Intact | Barbashina et al. [8] |
| 16 | 65/M | Frontal lobe | 1p/19q small interstitial deletions | Barbashina et al. [8] |
| 17 | 42/F | Temporal lobe | Intact | Barbashina et al. [8] |
| 18 | 59/F | L frontal lobe | Unknown | Vajtai et al. [9] |
| 19 | 39/F | Temporal lobe | Unknown | Wang et al. [11] |
| 20 | 1/M | Cervical SC | Unknown | Poliani et al. [14] |
| 21 | 60/M | L frontal lobe | Intact | Agarwal et al. [10] |
| 22 | 5/M | Thoracic SC | 1p whole-arm deletion | Scholz et al. [15] |
| 23 | 55/M | R temporal lobe & MB | Intact | Min et al. [12] |
| 24 | 54/F | Thoracic SC | Unknown | Ruppert et al. [5] |
| 25 | 48/M | Thoracic SC | 1p/19q whole-arm deletions | Present case |
Methods
The clinical, radiographic, and histological aspects of the patient’s presentation, evaluation, and treatment are presented. The cerebral tumor was stereotactically biopsied, while the spinal cord tumor tissue was obtained via thoracic laminectomy and open biopsy. Representative specimens from both tumors were formalin-fixed and frozen for sectioning (5 μm for all studies). Tissue from both tumors underwent hematoxylin and eosin (H&E) staining; and immunohistochemistry against MIB1 (Dako, Carpinteria, CA; 1:200). Additionally, tissue from the spinal cord tumor underwent immunohistochemistry against p53 (Dako; 1:1,000), GFAP (Dako; 1:200), synaptophysin (Zymed, S. San Francisco, CA; 1:100), and NeuN (Chemicon, Billerica, MA; 1:200). Fluorescent in situ hybridization (FISH) with probes targeting 1p36/1q25 and 19p13/19q13 (Abbott Laboratories) were performed in both tumors. For each tumor sample and chromosome, 200 nonoverlapping nuclei for red ‘R’ (marker) and green ‘G’ (reference) signals were evaluated. The frequencies of signal patterns for 1p (R) and 1q (G) on one slide, and for 19p (G) and 19q (R) on another slide, were noted. A signal pattern with red signals less than green signals was considered a deletion. The cutoffs for the signal pattern 1R2G was 12% for chromosome 1 and 14% for chromosome 19 as previously described [16]. In a later time, we performed immunohistochemistry analysis of IDH1 (clone H09, dilution 1:250 high pH, DIANOVA, Germany) on the brain tumor only because there was no more spinal cord tumor tissue available.
Results
Cerebral 1p/19q-deleted low-grade oligodendroglioma
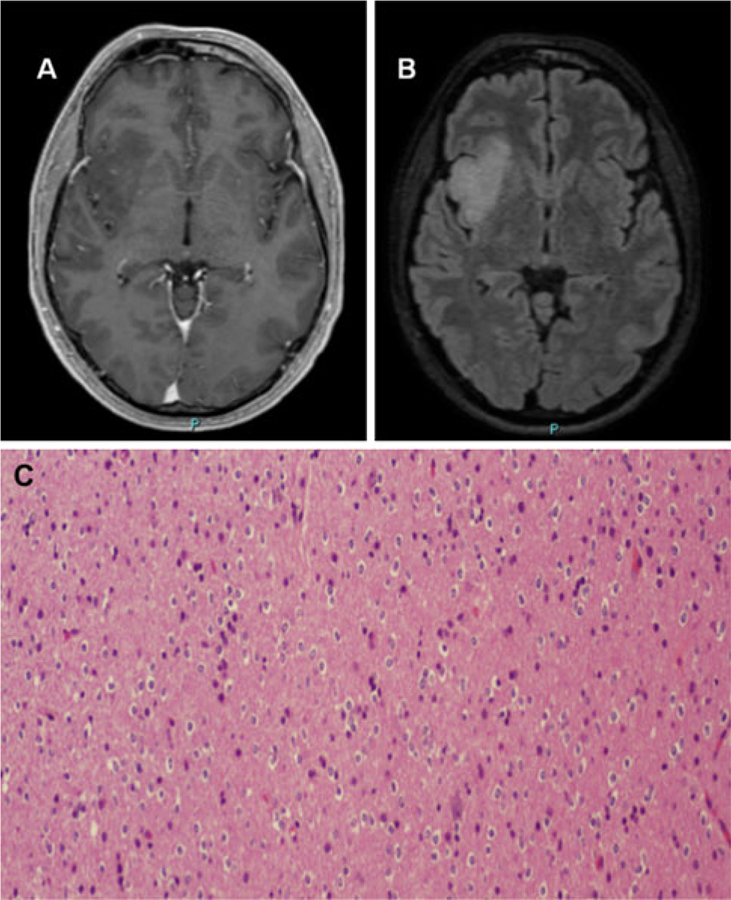
A 48-year-old man without significant past medical history experienced a generalized tonic-clonic seizure 2 weeks prior to our evaluation. His neurological examination was normal. His brain MRI showed a right insular nonenhancing mass, which appeared hypointense on T1-weighted images (Fig. 1a) and hyperintense on fluid attenuation inversion recovery (FLAIR) images (Fig. 1b), consistent with a low-grade primary brain tumor. Ten days later, he underwent stereotactic biopsy of the right insular lesion, which revealed a WHO grade II oligodendroglioma, characterized by cells with round nuclei and perinuclear halos of classic ‘fried-egg’ appearance and a discrete amount of branching capillaries (Fig. 1c). There were no mitoses, and the MIB-1 index was less than 2%. Fluorescence in situ hybridization (FISH) probes targeting 1p/1q and 19p/19q revealed chromosomal deletion of 1p and 19q (Fig. 2), a cytogenetic alteration present in 50–80% of grade II oligodendrogliomas that predicts a better prognosis [17–20]. His intracranial oligodendroglioma was also positive for IDH1 mutation. Based on the intracranial oligodendroglioma diagnosis with 1p/19q deletions, the patient received 14 monthly cycles of temozolomide; due to its insular location, the tumor had been deemed inoperable. He tolerated this therapy without difficulty, and his seizures remained well-controlled on levetiracetam. During this period, brain MRI scans showed a minor decrease in the size of the nonenhancing tumor.
Fig. 1.
Radiographic and histological characterization of the original cerebral lesion. T1-weighted post-gadolinium MRI a revealed a hypointense, nonenhancing mass of the right insula that appeared hyperintense on FLAIR images b, consistent with a low-grade glioma. Stereotactic biopsy of this lesion c revealed cells with round nuclei and perinuclear halos (‘fried-egg’ appearance), with a discrete amount of branching capillaries, consistent with a WHO grade II oligodendroglioma
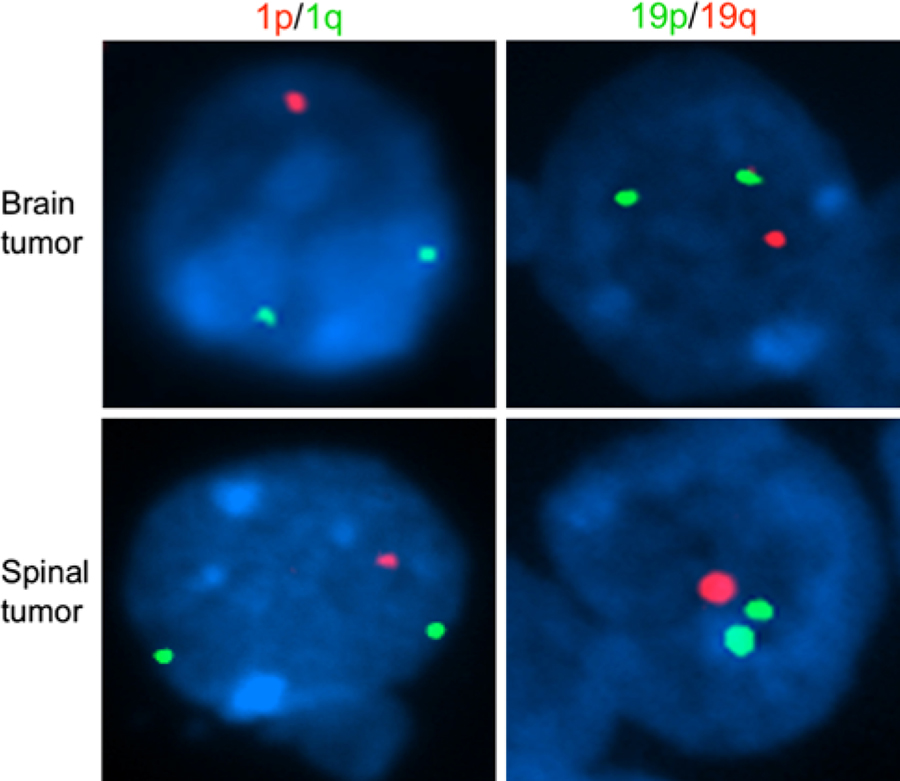
Fig. 2.
Fluorescent in situ hybridization (FISH) showing presence of 1p/19q deletion in both cerebral and spinal cord lesions. Upper panel FISH probes targeting chromosome arms 1p (red) and 1q (green) show loss of one copy of the 1p arm among nuclei of tumor cells from the cerebral lesion. Similarly, probes targeting chromosome arms 19p (green) and 19q (red) show concurrent loss of one copy of the 19q arm among tumor cell nuclei from the same cerebral lesion. Lower panel FISH probes targeting chromosome arms 1p (red) and 1q (green) show loss of one copy of the 1p arm among nuclei of tumor cells from the spinal cord lesion. Similarly, probes targeting chromosome arms 19p (green) and 19q (red) show concurrent loss of one copy of the 19q arm among tumor cell nuclei from the same spinal cord lesion
Spinal cord 1p/19q-deleted GTNI
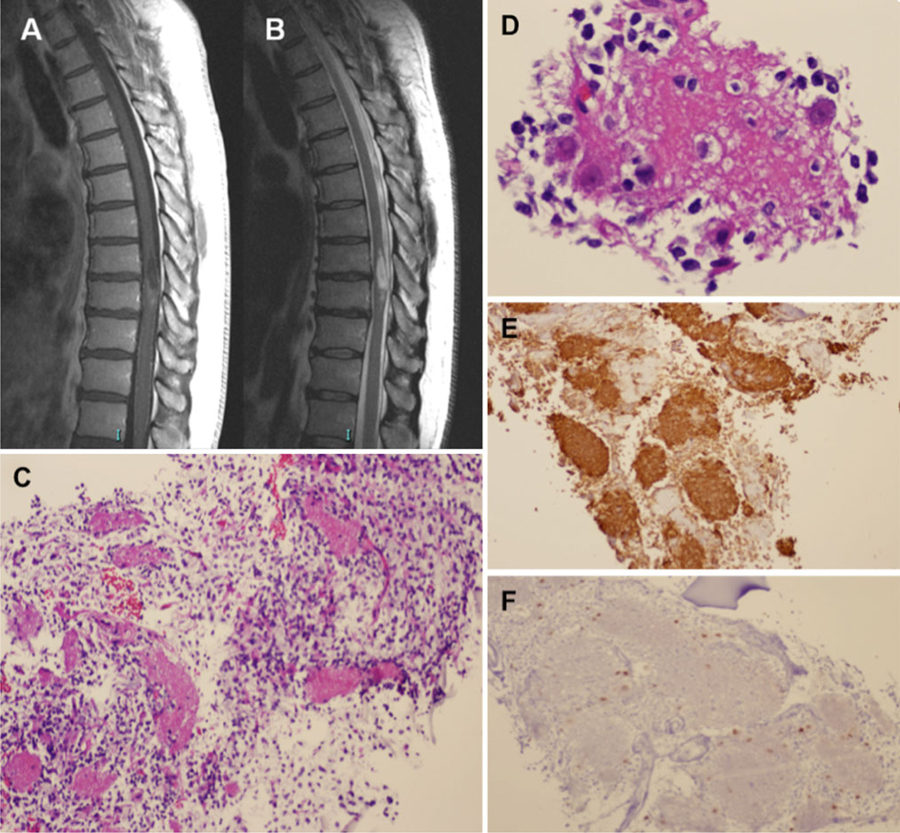
During these 14 cycles of temozolomide, his neurological status remained stable, with the exception of the emergence of progressive paresthesias along the entire plantar surface of the right foot and the left big toe. This symptom, which the patient was able to trace to before his cerebral oligodendroglioma diagnosis, prompted an MRI of his spine. This scan revealed an intramedullary lesion at the T8-T9 level, which appeared to have a cystic component with mild enhancement on T1-weighted post-gadolinium images (Fig. 3a); the lesion was hyperintense on T2-weighted images (Fig. 3b). The temozolomide regimen was ceased. One month later, he underwent a thoracic laminectomy with biopsy of the spinal cord lesion, which revealed a biphasic neoplasm consisting of a loose weave of fibrillary glial cells in association with neuropil-like islands (Fig. 3c). Within and around these islands, oligodendrocyte-like cells and mature-appearing neurons were present (Fig. 3d). The neuropil-like islands stained for synaptophysin (Fig. 3e), a neuronal marker. The glial component stained for GFAP. The p53 stain was negative, and the MIB-1 index was very low. Immunohistochemistry for NeuN labeled cells around the edges of the neuropil-like islands (Fig. 3f), as well as a few large mature-appearing neurons. FISH analysis, again targeting 1p/1q and 19p/19q, revealed chromosomal deletion of 1p and 19q (Fig. 2). The patient has recently completed radiation therapy (RT) to the spinal cord lesion, with concurrent temozolomide. He continues to be neurologically stable at 10 months post-RT, with only minor sensory deficits below T8. Moreover, his cerebral oligodendroglioma remains unchanged since the diagnosis of the spinal cord lesion. He is presently receiving no antitumoral agents and is being followed with serial imaging of the brain and spine.
Fig. 3.
Radiographic and histological characterization of spinal cord lesion. T1-weighted post-gadolinium MRI a revealed an intramedullary lesion at the T8-T9 level that demonstrated mild enhancement and a cystic component. The lesion was hyperintense on T2-weighted images b. Biopsy revealed a biphasic lesion consisting of a loose weave of fibrillary glial cells in association with neuropil-like islands c, with oligodendrocyte-like cells and mature-appearing neurons present around these islands d. The neuropil-like islands stained strongly for the neuronal marker synaptophysin (e), while immunohistochemistry for NeuN f labeled cells around the edges of these islands
Discussion
This case of a spinal cord GTNI with 1p/19q deletions is unique in several respects. It is just the third reported case of a spinal cord GTNI in an adult patient, with the only other cases occurring in a 44-year-old woman who underwent subtotal resection but later experienced recurrence with leptomeningeal dissemination [4] and in a 54-year-old woman who exhibited leptomeningeal dissemination at the time of diagnosis [5]. No chromosomal analysis for 1p/19q deletions was reported in these studies. Additionally, our case constitutes the first report of a GTNI in an individual with a separate CNS neoplasm. Finally, this case also represents the first report of a GTNI with 1p/19q deletions. While a previous study of eight adult GTNIs revealed a single case of microdeletions at 1p36 and 1q13 [8], none of these tumors exhibited large-scale 1p/19q deletions tantamount to the deletions observed in our case. Because our patient’s cerebral tumor also had 1p/19q deletions, this observation further supports a connection between these two lesions. Of note, two pediatric GTNI cases [13, 15], both occurring in the spinal cord, exhibited chromosomal deletion of 1p but not 19q, though the significance of this deletion in these cases is unclear.
Since the initial description of GTNI by Teo et al. [3], there has been considerable debate regarding its histogenesis, especially its relationship to oligodendrogliomas [8]. While the glial background of GTNIs classically exhibits features characteristic of astrocytoma, one of the four tumors from this initial study contained a minor oligodendroglioma-like component negative for the astrocytic marker GFAP and the neuronal markers synaptophysin, NeuN, and Hu [3]. Additionally, Perry et al. [21] reported three cases of oligodendrogliomas with neurocytic differentiation and 1p/19q chromosomal deletions by FISH analysis, further highlighting a potential relationship between oligodendrogliomas and GTNIs. Oligodendrocytes and particular neuronal subpopulations have also been shown to arise from a common developmental precursor by means of OLIG expression [22]. However, as noted by Barbashina et al. [8], this discussion is complicated by the observation that conventional oligodendrogliomas can express neuronal markers, such as synaptophysin [23].
In their analysis of 1p/19q deletion status in eight cases of GTNI, Barbashina et al. [8] conclude that the absence of whole-arm 1p/19q deletions in this group of tumors distances GTNI from conventional oligodendrogliomas and variants with neurocytic differentiation [21]. While this conclusion is reasonable based on their findings, our case of a GTNI in a patient with an oligodendroglioma, with both lesions harboring 1p/19q co-deletions, clearly calls for a reexamination. Our case suggests that GTNIs, despite their typically astrocytic background histology, may exhibit a genetic relationship to oligodendroglioma in a minority of cases, as implied by this patient’s concomitant cerebral oligodendroglioma and the presence of the 1p/19q deletion in both tumors. Because of insufficient spinal cord tumor tissue, we could not perform more detailed analysis of these 1p/19q deletions, other markers such as IDH1, or molecular profiling. It is possible that these two lesions could have arisen completely independently, presumably as the result of some predisposing genetic lesion affecting a common oligodendroglial/neuronal precursor. A less likely hypothesis is that one lesion represents CSF dissemination of the other lesion (with either tumor potentially serving as the primary). Notably, the GTNI was an intramedullary, non-superficial lesion that produced lower limb paresthesias, making the notion that it arose from CSF seeding by the cerebral tumor highly improbable. Overall, regardless of the actual pathological mechanism, our case reestablishes a potential genetic kinship between GTNI and oligodendroglioma that warrants further investigation.
Acknowledgements
NIH Intramural Research Program (1ZIDBC011098-02). T.J.F. is a Fellow in the Clinical Research Training Program, a public–private partnership supported jointly by the NIH and Pfizer Inc (via a grant to the Foundation for NIH from Pfizer Inc). F.M.I. is supported by the National Cancer Institute’s Clinical Investigator Development Program and the NIH Intramural Program (1ZIABC011347-01 and 1ZIABC011348-01).
Contributor Information
Tyler J. Fraum, Neuro-Oncology Branch, National Institutes of Health, 9030 Old Georgetown Rd., Room 221, Bethesda, MD 20892-8202, USA, Duke University School of Medicine, Durham, NC, USA
Stephanie Barak, Laboratory of Pathology, National Cancer Institute, NIH, 10 Center Drive – Building 10, Room 2A10, Bethesda, MD 20892, USA.
Svetlana Pack, Laboratory of Pathology, National Cancer Institute, NIH, 10 Center Drive – Building 10, Room 2N115, MSC1500, Bethesda, MD 20892, USA.
Russell R. Lonser, Surgical Neurology Branch, National Institute of Neurological Disorders and Stroke, NIH, 10 Center Drive – Building 10, Magnuson, CC, Room 3D20, Bethesda, MD 20892, USA
Howard A. Fine, Neuro-Oncology Branch, National Institutes of Health, 9030 Old Georgetown Rd., Room 235, Bethesda, MD 20892-8202, USA
Martha Quezado, Laboratory of Pathology, National Cancer Institute, NIH, 10 Center Drive – Building 10, Room 2A10, Bethesda, MD 20892, USA.
Fabio M. Iwamoto, Neuro-Oncology Branch, National Institutes of Health, 9030 Old Georgetown Rd., Room 221, Bethesda, MD 20892-8202, USA
References
- 1.Allende DS, Prayson RA (2009) The expanding family of glioneuronal tumors. Adv Anat Pathol 16:33–39. doi: 10.1097/PAP.0b013e3181915e3b [DOI] [PubMed] [Google Scholar]
- 2.Prayson RA, Abramovich CM (2000) Glioneuronal tumor with neuropil-like islands. Hum Pathol 31:1435–1438 [PubMed] [Google Scholar]
- 3.Teo JG, Gultekin SH, Bilsky M, Gutin P, Rosenblum MK (1999) A distinctive glioneuronal tumor of the adult cerebrum with neuropil-like (including “rosetted”) islands: report of 4 cases. Am J Surg Pathol 23:502–510 [DOI] [PubMed] [Google Scholar]
- 4.Harris BT, Horoupian DS (2000) Spinal cord glioneuronal tumor with “rosetted” neuropil islands and meningeal dissemination: a case report. Acta Neuropathol 100:575–579 [DOI] [PubMed] [Google Scholar]
- 5.Ruppert B, Welsh CT, Hannah J, Giglio P, Rumboldt Z, Johnson I, Fortney J, Jenrette JM, Patel S, Scheithauer BW (2010) Glioneuronal tumor with neuropil-like islands of the spinal cord with diffuse leptomeningeal neuraxis dissemination. J Neurooncol 104((2):529–533. doi: 10.1007/s11060-010-0505-1 [DOI] [PubMed] [Google Scholar]
- 6.Keyvani K, Rickert CH, von Wild K, Paulus W (2001) Rosetted glioneuronal tumor: a case with proliferating neuronal nodules. Acta Neuropathol 101:525–528 [DOI] [PubMed] [Google Scholar]
- 7.Bisson EF, Pendlebury WW, Horgan MA (2005) Glioneuronal tumor with unique imaging and histologic features. J Neurooncol 72:89–90. doi: 10.1007/s11060-004-2277-y [DOI] [PubMed] [Google Scholar]
- 8.Barbashina V, Salazar P, Ladanyi M, Rosenblum MK, Edgar MA (2007) Glioneuronal tumor with neuropil-like islands (GTNI): a report of 8 cases with chromosome 1p/19q deletion analysis. Am J Surg Pathol 31:1196–1202. doi: 10.1097/PAS.0b013e3180335f65 [DOI] [PubMed] [Google Scholar]
- 9.Vajtai I, Reinert MM (2007) Malignant glioneuronal tumor of the adult cerebrum with neuropil-like islands involving “proliferating nodules”: confirmatory report of an unusual variant. Acta Neuropathol 113:711–713. doi: 10.1007/s00401-007-0219-4 [DOI] [PubMed] [Google Scholar]
- 10.Agarwal S, Suri V, Rishi A, Shukla B, Garg A, Sharma MC, Sinha S, Sarkar C (2009) Glioneuronal tumor with neuropil-like islands: a new entity. Neuropathology 29:96–100. doi: 10.1111/j.1440-1789.2008.00933.x [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Fan QH, Yu MN, Zhou ZS, Song GX, Zhang WM (2007) Glioneuronal tumor with neuropil-like islands and rosettes: report of a case. Zhonghua Bing Li Xue Za Zhi 36:788–789 [PubMed] [Google Scholar]
- 12.Min HS, Lee SH, Yoo H, Myung J, Hong EK, Park SH (2010) Cytogenetic study of glioneuronal tumor with neuropil-like islands: a case report. Neuropathology 30:420–426. doi: 10.1111/j.1440-1789.2009.01066.x [DOI] [PubMed] [Google Scholar]
- 13.Rickert CH, Jasper M, Sepehrnia A, Jeibmann A (2006) Rosetted glioneuronal tumour of the spine: clinical, histological and cytogenetic data. Acta Neuropathol 112:231–233. doi: 10.1007/s00401-006-0091-7 [DOI] [PubMed] [Google Scholar]
- 14.Poliani PL, Sperli D, Valentini S, Armentano A, Bercich L, Bonetti MF, Corriero G, Brisigotti M, Quattrone A, Lanza PL (2009) Spinal glioneuronal tumor with neuropil-like islands and meningeal dissemination: histopathological and radiological study of a pediatric case. Neuropathology 29:574–578. doi: 10.1111/j.1440-1789.2008.00988.x [DOI] [PubMed] [Google Scholar]
- 15.Scholz M, Hoischen A, Radlwimmer B, Weber RG, Harders A, Reifenberger G, Riemenschneider MJ (2009) Rosetted glioneuronal tumor of the spine with overtly anaplastic histological features. Acta Neuropathol 117:591–593. doi: 10.1007/s00401-009-0510-7 [DOI] [PubMed] [Google Scholar]
- 16.Reddy KS (2008) Assessment of 1p/19q deletions by fluorescence in situ hybridization in gliomas. Cancer Genet Cytogenet 184:77–86. doi: 10.1016/j.cancergencyto.2008.03.009 [DOI] [PubMed] [Google Scholar]
- 17.Iwamoto FM, Nicolardi L, Demopoulos A, Barbashina V, Salazar P, Rosenblum M, Hormigo A (2008) Clinical relevance of 1p and 19q deletion for patients with WHO grade 2 and 3 gliomas. J Neurooncol 88:293–298. doi: 10.1007/s11060-008-9563-z [DOI] [PubMed] [Google Scholar]
- 18.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, Flynn H, Passe S, Felten S, Brown PD, Shaw EG, Buckner JC (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796 [DOI] [PubMed] [Google Scholar]
- 19.Kim YH, Nobusawa S, Mittelbronn M, Paulus W, Brokinkel B, Keyvani K, Sure U, Wrede K, Nakazato Y, Tanaka Y, Vital A, Mariani L, Stawski R, Watanabe T, De Girolami U, Kleihues P, Ohgaki H (2010) Molecular classification of low-grade diffuse gliomas. Am J Pathol 177:2708–2714. doi: 10.2353/ajpath.2010.100680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki H, Zlatescu MC, Betensky RA, Johnk LB, Cutone AN, Cairncross JG, Louis DN (2002) Histopathological-molecular genetic correlations in referral pathologist-diagnosed low-grade “oligodendroglioma”. J Neuropathol Exp Neurol 61:58–63 [DOI] [PubMed] [Google Scholar]
- 21.Perry A, Scheithauer BW, Macaulay RJ, Raffel C, Roth KA, Kros JM (2002) Oligodendrogliomas with neurocytic differentiation. A report of 4 cases with diagnostic and histogenetic implications. J Neuropathol Exp Neurol 61:947–955 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Q, Anderson DJ (2002) The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109:61–73 [DOI] [PubMed] [Google Scholar]
- 23.Wharton SB, Chan KK, Hamilton FA, Anderson JR (1998) Expression of neuronal markers in oligodendrogliomas: an immunohistochemical study. Neuropathol Appl Neurobiol 24: 302–308 [DOI] [PubMed] [Google Scholar]