NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression (original) (raw)
Abstract
Pluripotent cells possess the ability to differentiate into any cell type. Commitment to differentiate into specific lineages requires strict control of gene expression to coordinate the downregulation of lineage inappropriate genes while enabling the expression of lineage-specific genes. The nucleosome remodelling and deacetylation complex (NuRD) is required for lineage commitment of pluripotent cells; however, the mechanism through which it exerts this effect has not been defined. Here, we show that histone deacetylation by NuRD specifies recruitment for Polycomb Repressive Complex 2 (PRC2) in embryonic stem (ES) cells. NuRD-mediated deacetylation of histone H3K27 enables PRC2 recruitment and subsequent H3K27 trimethylation at NuRD target promoters. We propose a gene-specific mechanism for modulating expression of transcriptionally poised genes whereby NuRD controls the balance between acetylation and methylation of histones, thereby precisely directing the expression of genes critical for embryonic development.
Keywords: chromatin, embryonic stem cell, histone, NuRD, polycomb-repressive complex 2
Introduction
Precise control of gene expression is essential both for embryonic stem (ES) cell differentiation and for early embryonic development. Lineage-specific genes must be expressed in a precise temporal and spatial fashion, but it is equally important that expression of other genes is repressed. In short, it is the balance between transcription of lineage appropriate genes and repression of inappropriate genes that allows progression through development.
Regulation of gene expression is intimately linked to chromatin state, which in turn is heavily influenced by the presence of post-translational modifications of the histone proteins contained within the nucleosome. These modifications include acetylation, methylation, phosphorylation, ubiquitylation and sumoylation at specific residues both on the histone globular domain and on the histone N-terminal tails (Berger, 2007; Kouzarides, 2007). The relationship between specific histone modifications and transcriptional state has been well established. For example, acetylation of histone tails is generally associated with active transcription, while methylation may be linked to either activation or silencing of transcription depending on which histone residue is modified (Jenuwein and Allis, 2001; Rice and Allis, 2001; Roh et al, 2005; Kouzarides, 2007; Shahbazian and Grunstein, 2007). In some cases, such as at H3K27 and H3K9, acetylation or methylation can occur at the same histone residue and it is the balance between these opposing modifications that determines the transcriptional status of that region (Rice and Allis, 2001; Tie et al, 2009; Jung et al, 2010; Pasini et al, 2010b).
The nucleosome remodelling and deacetylation (NuRD) complex is a transcriptional co-repressor essential for developmental transitions in early embryogenesis as well as for ES cell function (reviewed in McDonel et al, 2009). In the absence of Mbd3, which encodes a core structural component of NuRD, the complex does not form and embryonic development stalls at the implantation stage, with the mutant embryos failing to form differentiated cell types (Zhang et al, 1999; Hendrich et al, 2001; Kaji et al, 2006, 2007). ES cells lacking NuRD are viable but are unable to exit self-renewal and commit to differentiation upon withdrawal of LIF (Kaji et al, 2006). NuRD components have been reported to interact with Oct4, a protein essential for the maintenance of pluripotency in ES cells (Liang et al, 2008; Pardo et al, 2010; van den Berg et al, 2010), despite the fact that several NuRD components are dispensable for pluripotency (McDonel et al, 2009). NuRD function is also important for homeostasis of both haematopoietic and epithelial stem cells (Williams et al, 2004; Kashiwagi et al, 2007). Aberrant gene expression patterns have been demonstrated in embryonic and somatic cell types in the absence of a functional NuRD complex (Kaji et al, 2006, 2007; Yoshida et al, 2008). Taken together, these findings suggest that NuRD-mediated gene regulation is required for stem cell fate decisions both in culture and during embryonic development.
The NuRD complex has been extensively characterised biochemically and contains multiple protein subunits, including the class I histone deacetylases HdacI and II, and the ATP-dependent chromatin remodelling component Mi2β (reviewed in McDonel et al, 2009). Nevertheless, the precise mechanism through which NuRD controls gene expression is unclear. While a reduction of histone H3K27 trimethylation was found in plants lacking the NuRD component PICKLE (Zhang et al, 2008a) and knockdown of Mi2 in mammalian cells was shown to be associated with a decrease of H3K27 methylation at one target locus (Morey et al, 2008), no global changes in histone modifications have been detected upon loss of NuRD in mammalian cells (Kaji et al, 2006). As yet, no direct link between the action of this histone deacetylase containing complex and the methylation state of H3K27 has been demonstrated.
Like NuRD, Polycomb group (PcG) complex proteins repress transcription in ES cells and during multiple developmental programs. Biochemically, Polycomb-Repressive Complex 2 (PRC2) acts to di- and tri-methylate H3K27 via the methyltransferase activity of Ezh2 (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Muller et al, 2002). This may then lead to the recruitment of PRC1 which ubiquitinates histone H2A, thereby silencing transcription of target genes (Muller and Verrijzer, 2009; Christophersen and Helin, 2010). In the absence of PcG proteins, both early embryonic development and ES cell differentiation are disrupted, although ES cells remain pluripotent (Faust et al, 1995; O’Carroll et al, 2001; Voncken et al, 2003; Pasini et al, 2004; Isono et al, 2005; Boyer et al, 2006). Polycomb targets have been extensively identified and include genes with ‘bivalent’ modifications, that is, a combination of H3K4 trimethylation, associated with active transcription, and H3K27 trimethylation, a repressive transcriptional mark. It has been proposed that bivalent genes in ES cells are poised for transcription and may be activated or silenced by the removal of the repressive or active marks, respectively (Azuara et al, 2006; Bernstein et al, 2006; Mikkelsen et al, 2007).
While there is some evidence for an in-vivo interaction between PcG proteins and the NuRD complex in various organisms (Kehle et al, 1998; Unhavaithaya et al, 2002; Morey et al, 2008; Aichinger et al, 2009), the precise nature of this interaction has not been characterised. Nevertheless, the importance of a balance between the acetylation and methylation state of H3K27 has been shown in both mammalian cells and flies (Tie et al, 2009; Jung et al, 2010; Pasini et al, 2010b) and could provide the link between these two complexes in stem cell function.
By comparing levels of specific chromatin modifications in ES cells with or without functional NuRD complex, we demonstrate the role played by NuRD in regulating the balance between acetylation and methylation state of H3K27. We propose a two-step model for repression of gene expression through the combined action of the deacetylase activity of NuRD and the methlytransferase activity of PRC2, which provides a molecular mechanism underlying NuRD-mediated lineage commitment of ES cells. Moreover, we illustrate a new level of complexity in transcriptional regulation through PcG proteins by showing that PRC2 can be directed to act at specific genes by NuRD.
Results
Gene expression changes in absence of Mbd3/NuRD in ES cells
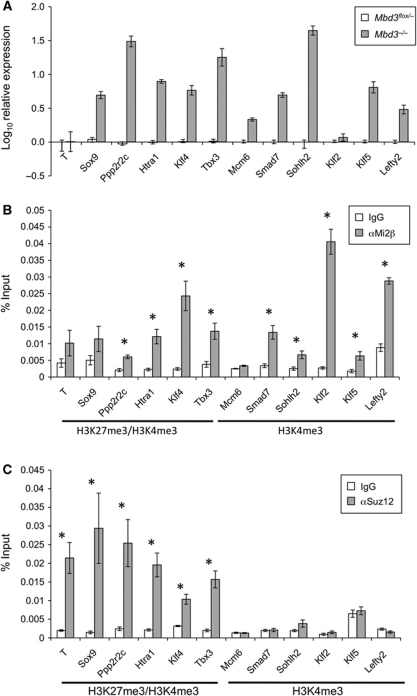
To obtain a global view of NuRD-mediated transcriptional regulation, gene expression profiles of wild-type and _Mbd3_-null ES cells were compared by microarray analysis. A surprisingly large number of genes was found to be significantly downregulated (839 genes, P<0.01) while 531 genes showed significant derepression (P<0.01) in Mbd3 −/− compared with wild-type ES cells (Supplementary Table 1). While many of these changes in gene expression will be caused indirectly by a lack of NuRD, these numbers indicate that NuRD is likely to activate as well as silence gene transcription in ES cells. To investigate the means by which NuRD acts to repress transcription, we decided to focus on the upregulated genes identified in this study. Expression changes were verified for a subset of upregulated and control genes by quantitative RT–PCR in both Mbd3 −/− and Mbd3 flox/− ES cells (Figure 1A).
Figure 1.
Identification of direct gene targets for NuRD and PRC2. (A) Quantitative RT–PCR comparing transcript levels in Mbd3 −/− ES cells to those in wild-type ES cells. Results are plotted as log10 fold change relative to wild-type levels. Error bars indicate standard error of the mean (s.e.m.). (B) Chromatin IP in wild-type cells for either Mi2β or IgG control, with qPCR for proximal promoter regions of genes shown. Results are plotted as percentage of input DNA. Histone signatures (according to Mikkelsen et al, 2007) are indicated underneath. Asterisks denote loci at which ChIP for Mi2β is significant with respect to IgG control (P<0.005). (C) Chromatin IP in wild-type cells for either Suz12 or IgG control, plotted as percentage of input DNA. Histone signatures are indicated underneath. Asterisks denote loci where ChIP for Suz12 is significant with respect to IgG control (P<0.005). Error bars indicate s.e.m.
Although NuRD has long been known to be a transcriptional silencer, only 0.2% of the genes upregulated in Mbd3 −/− ES cells show hallmarks of transcriptionally inactive chromatin, such as H3K27 trimethylation, in wild-type ES cells (according to Mikkelsen et al, 2007). In contrast, 17% of upregulated genes in wild-type cells are associated with ‘bivalent’ chromatin, while 64% are associated with H3K4me3 but not with H3K27me3. The proportions of genes with either H3K4me3 alone, or with both H3K4me3 and H3K27me3 that are upregulated in the absence of NuRD are similar to those seen on a genome-wide scale in ES cells (Mikkelsen et al, 2007). This indicates a lack of specificity towards these particular histone modifications at loci targeted by NuRD. While the bivalent mark has been associated with poised genes, H3K4me3 marks active genes (Kouzarides, 2007; Stock et al, 2007). Therefore, rather than functioning as a transcriptional silencer, our analysis indicates that NuRD acts to modulate the output of both ‘poised’ and actively transcribed genes in ES cells.
NuRD binds directly to loci with H3K4me3 and H3K4me3/H3K27me3
Chromatin immunoprecipitation (ChIP) was carried out using an antibody specific to Mi2β, a defining component of the NuRD complex, to determine which of the misregulated genes were direct targets of NuRD (Figure 1B). ChIP profiles for Mi2β at physiologically relevant target genes using this antibody are highly similar as those for a tagged protein (unpublished observations). In addition, ChIP using antibodies to other components of the NuRD complex, Mta2 and Hdac1, as well as an Avi-tagged Mbd3 showed comparable binding properties to Mi2β (Supplementary Figure S1). We have seen that Mi2β is generally associated with chromatin, but that regions of specific enrichment can be identified (NR and BH, unpublished observations). Therefore, we focussed on regions of the genome close to the transcription start sites of differentially expressed genes. Genes at which Mi2β was found to be enriched include those with both H3K27me3/H3K4me3 (e.g., Ppp2r2c, Htra1, Klf4 and Tbx3) and H3K4me3 only-associated promoters (e.g., Smad7, Sohlh2, Klf2, Klf5 and Lefty2) (Figure 1B).
To obtain a more global view of NuRD-associated regions in ES cells, we performed ChIP followed by high-throughput sequencing (ChIP-seq) for Mi2β in wild-type ES cells. The results obtained are in good agreement with our ChIP-qPCR data; however, the affinity of antibodies to native Mi2β is relatively low and the high-throughput data are inexhaustive (Supplementary Table 2).
PRC2, like NuRD, is a transcriptional repressor complex required for gene silencing in ES cells. PRC2 acts by directing methyltransferase activity to di- and tri-methylate H3K27. ChIP assays performed with an antibody to Suz12, a core component of the PRC2 complex, confirmed that binding of PRC2 is restricted to regions that are enriched in H3K27me3 (Figure 1C). NuRD components were found at a subset of bivalent promoters as well as those with only H3K4 trimethylation, revealing that while some overlap is observed in target loci between NuRD and PRC2, the two complexes target distinct sets of genes (Figure 1B and C).
H3K27 is the focus of histone modification by NuRD
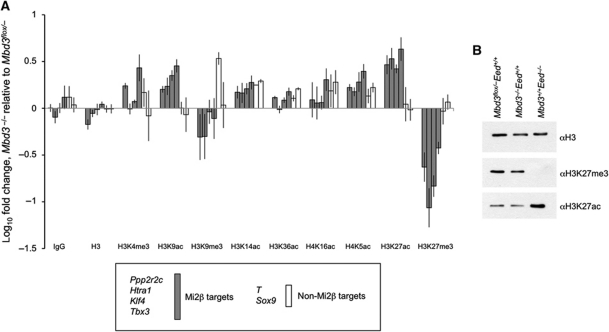
NuRD is a chromatin remodelling complex exhibiting deacetylase activity. We therefore, expected that loss of regulation of gene expression in the absence of NuRD would be reflected in changes to chromatin state, particularly in histone acetylation levels. To test this hypothesis, we used ChIP to compare levels of various histone modifications between Mbd3 flox/− and Mbd3 −/− cells for four NuRD target genes and two non-targets, all associated with bivalent chromatin domains. Promoter-proximal regions were assayed by qPCR from chromatin purified with antibodies specific to either acetylation or methylation of histones H3 and H4, and fold change determined relative to wild type (Figure 2A). In general, there was a strong association between an increase in levels of marks of active transcription (e.g., histone acetylation), a corresponding decrease in repressive marks (H3K27me3 and H3K9me3) and increased transcription levels in _Mbd3_-null cells (Figures 1A and 2A). In contrast, no clear pattern in changes to H3K4me3 levels was apparent between Mbd3 flox/− and Mbd3 −/− ES cells. Importantly, comparison of ChIP levels for the same regions using an antibody specific to histone H3 showed no overall change, indicating that the differences detected for specific histone modifications were not due to variation in nucleosome occupancy.
Figure 2.
Comparison of multiple histone modifications between Mbd3 −/− and wild-type ES cells. (A) ChIP for histone modifications. Enrichment at promoter regions of genes for histone modifications, bulk histone H3 levels or IgG control plotted as null value relative to wild type. Error bars indicate s.e.m. (B) Whole histone extracts from wild-type, Mbd3 −/− and Eed mutant ES cells blotted with antibodies for histone H3 (loading control), H3K27me3 and H3K27ac to show relative levels.
While a correlation between an increase in transcription and an increase in chromatin marks of active transcription was not unexpected, the most profound changes seen in Mbd3 −/− ES cells are the increase in H3K27 acetylation and loss of H3K27 trimethylation (Figure 2A). These changes are localised specifically to NuRD target genes, but not to the non-targets assayed (Figure 2A). A similar effect was seen for H3K9 acetylation and trimethylation, although the magnitude of changes observed was less striking. These reciprocal changes in methylation and acetylation at H3K27, and to a lesser extent at H3K9, in the absence of Mbd3 led us to hypothesise that NuRD acts through deacetylation of these residues to control chromatin state and thereby the transcriptional status of specific genes.
PRC2 is required to maintain global H3K27 methylation levels in ES cells (Montgomery et al, 2005). Similarly, we find that NuRD is required to maintain H3K27Me3 levels at targeted bivalent genes (Figure 2A). To determine whether NuRD is also required for global H3K27 trimethylation levels, we next compared H3K27 acetylation and trimethylation levels in bulk histones prepared from Mbd3 −/− and Eed −/− ES cells. In the absence of Eed, we observed a genome-wide loss of H3K27 trimethylation and an increase in acetylation levels (Figure 2B), consistent with published reports (Tie et al, 2009; Pasini et al, 2010b). In contrast, no changes in global levels of either modification were detectable by western blot in bulk histones extracted from _Mbd3_-null lines, consistent with our hypothesis that NuRD controls H3K27 modifications only at specific target genes.
Reciprocal changes in H3K27 acetylation and methylation are common to bivalent NuRD target loci
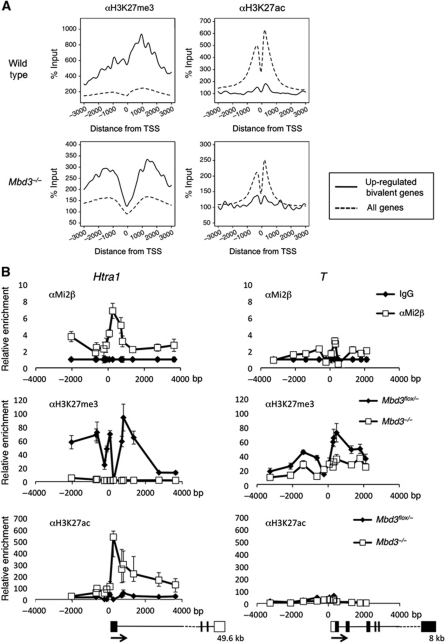
To address the extent to which this switch between acetylation and methylation status at H3K27 correlates with the action of NuRD in ES cells, we performed ChIP-seq for H3K27ac and H3K27me3 in parental and Mbd3 −/− ES cells. Full details of genomic regions associated with each modification in wild-type and null cells are listed in Supplementary Table 3.
Comparing average ChIP-seq profiles for wild-type samples, bivalent genes displaying altered expression in the absence of Mbd3 also showed a propensity for increased trimethylation and decreased acetylation at H3K27 relative to the average levels for all genes in the sample (Figure 3A; Supplementary Figure S2; Supplementary Table 3). In the absence of NuRD, differentially expressed bivalent genes showed a loss of H3K27 trimethylation, particularly around the transcription start site, combined with a reciprocal increase in the relative abundance of H3K27 acetylation (Figure 3A). While these changes in H3K27 acetylation and trimethylation are apparent across the entire gene body and at the transcription termination site (Supplementary Figure S2), they are most pronounced close to the transcription start site (Figure 3A). The same effect was seen when the comparison was repeated using Mi2β targets as determined by ChIP-seq (Supplementary Table 2; Supplementary Figure S3). Here, an overall decrease in trimethylation coincident with an increase in acetylation at H3K27 was evident. These results are in agreement with the hypothesis that these two histone marks act in opposition and that their relative levels are controlled by NuRD activity.
Figure 3.
ChIP-seq analysis, reciprocal changes of H3K27me3 and H3K27ac levels in the absence of NuRD. (A) ChIP-seq for H3K27me3 or H3K27ac in wild-type or Mbd3 −/− cells: average signal profiles, normalised to input, are shown for upregulated, bivalent genes (solid line) relative to all RefSeq (Pruitt et al, 2007) genes in each data set (dotted line) for a 6-Kb region spanning the transcription start site (TSS). Distance from TSS is indicated in base pairs. (B) ChIP for H3K27me3 and H3K27ac in wild-type and _Mbd3_-null cells and for Mi2β in wild-type cells at one target gene (Htra1) and one non-target gene (T). Data are shown relative to IgG control, distance from TSS is measured in base pairs as the mid point of PCR product. Outlines of the gene structures are indicated, with the total length of the transcriptome indicated below (not to scale). Error bars indicate s.e.m.
We sought to confirm the patterns revealed by the ChIP-seq data at specific, bivalent loci for a NuRD target (Htra1) and a non-target (T, also known as Brachyury). T expression was not altered in _Mbd3_-null cells, whereas Htra1 increases by ∼10-fold in null cells (Figure 1A; Supplementary Table 1). Detailed binding profiles for αH3K27ac, αH3K27me3 and αMi2β were compared across a ∼6 Kb region spanning the transcription start sites for these genes in both Mbd3 flox/− and Mbd3 −/− ES cells (Figure 3B). Mi2β ChIP supports the hypothesis that NuRD binds across the Htra1 promoter region and within the body of the gene, but is specifically enriched close to the transcription start site. Mi2β, however, is present at very low levels across the equivalent region of the T promoter (Figure 3B). H3K27 trimethylation is detected across both the Htra1 and T loci in wild-type cells, while very little H3K27 acetylation is present at either locus. In contrast, ES cells lacking Mbd3 display an absence of H3K27 trimethylation and a significant increase in H3K27 acetylation at Htra1. These changes are not evident at the T locus, suggesting that the effect is specific to NuRD target genes. At Htra1, a strong peak of Mi2β binding is observed close to the transcription start site that coincides with the peak of H3K27 acetylation present in Mbd3 −/− cells. Thus, we conclude that NuRD is targeted to a subset of bivalent genes where it is required to prevent H3K27 acetylation and, indirectly, to maintain H3K27 methylation.
Gene-specific recruitment of PRC2 is dependent on NuRD
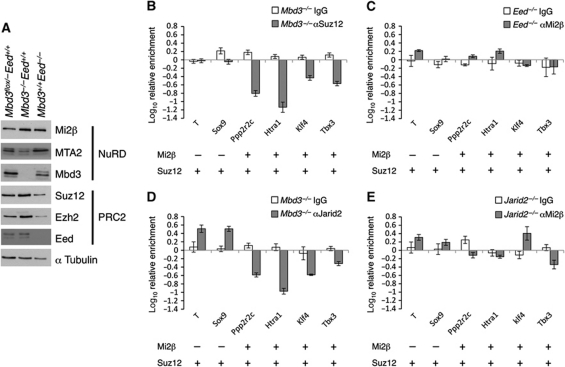
Thus far, our data indicate that NuRD and PRC2 work in concert at bivalent NuRD target promoters to maintain H3K27 in a deacetylated, trimethylated state. We next addressed the nature of the interplay between NuRD and PRC2 at their target genes. We were unable to detect a direct protein–protein interaction between the two complexes in ES cells by immunoprecipitation (Supplementary Figure S4), suggesting that neither complex is physically recruited by the other to specific chromatin sites. Further, levels of PRC2 component proteins were essentially unchanged in Mbd3 −/− ES cells, and NuRD subunits were present at approximately wild-type levels in Eed −/− ES cells (Figure 4A). These results do not support an interdependence at the level of component protein abundance or stability between the two complexes (Figure 4A).
Figure 4.
Interdependency between NuRD and PRC2 chromatin interactions. (A) Western blot showing relative levels of NuRD and PRC2 components in wild-type, Mbd3 −/− and Eed mutant cells. Alpha tubulin is used as loading control. (B) ChIP for Suz12 in _Mbd3_-null cells relative to wild type. (C) ChIP for Mi2β in Eed mutant cells relative to wild type. Association of Mi2β and Suz12 in wild-type cells at each gene is indicated below each panel. (D) ChIP for Jarid2 in _Mbd3_-null cells relative to wild type. (E) ChIP for Mi2β in Jarid2 −/− cells relative to wild type. ChIP data shown is for bivalent genes only. Error bars indicate s.e.m.
We next asked whether NuRD activity was required for PRC2 to be physically recruited to its target loci. ChIP for Suz12 in Mbd3 flox/− and Mbd3 −/− cells revealed a loss of PRC2 at NuRD target genes in the absence of NuRD, whereas Mi2β levels were not significantly changed at its target loci in Eed mutant ES cells (Figure 4B and C). ChIP for Jarid2, which directs PRC2 to target loci (Peng et al, 2009; Shen et al, 2009; Landeira et al, 2010; Pasini et al, 2010a) also revealed a dependency on NuRD for its recruitment while no change was seen in Mi2β binding in _Jarid2_-null ES cells (Shen et al, 2009; Figure 4D and E). This observation supports a model in which recruitment of PRC2 at NuRD target loci is dependent on either the presence or activity of NuRD, but in which the loss of PRC2 has no effect on Mi2β recruitment.
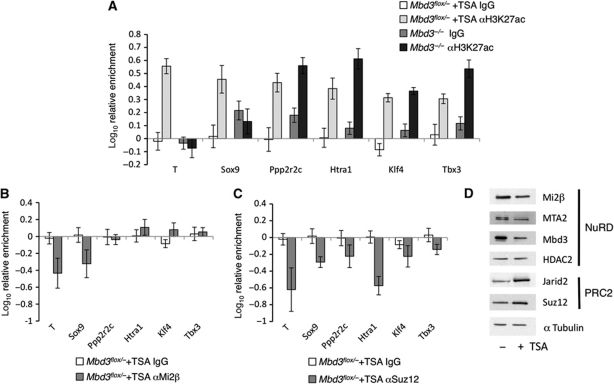
We next sought to determine whether it is the physical presence or deacetylase activity of NuRD that is responsible for this gene-specific recruitment of PRC2. To address this question, we performed ChIP experiments in the presence or absence of Trichostatin A (TSA), a potent inhibitor of histone deacetylase activity. Mbd3 flox/− ES cells were exposed to 0.1 mM TSA for 2 h. This treatment resulted in increased levels of H3K27 acetylation, which were comparable to those in Mbd3 −/− ES cells but which preceded gene expression changes (Figure 5A; Supplementary Figure S5). In the presence of TSA, acetylation levels were increased at all genes tested, in contrast to the gene-specific increase in acetylation of H3K27 observed in Mbd3 −/− ES cells. Exposure to TSA had little effect on the presence of Mi2β at previously identified target genes (Figure 5B) although Suz12 recruitment was reduced at all loci. This is unlikely to be a consequence of changes in protein abundance since levels of some NuRD component proteins decreased slightly in response to exposure to TSA, while PRC2 components showed a slight increase in abundance (Figure 5D). The observed dependency on acetylation levels for recruitment of PRC2 is consistent with previous reports that Suz12 occupancy shows an inverse correlation with the presence of H3K27ac at target genes (Pasini et al, 2010b).
Figure 5.
Effect of acetylation on recruitment of PRC2 to chromatin. (A) ChIP for H3K27ac and IgG control in wild-type cells treated with TSA and in Mbd3 −/− cells without TSA treatment. (B) Relative enrichment for Mi2β and IgG control in wild-type cells treated with TSA compared with untreated. (C) Relative enrichment for Suz12 and IgG control in wild-type cells treated with TSA compared with untreated. Error bars show s.e.m. (D) Western blot of nuclear extracts prepared from wild-type ES cells with or without TSA treatment. Alpha tubulin is used as a loading control.
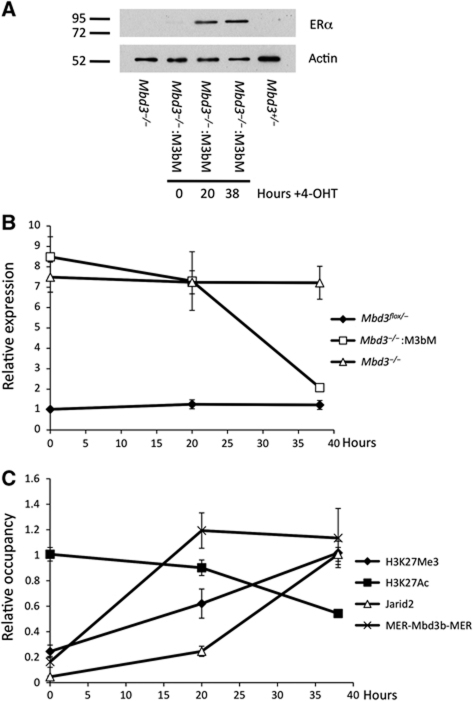
ES cells maintained for many passages in the absence of Mbd3 or PRC2 components may have undergone adaptive changes in response to prolonged culture. To determine whether PRC2 recruitment is immediately responsive to the presence of the NuRD complex, we made use of a tamoxifen-inducible Mbd3 system. The Mbd3b protein isoform was fused to the two copies of the mouse oestrogen receptor (MER-Mbd3b-MER), and expressed in _Mbd3_-null ES cells. In the absence of 4-hydroxytamoxifen, the protein is localised to the cytoplasm and the cells lack functional NuRD complex. Upon addition of tamoxifen, the protein translocates into the nucleus, restoring silencing of the NuRD/PRC2 target gene Htra1 (Figure 6A and B). Recruitment of MER-Mbd3b-MER to the Htra1 locus is observed by 20 h of tamoxifen treatment. In addition, over the same time period, H3K27 acetylation levels decrease and H3K27 trimethylation levels increase (Figure 6C). Coincident with these changes in H3K27 modification and gene expression, there is a recruitment of Jarid2 to the Htra1 locus. This experiment demonstrates that H3K27 modification status and PRC2 localisation at the Htra1 locus are highly responsive to the presence or absence of a functional NuRD complex. Together with the lack of Suz12 binding at NuRD-specific genes in Mbd3 −/− ES cells (Figure 4B), we conclude that NuRD-dependent deacetylation of H3K27 is required for PRC2 recruitment and subsequent trimethylation at NuRD target loci.
Figure 6.
Responsiveness of PRC2 recruitment to presence of NuRD. (A) Western blot of nuclear extracts prepared from wild-type, Mbd3 −/− and MER-Mbd3b-MER Mbd3 −/− cells after addition of 4-hydroxytamoxifen for indicated time. Alpha tubulin acts as a loading control. (B) Expression levels of Htra1 in wild-type, Mbd3 −/− and MER-Mbd3b-MER Mbd3 −/− cells over the indicated 4-hydroxytamoxifen time course relative to Mbd3 −/− levels. (C) Occupancy levels for H3K27ac, H3K27me3, Jarid2 and Mer-Mbd3b-MER at the Htra1 locus over the 4-hydroxytamoxifen time course in MER-Mbd3b-MER Mbd3 −/− ES cells. Levels are shown relative to highest point for each protein.
Overlap of NuRD and PRC function in ES cells
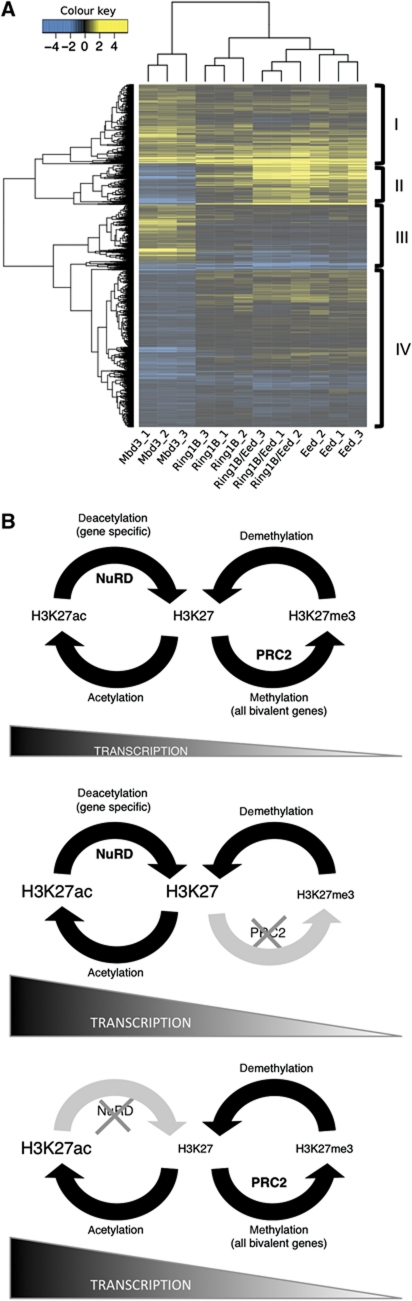
Both NuRD and PRC regulate gene expression and both are required for early embryonic development. However, the functions of these two complexes do not overlap completely as NuRD is required for lineage commitment of both early embryonic cells and ES cells, whereas ES cells or embryos lacking PRC2 components show some capacity for differentiation (Faust et al, 1995; O’Carroll et al, 2001; Pasini et al, 2004; Shen et al, 2009; Leeb et al, 2010). In order to assess more globally the degree to which NuRD- and PRC2-dependent transcriptional regulation overlaps in ES cells, we compared the gene expression changes found in Mbd3 −/− ES cells with those seen in ES cells lacking components of the PRCs (Leeb et al, 2010). Using a significance cutoff of P<0.05, we are able to identify 1879 genes differentially expressed in Mbd3 −/− ES cells also present on the microarray platform used in the Leeb et al study. Of these, ∼23% (436, Figure 7A, group I) were misregulated both in the Mbd3 and Polycomb mutant ES cells, consistent with the observed synergy in gene regulation between the NuRD and Polycomb systems at a large proportion of target loci (Figure 7A). This subset of genes includes several encoding proteins important for embryonic development and key components of signalling pathways such as β-Catenin, Sfrp1, Tbx3, Tgfβ and Wnt7b (Niwa et al, 2009; Pera and Tam, 2010; Kelly et al, 2011; Supplementary Table 4).
Figure 7.
Overlap of NuRD and PRC function in gene regulation. (A) Expression patterns of genes differentially expressed between wild-type and Mbd3 −/− ES cells (P<0.05) were assessed in data from Leeb et al (2010) describing gene expression changes in PRC mutant ES cells. A total of 2907 probe sets were found to be differentially expressed between the reference and Mbd3 mutant samples, mapping to 1967 unique genes using the re-annotated array information provided by Leeb et al. Of these, 1879 genes were also present on the Leeb microarray platform, although not necessarily differentially expressed. The log2 fold change of these genes relative to their respective wild-type samples is illustrated in the figure. Genes showing reduced expression compared with wild-type cells are indicated in blue; those showing increased expression are indicated in yellow. Mbd3_1, Mbd3_2 and Mbd3_3 indicate results from three different Mbd3 −/− ES cell samples corresponding to two independently derived ES cell lines. Ring1B_1-3, Eed_1-3 and Ring1B/Eed_1-3 represent results from three replicates each of Ring1B-null ES cells, Eed-null ES cells and Ring1B/Eed-double null ES cells, respectively (Leeb et al, 2010). Where multiple probes map to the same gene, only the probe displaying the largest deviation from the reference sample is included here. The four main gene clusters are indicated on the right hand side (I–IV). (B) Two-step model for control of transcription illustrating relationship between acetylation status, here controlled by NuRD, and methylation of H3K27 by PRC2. Top panel: in wild-type cells, PRC2 and NuRD function fully. NuRD directs deacetylation of specific genes, which then become available for trimethylation by PRC2. Middle panel: in cells lacking functional PRC2, for example, Eed−/− cells, H3K27 trimethylation is lost genome-wide, the balance moves towards the acetylation/deacetylation cycle and transcription of bivalent genes increases overall. Bottom panel: in the absence of NuRD, deacetylation fails to occur at specific loci only. At these genes, there is an increase in acetylation, a subsequent reduction in trimethylation at H3K27 through loss of substrate for PRC2 and an increase in transcription.
To identify more directly the extent to which PRC2 localisation is dependent on the action of NuRD, we performed ChIP-seq for Suz12 in Mbd3 −/− ES cells. Of the 1928 high confidence Suz12 binding peaks we identified in Mbd3 Flox/− ES cells, 330 of these were not found in Mbd3 −/− ES cells (17.1%; Supplementary Table 5). Consistent with being PRC2 targets, these genes were generally enriched for trimethylation and depleted for acetylation at H3K27 (Supplementary Figure S6). In contrast, this same set of genes show a relative decrease in H3K27Me3 levels and a corresponding increase in H3K27Ac levels in Mbd3-null ES cells. Notably, the loss of methylation is most pronounced close to the transcription start sites (Supplementary Figure S6). These data strongly support a model whereby NuRD and PRC2 control expression through a balance between acetylation and methylation status of H3K27.
Of the genes displaying NuRD-dependent peaks of Suz12 binding, 133 were also identified as Mi2β targets by ChIP-seq (Table I). This group is highly enriched for genes, which have functions critical for transcriptional regulation (Table II). Many of these transcription factors have known functions in stem cell maintenance or pluripotency (e.g., Tbx3, Klf4 and Foxd3; Hanna et al, 2002; Niwa et al, 2009) or early embryogenesis and differentiation processes (e.g., Gata5, Sfrp1 and Smarcd3; Lessard et al, 2007; Lou et al, 2011). Thus, while the number and identity of genes for which NuRD function appears to be required for PRC2 binding represents a relatively small proportion of either total NuRD or PRC2 targets, many of the genes targeted for joint regulation by both PRC2 and NuRD are known to be critical for development and highlights the importance of such a mechanism in vivo. Based upon these findings, we conclude that the mechanism described here underlies the regulation of a significant subset of PRC and NuRD target genes in ES cells, and that this interaction may be responsible for the shared biological functions of the two silencing complexes.
Table 1. Mi2β target genes showing Mbd3-dependent Suz12 binding.
| 1700029F12Rik | E030030I06Rik | Klf4 | Pth1r |
|---|---|---|---|
| 1700067P10Rik | Edn2 | Lhx5 | Pthlh |
| 2810030E01Rik | Efna2 | Lingo1 | Ptprd |
| 6030419C18Rik | Egr4 | Lphn2 | Ptrf |
| 8430427H17Rik | Ezr | Ly6c2 | Rap1gap2 |
| 9030425E11Rik | Fam129c | Lyzl4 | Rem1 |
| A930004D18Rik | Fam92b | Man1c1 | Rnf220 |
| Abcc5 | Fosl2 | Mapk15 | Ror2 |
| Abhd15 | Foxd3 | Mapt | RP23-197C15.2 |
| AC131780.10 | Gata5 | Mcf2l | Rufy4 |
| AC131780.6 | Gbx1 | Mlxipl | Scarf2 |
| AC140264.1 | Gbx2 | Nav2 | Sdk2 |
| AC140331.1 | Gm5089 | Ncam1 | Sfrp1 |
| Agpat9 | Gm5607 | Ncoa2 | Slc22a21 |
| AL837506.1 | Gm5627 | Nebl | Smarcd3 |
| Ankrd56 | Gm6020 | Nfil3 | Sox21 |
| Ano4 | Gm6304 | Nkx6-2 | Tbca |
| Astn2 | Gpm6b | Nmnat2 | Tbx3 |
| Atxn1 | Gpr150 | Npas2 | Tcf4 |
| Bach2 | Gpr83 | Nr2f2 | Tcte2 |
| Bahcc1 | Gramd1b | Ntn1 | Thpo |
| Brunol4 | Grb10 | Olfm2 | Tmtc1 |
| Camta1 | Grik3 | Olig3 | Unc5b |
| Casz1 | Grik4 | Ovol1 | Unc5d |
| Cbfa2t3 | Grin2a | P4ha2 | Zbtb10 |
| Cbln4 | Hmga2 | Palld | Zbtb7c |
| Cdk6 | Hrh2 | Pcdhga11 | Zeb2 |
| Clstn1 | Hs3st3b1 | Pde8a | Zfhx3 |
| Cnih3 | Htra1 | Pdgfa | Zfp36l2 |
| Crem | Igsf21 | Pou2f1 | Zfp423 |
| Crlf1 | Irx2 | Ppp2r2c | Zfp804a |
| Csf2ra | Kazald1 | Prickle1 | |
| Cux1 | Kcnh3 | Prkar1b | |
| Dgkz | Kcnk3 | Prkcdbp |
Table 2. Gene ontology analysis of NuRD-dependent Suz12 targets.
| GOMFID | _P_-value | Counta | Sizeb | Term |
|---|---|---|---|---|
| GO:0003700 | <0.01 | 23 | 738 | Sequence-specific DNA binding transcription factor activity |
| GO:0043565 | <0.01 | 14 | 430 | Sequence-specific DNA binding |
| GO:0030528 | <0.01 | 11 | 286 | Transcription regulator activity |
| GO:0016564 | <0.01 | 9 | 206 | Transcription repressor activity |
| GO:0000975 | <0.01 | 9 | 209 | Regulatory region DNA binding |
| GO:0016563 | <0.01 | 9 | 335 | Transcription activator activity |
| GO:0010843 | <0.01 | 7 | 189 | Promoter binding |
| GO:0008270 | <0.01 | 18 | 1358 | Zinc ion binding |
| GO:0005488 | <0.01 | 18 | 2712 | Binding |
| GO:0005515 | <0.01 | 23 | 3229 | Protein binding |
| GO:0005216 | <0.01 | 7 | 367 | Ion channel activity |
| GO:0015267 | <0.01 | 7 | 387 | Channel activity |
| GO:0003677 | <0.01 | 11 | 1076 | DNA binding |
| GO:0060089 | <0.01 | 23 | 2850 | Molecular transducer activity |
| GO:0003690 | <0.01 | 4 | 140 | Double-stranded DNA binding |
| GO:0003702 | <0.01 | 4 | 148 | RNA polymerase II transcription factor activity |
| GO:0005102 | <0.01 | 10 | 877 | Receptor binding |
| GO:0003705 | 0.01 | 3 | 82 | Sequence-specific enhancer binding RNA polymerase II transcription factor activity |
| GO:0035257 | 0.01 | 3 | 82 | Nuclear hormone receptor binding |
| GO:0008022 | 0.01 | 4 | 169 | Protein C-terminus binding |
| GO:0030165 | 0.01 | 3 | 94 | PDZ domain binding |
| GO:0016566 | 0.01 | 3 | 97 | Specific transcriptional repressor activity |
| GO:0008134 | 0.01 | 5 | 299 | Transcription factor binding |
| GO:0008201 | 0.01 | 3 | 104 | Heparin binding |
| GO:0046872 | 0.01 | 23 | 3246 | Metal ion binding |
| GO:0043167 | 0.01 | 23 | 3297 | Ion binding |
| GO:0005179 | 0.01 | 3 | 118 | Hormone activity |
| GO:0003682 | 0.01 | 4 | 217 | Chromatin binding |
| aNumber of genes from the test set (_n_=133) corresponding to this GO term. Only those categories containing three or more genes, and with a _P_-value ⩽0.01 are shown. | ||||
| bNumber of total genes corresponding to this GO term. |
Discussion
NuRD-mediated gene regulation has been shown to facilitate developmental transitions in early mouse embryos, lineage commitment in ES cells and developmental decisions in haematopoietic and epithelial stem cells (Kaji et al, 2006, 2007; Kashiwagi et al, 2007; Yoshida et al, 2008). Using a combination of expression analysis and ChIP experiments, we demonstrate for the first time a mechanism by which NuRD regulates the transcription of specific genes in ES cells. We show that NuRD-dependent deacetylation of H3K27 makes NuRD target genes available for further repressive action by the PRC2. Our data support a model for a two-step process for transcriptional silencing of bivalent genes (Figure 7B) centering on a switch between the acetylation and methylation status of H3K27.
Our model is consistent with previous reports that describe both reciprocity between H3K27 acetylation and methylation and correlation of H3K27 modification status with transcription (Tie et al, 2009; Jung et al, 2010; Pasini et al, 2010b). The status of H3K27 can be reversibly modified by a combination of enzyme activities (Figure 7B; Tie et al, 2009; Pasini et al, 2010b), allowing an exquisitely responsive yet robust mechanism of gene regulation. Repressive modification (di- and trimethylation) is carried out at H3K27 by histone methyltransferase activity (i.e., by Ezh2 in the PRC2 complex) and removed by the action of histone demethylases (such as trithorax group proteins). Histone H3K27 may be acetylated to establish active transcription (by histone acetyltransferases) or deacetylated (by histone deacetylases) as a further level of regulation. The identity of some of these enzymes has been determined (Tie et al, 2009; Pasini et al, 2010b). Regulation of transcriptional status in the case of H3K27 may therefore be regarded as two interconnected cycles, with methylation/demethylation establishing the silencing of a gene, and acetylation/deacetylation controlling full activation of transcription (Figure 7B).
To date, models for transcriptional regulation revolving around H3K27 status have lacked any mechanism for locus specificity. For instance, loss of Suz12 in ES cells results in loss of H3K27 trimethylation, and an increase in H3K27 acetylation on a global scale (Pasini et al, 2010b). Similarly, knockdown experiments to deplete cells of the histone acetyltransferases Cbp (Crebbp) or p300 (Ep300) resulted in changes in H3K27 acetylation levels that were detectable in western blots of bulk histones (Pasini et al, 2010b). This view is in contrast to the activity of NuRD described here, in which transcription and H3K27 acetylation of a relatively small number of genes are affected (Figure 2). Here, we show that the balance between acetylation and methylation status of H3K27 can be controlled at a fine level by NuRD, which indirectly affects the activity of PRC2 by controlling substrate availability at NuRD target genes. This presumably takes the form of making unmodified H3K27 available for monomethylation prior to di- and trimethylation by the PRC2 complex.
Based on our model (Figure 7B), several mechanistic predictions are possible. In the event of PRC2 being removed from the system (Figure 7B, middle panel), H3K27me3 will be lost and H3K27 acetylation will increase globally, a fact that has been established in other studies (Tie et al, 2009; Jung et al, 2010; Pasini et al, 2010b) and confirmed here (Figure 2B). The resulting shift from a silent methyl mark to that of acetylation is accompanied by a general upregulation of bivalent genes in the absence of PRC2. A lack of methylation at H3K27 would not necessarily affect recruitment of NuRD to the chromatin as the acetylation/deacetylation element of the cycle remains in balance and substrate levels will be unaffected (Figure 7B), an hypothesis that we confirmed experimentally for the NuRD component Mi2β (Figure 4C, E).
As NuRD acts in a gene-specific manner, its loss affects acetylation levels of H3K27 at direct target genes only. Here, accumulation of H3K27 in an acetylated state would result in localised loss of substrate for PRC2 and subsequent reduction in occupancy only at specific gene loci (Figures 4B, D and 7B, bottom panel). We propose that deacetylation of H3K27 is required for PRC2 function, and that the action of NuRD enables PRC2 to silence specific loci. Pasini et al (2010b) proposed that the function of PcG proteins is to prevent acetylation of H3K27. According to our model, the action of enzymes which affect the methylation status of this histone residue will inevitably affect the availability of substrate for enzymes which control the acetylation status, and vice versa. Since it is the balance between the activation cycle (acetylation/deacetylation) and the silencing cycle (methylation/demethylation) that ultimately determine transcriptional status, an increase in influence of either one will necessarily have an affect on the other.
While a requirement for NuRD in ES cell differentiation has been established (Kaji et al, 2006), the identities of specific gene loci targeted by the complex have remained largely unexplored. In this study, we have shown a correlation between promoter occupancy and gene expression levels using a combination of ChIP and microarray analysis in Mbd3 −/− ES cells. Based on our expression results, there appears to be no bias towards target genes with H3K4me3, H3K27me3 or bivalent histone modifications. In fact, while NuRD is traditionally viewed as a transcriptional co-repressor, the majority of genes expressed in its absence exhibit regions of H3K4me3 at their promoter regions, indicative of actively transcribed genes. This is consistent with a recent study in which a number of different histone deacetylases were found to associate globally with actively transcribed genes in human T cells (Wang et al, 2009). Therefore, rather than acting as a conventional silencer, our data are consistent with NuRD functioning as a modulator of transcriptional activity at a large proportion of its target genes. Indeed, we have recently shown that NuRD controls the dynamic range of expression levels of some actively transcribed genes in ES cells to facilitate lineage commitment (Reynolds et al, submitted).
Interactions between the Mi2 and PcG family proteins have been reported to act either in parallel with each other, for example, at sites of DNA damage (Chou et al, 2010), antagonistically, for example, in the control of cell identity in plants (Aichinger et al, 2009), or in synergy with each other, as in the case of Drosophila dMi-2 which has been shown to specify sites of Polycomb-mediated repression in vivo (Kehle et al, 1998). How these interactions result in transcriptional changes has not been determined. Using gene expression profiling in ES cells, we show the extent of functional overlap of the NuRD- and PRC-dependent gene expression programmes. We further provide the first clear evidence for a mechanism by which these important regulators may interact to control gene expression in mammalian cells, explaining observations made previously in both plants and human leukaemic cells (Morey et al, 2008; Zhang et al, 2008a). Uniquely, we show that NuRD targets the action of the PRC2 complex to specific gene loci via H3H27 deacetylase activity. We further show that NuRD activity is required for the recruitment of PRC2 to a set of developmentally important transcriptional regulators in ES cells, providing a mechanism for targeting PRC2 activity to specific loci. In summary, here we provide evidence that NuRD and PcG act together to both dictate and reinforce transcriptional silencing in ES cells.
Materials and methods
ES cell culture
ES cells were grown in self-renewing conditions, that is, in the presence of serum and murine LIF. _Mbd3_-null and _Eed_-null ES cell lines have been described previously (Silva et al, 2003; Kaji et al, 2006). _Eed_-null ES cells were cultured using a layer of feeder cells which were depleted from ES cells upon harvest by taking advantage of their differential attachment properties and allowing the feeders to attach to non-gelatinised plates. TSA (Sigma) was added to normal ES cell media to a final concentration of 0.1 mM for 2 h prior to harvesting and further analysis. An expression construct in which the Mbd3b coding region was fused at both N- and C-termini to mouse oestrogen receptor (MER) domains (Verrou et al, 1999) was transfected into Mbd3 −/− ES cells. Clones in which uninduced cells showed neither nuclear localisation of the MER-Mbd3b-MER protein by immunofluorescence nor silencing of known NuRD target genes were selected for further analysis. Responsiveness to tamoxifen was measured both by immunofluorescence and quantitative RT–PCR for NuRD responsive genes.
RNA isolation and gene expression analysis
Total RNA was prepared from _Mbd3-_null or parental ES cell lines using Trizol (Life Technologies) according to the manufacturer's protocol and hybridised to Mouse WG-6 Expression arrays (Illumina, Inc). Fluorescence data were processed using software packages from v2.7 of the Bioconductor project (Gentleman et al, 2004). Data files were handled using beadarray v2.0.0 (Dunning et al, 2007) and hybridisation quality assessed with arrayQualityMetrics v3.2.0 (Kauffmann et al, 2009). Normalisation was performed using the ssn algorithm from lumi v2.2.0 (Du et al, 2008). Microarray probe sets with an interquartile range <0.4 were removed, and for those remaining differential expression was calculated using limma v3.6.0 (Smyth, 2005). The resulting _P_-values were corrected for multiple testing using the Benjamini and Hochberg (1995) approach, and probes with P<0.01 were deemed significant. To improve the interpretation of the data, microarray probe identifiers were re-annotated as outlined in Barbosa-Morais et al (2010). Published primary microarray data from Eed −/− ES cells (Leeb et al, 2010) were re-analysed using RMA (Irizarry et al, 2003) prior to comparison with data obtained in this study from Mbd3 −/− ES cells.
Quantitative real-time PCR
Gene expression levels were verified by quantitative PCR carried out in triplicate using cDNA from total RNA prepared from ES cell lines with Fast SYBR green Master Mix (Life Technologies) on the 7900HT Fast Real-Time PCR System (Life Technologies). Primers to either β_-actin_ or Pp1A were used as endogenous controls and relative values calculated using the ΔCt method. Primer sequences are listed in Supplementary Table 6.
Chromatin immunoprecipitation
ChIP was carried out using antibodies to endogenous proteins listed in Supplementary Table 7 and standard ChIP protocols. In brief, cells were fixed using 1% formaldehyde for 10 min at room temperature and fixation quenched with 150 mM glycine. For Mi2β ChIP-seq, formaldehyde fixation was preceded by 45 min incubation in 2 mM DSG (disuccinimidyl glutarate (Sigma); Wang et al, 2009). Chromatin was sheared by sonication to an average fragment size between 200 and 300 bp using a Bioruptor sonication instrument (Diagenode). Immunoprecipitation was performed using Protein A sepharose, pre-blocked with fish gelatin and single-stranded herring sperm DNA. Quantitative PCR of ChIP DNA was carried out as above using gene-specific primers listed in Supplementary Table 6. All qPCR was carried out in triplicate for at least three biological replicates. Analysis was carried out either as percentage of input DNA or as ΔCt values in order to compare relative levels between samples, using primers to the promoters of Actin or Cdx2 (Supplementary Table 6b) as endogenous controls. _P_-values were calculated by _T_-test analysis.
ChIP-seq analysis
For sequencing of ChIP DNA, samples from three individual ChIP experiments for each cell line were verified by qPCR before pooling for library construction. Sequencing was carried out on the Illumina GAIIx at a read length of 36 bp. These were mapped to build mm9 of the mouse reference genome using v0.12.7 of the Bowtie short-read aligner (Langmead, 2010). Redundant reads were removed, and those from the two biological replicates were merged into a single data set. MACS (Zhang et al, 2008b) and CCAT (Xu et al, 2010) peak-finding programs were used to identify histone-modified regions by comparing both Mbd3-null and parental samples to their respective input DNA controls. MACS was used to identify peaks in Mi2β and Suz12 ChIP-seq data. Equal numbers of reads from ChIP and control samples were used for each comparison, and regions that were determined to be significantly enriched by both peak callers were included in downstream analyses. Annotation of modified regions was performed with PeakAnalyzer (Salmon-Divon et al, 2010) and CEAS (Shin et al, 2009).
The Refseq gene model annotation was used to define transcription start and end sites. The CEAS tool (Shin et al, 2009) was used to extract the signal surrounding every relevant genomic region. A region comprising three kilobases upstream and downstream of each feature was divided into 120 bins (50 bp each). The median signal of each bin was calculated, and then averaged across all features. The resulting average profiles were normalised to input signal level. ChIP-seq and expression data have been deposited in the ArrayExpress database under accession numbers E-MTAB-888 and E-MTAB-889, respectively.
Western blotting
Nuclear extracts were prepared as described previously (Kaji et al, 2006), subjected to SDS–PAGE and probed with antibodies listed in Supplementary Table 4.
Acid extraction of bulk histones was achieved by washing nuclei in 0.1 M HCl according to the standard protocols.
Supplementary Material
Supplementary Information
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Review Process File
Acknowledgments
We thank Laura O’Neill for the kind gift of antibodies, Stuart Orkin for the Jarid2 −/− ES cell line, Jose Silva for the Eed null ES cell line, Elly Tanaka for an MER-MER plasmid, and Ernest Laue for comments on the manuscript. Microarray experiments were carried out at Cambridge Genomics Services, Department of Pathology, University of Cambridge. ChIP DNA libraries were prepared by Maike Paramor of the Gene Service Facility, CSCR, University of Cambridge and sequencing was carried out at EMBL Genomics Core Facility, Heidelberg, Germany. This work was supported by a Wellcome Trust Senior Research Fellowship in the Basic Biomedical Sciences to BH.
Author contributions: NR, GB and AH-A prepared cells and performed ChIP experiments; MS-D analysed the ChIP-seq data and HD analysed the microarray data in consultation with PB; DL made RNA for microarray hybridisation and performed expression analysis by qRT–PCR; AB provided reagents prior to their publication; BH, PB and NR designed the experiments and NR and BH wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aichinger E, Villar CB, Farrona S, Reyes JC, Hennig L, Kohler C (2009) CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, Darot JF, Ritchie ME, Lynch AG, Tavare S (2010) A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res 38: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57: 289–300 [Google Scholar]
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science (New York, NY) 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA 107: 18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophersen NS, Helin K (2010) Epigenetic control of embryonic stem cell fate. J Exp Med 207: 2287–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM (2008) Lumi: a pipeline for processing Illumina microarray. Bioinformatics 24: 1547–1548 [DOI] [PubMed] [Google Scholar]
- Dunning MJ, Smith ML, Ritchie ME, Tavare S (2007) Beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23: 2183–2184 [DOI] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T (1995) The eed mutation disrupts anterior mesoderm production in mice. Development (Cambridge, England) 121: 273–285 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA (2002) Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev 16: 2650–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A (2001) Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev 15: 710–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Fujimura Y, Shinga J, Yamaki M, O-Wang J, Takihara Y, Murahashi Y, Takada Y, Mizutani-Koseki Y, Koseki H (2005) Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol Cell Biol 25: 6694–6706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jung HR, Pasini D, Helin K, Jensen ON (2010) Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics 9: 838–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B (2006) The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol 8: 285–292 [DOI] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B (2007) Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development (Cambridge, England) 134: 1123–1132 [DOI] [PubMed] [Google Scholar]
- Kashiwagi M, Morgan BA, Georgopoulos K (2007) The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development (Cambridge, England) 134: 1571–1582 [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W (2009) arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25: 415–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282: 1897–1900 [DOI] [PubMed] [Google Scholar]
- Kelly KF, Ng DY, Jayakumaran G, Wood GA, Koide H, Doble BW (2011) beta-Catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell 8: 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L et al. (2010) Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 12: 618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B (2010) Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics, Chapter 11: Unit 11.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A (2010) Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR (2007) An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 10: 731–739 [DOI] [PubMed] [Google Scholar]
- Lou X, Deshwar AR, Crump JG, Scott IC (2011) Smarcd3b and Gata5 promote a cardiac progenitor fate in the zebrafish embryo. Development 138: 3113–3123 [DOI] [PubMed] [Google Scholar]
- McDonel P, Costello I, Hendrich B (2009) Keeping things quiet: Roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol 41: 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T (2005) The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 15: 942–947 [DOI] [PubMed] [Google Scholar]
- Morey L, Brenner C, Fazi F, Villa R, Gutierrez A, Buschbeck M, Nervi C, Minucci S, Fuks F, Di Croce L (2008) MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol Cell Biol 28: 5912–5923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Muller J, Verrijzer P (2009) Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 19: 150–158 [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K (2009) A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460: 118–122 [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J (2010) An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K (2010a) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, Helin K (2010b) Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 38: 4958–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF, Tam PP (2010) Extrinsic regulation of pluripotent stem cells. Nature 465: 713–720 [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol 13: 263–273 [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K (2005) Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev 19: 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Divon M, Dvinge H, Tammoja K, Bertone P (2010) PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M (2007) Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan G, Lee Y, Orkin SH (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS (2009) CEAS: cis-regulatory element annotation system. Bioinformatics 25: 2605–2606 [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N (2003) Establishment of histone H3 methylation on the inactive x chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry W, Huber W (eds) Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer. pp 397–420 [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC (2002) MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111: 991–1002 [DOI] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrou C, Zhang Y, Zürn C, Schamel WW, Reth M (1999) Comparison of the tamoxifen regulated chimeric Cre recombinases MerCreMer and CreMer. Biol Chem 380: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M (2003) Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA 100: 2468–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K (2004) The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20: 719–733 [DOI] [PubMed] [Google Scholar]
- Xu H, Handoko L, Wei X, Ye C, Sheng J, Wei CL, Lin F, Sung WK (2010) A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics 26: 1199–1204 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, Georgopoulos K (2008) The role of the chromatin remodeler Mi-2{beta} in hematopoietic stem cell self-. Genes Dev 22: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rider SD Jr, Henderson JT, Fountain M, Chuang K, Kandachar V, Simons A, Edenberg HJ, Romero-Severson J, Muir WM, Ogas J (2008a) The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J Biol Chem 283: 22637–22648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008b) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev 13: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Review Process File