Genome-Wide Profiling of Liver X Receptor, Retinoid X Receptor, and Peroxisome Proliferator-Activated Receptor α in Mouse Liver Reveals Extensive Sharing of Binding Sites (original) (raw)
Abstract
The liver X receptors (LXRs) are nuclear receptors that form permissive heterodimers with retinoid X receptor (RXR) and are important regulators of lipid metabolism in the liver. We have recently shown that RXR agonist-induced hypertriglyceridemia and hepatic steatosis in mice are dependent on LXRs and correlate with an LXR-dependent hepatic induction of lipogenic genes. To further investigate the roles of RXR and LXR in the regulation of hepatic gene expression, we have mapped the ligand-regulated genome-wide binding of these factors in mouse liver. We find that the RXR agonist bexarotene primarily increases the genomic binding of RXR, whereas the LXR agonist T0901317 greatly increases both LXR and RXR binding. Functional annotation of putative direct LXR target genes revealed a significant association with classical LXR-regulated pathways as well as peroxisome proliferator-activated receptor (PPAR) signaling pathways, and subsequent chromatin immunoprecipitation-sequencing (ChIP-seq) mapping of PPARα binding demonstrated binding of PPARα to 71 to 88% of the identified LXR-RXR binding sites. The combination of sequence analysis of shared binding regions and sequential ChIP on selected sites indicate that LXR-RXR and PPARα-RXR bind to degenerate response elements in a mutually exclusive manner. Together, our findings suggest extensive and unexpected cross talk between hepatic LXR and PPARα at the level of binding to shared genomic sites.
INTRODUCTION
The liver plays a central role in the control of whole-body lipid homeostasis, and hepatic lipid metabolism is continuously adjusted to fit the needs of the organism. This adaptation requires major adjustments in the hepatic metabolic gene program, including a strong upregulation of lipogenic gene expression in the fed state, whereas in the fasting state, the expression of genes involved in fatty acid oxidation as well as ketogenesis and hepatic glucose production is highly induced. Class II nuclear receptors (NRs), i.e., NRs forming heterodimers with retinoid X receptor (RXR), play a key role in coordinating these changes. They include the liver X receptor (LXR) (29, 57) and peroxisome proliferator-activated receptor (PPAR) (32, 34, 41) families as well as farnesoid X receptor (FXR) (44, 55, 88), pregnane X receptor (PXR) (5, 33), vitamin D receptor (VDR) (43), constitutive androstane receptor (CAR) (3, 9, 23), and retinoic acid receptors (RARs) (13).
The LXR family consists of the two subtypes, LXRα (NR1H3) and LXRβ (NR1H2), both of which form obligate heterodimers with RXR. LXR-RXR heterodimers are reported to bind to LXR response elements (LXREs) that consist of a direct repeat of the core sequence 5′-AGGTCA-3′ spaced by 4 nucleotides (DR4) (2, 72, 76, 79, 92). LXRs are activated by oxidized cholesterol derivatives and play an important role in the regulation of cholesterol homeostasis in the liver. Thus, pharmacological activation of LXR leads to the induction of several genes implicated in reverse cholesterol transport and mobilization of cholesterol, such as the ATP binding cassette (ABC) transporter genes Abca1, Abcg1, Abcg5, and Abcg8 and the apolipoprotein E gene (ApoE) (12, 31, 37, 60, 63, 84). Furthermore, a recent genome-wide study of LXR in human hepatoma cells showed that LXR also downregulates expression of the cholesterologenic genes for lanosterol 14α-demethylase (Cyp51A1) and squalene synthase (Fdst1) (89). Moreover, LXR activation induces triglyceride synthesis partly through induction of the lipogenic transcription factors sterol regulatory element-binding protein 1c (SREBP-1c) (42, 61, 95) and carbohydrate response element-binding protein (ChREBP) (8) but also by direct activation of genes encoding lipogenic enzymes such as fatty acid synthase (Fasn), stearoyl coenzyme A (CoA) desaturase (Scd1), and acetyl-CoA carboxylase 1 (Acaca) (11, 30, 78). Consistent with this, pharmacological activation of LXR in vivo results in hepatic steatosis and hypertriglyceridemia (70). Recently, activation of hepatic LXR has also been shown to modulate the hepatic acute-phase response through repression of haptoglobin gene (Hp) and serum amyloid A gene (Saa1) expression (85). Due to the beneficial activation of reverse cholesterol transport by LXR agonists (50, 63), considerable effort has been spent on developing LXR-activating drugs to treat conditions involving excess cholesterol. However, the use of LXR agonists is hampered by the concomitant induction of lipogenic genes leading to hypertriglyceridemia and liver steatosis (70).
Members of the PPAR family are other key players in the regulation of hepatic lipid metabolism. All members of the family bind as heterodimers with RXR to direct repeats spaced by 1 nucleotide (DR1) (34, 52, 81), but they display significant subtype specificity in their ability to activate target genes (7, 14, 51). PPARα, the most prominent PPAR subtype in the liver, is particularly important for hepatic induction of β-oxidation and ketogenesis in response to fasting (32, 41). However, PPARα is also activated in response to a high-fat diet (56), and some data indicate that PPARα plays a role in induction of hepatic lipogenesis and cholesterol metabolism (35, 53, 82). The two other subtypes, PPARδ and PPARγ, are expressed at lower levels in murine liver. PPARγ plays a role in the induction of the lipogenic gene program during high-fat diet exposure and in various obesity models (4, 49, 56), whereas PPARδ seems to be involved in lipoprotein metabolism (67).
Since both PPAR-RXR and LXR-RXR heterodimers are considered permissive (34, 38, 69, 73, 92), i.e., activated by agonists of either heterodimeric receptors, treatment with RXR agonists has the potential to activate lipogenesis through activation of the LXR-RXR and PPARγ-RXR heterodimers as well as lipid catabolism through the activation of PPARα/δ-RXR. Notably however, treatment with RXR agonists such as bexarotene, which is used in the clinic to treat various T-cell disorders (15–17), leads to hypertriglyceridemia in humans (48, 64, 77), and we recently demonstrated that treatment of mice with bexarotene leads to hypertriglyceridemia and hepatic steatosis in an LXR-dependent manner (38).
Recent breakthroughs in DNA sequencing technologies have made it possible to generate detailed genome-wide maps of transcription factor binding sites in a given cell type or tissue. These global maps provide valuable tools for identification of novel transcription factor binding sites and for establishing putative “transcription factor-target gene” relationships. Furthermore, comparison of different global genome-wide binding profiles can identify genomic regions occupied by several transcription factors to establish novel putative transcription factor interactions and synergies (21, 74, 87). Thus, in order to explore the architecture of the hepatic LXR and RXR gene regulatory network and to assess how this network is affected by LXR and RXR agonists, we performed LXR and RXR chromatin immunoprecipitation-sequencing (ChIP-seq) on the livers from LXR- and RXR agonist-treated mice. This revealed extensive overlapping binding patterns throughout the murine genome and significant association with known LXR-regulated pathways. We find that bexarotene treatment greatly increases the extent of RXR binding, whereas treatment with the LXR agonist T0901317 leads to markedly increased occupancy of both LXRs and RXRs. Interestingly, functional annotation of LXR-regulated genes revealed significant association with PPAR signaling pathways, and subsequent ChIP-seq mapping of PPARα binding revealed PPARα binding to 71 to 88% of the shared LXR-RXR binding sites. The combination of sequence analysis of shared binding regions and sequential ChIP on selected sites indicate that LXR and PPARα bind to degenerate response elements in a mutually exclusive manner in mouse liver. Taken together, our findings point to unexpected functional cross talk between LXR-RXR and PPARα-RXR signaling pathways at the level of binding to chromatin.
MATERIALS AND METHODS
Animal experiments.
Mouse experiments with chronic ligand treatment were performed as previously described (38). Briefly, wild-type or LXRα/β-deficient female C57BL6 mice (13 weeks of age) (for ChIP-seq, n = 1 per condition) (1) were treated by oral gavage once daily for 14 days with the RXR agonist bexarotene (100 mg/kg body weight [mpk], in 1% carboxymethyl cellulose), the LXR agonist T0901317 (T09) (30 mpk) or vehicle alone. Vehicle- and agonist-treated animals were sacrificed at the same time of the day (1 p.m.), and the livers were removed immediately and snap-frozen in liquid nitrogen. For sequential chromatin immunoprecipitation (ChIP-reChIP) experiments, wild-type male C57BL6 mice were treated with 30 mpk T0901317 24 h and 8 h before they were euthanized.
To assess LXR-PPAR cross talk, 13-week-old female C57BL6 mice were treated by oral gavage with PPARα agonist fenofibrate (FF) (200 mpk), LXR agonist T0901317 (T09) (30 mpk), both FF and T09 (200 and 30 mpk, respectively) or vehicle once daily for 5 days. On the day of sacrifice, mice were forced to fast for 4 h. Blood was collected by retro-orbital venipuncture while the mice were given isoflurane anesthesia. The mice were euthanized by cervical dislocation, and their livers were harvested, flash frozen, and stored at −80°C until required.
Plasma and hepatic lipid analyses.
Retro-orbital blood samples were drawn in EDTA-coated tubes at sacrifice. Plasma was separated by low-speed centrifugation and kept at 4°C and used within 3 days for biochemical analysis. Plasma concentrations of total cholesterol (TC) and triglycerides (TG) were determined by enzymatic assays using commercially available reagents, and TC and TG from frozen liver tissue (50 mg) were assayed as previously described (38).
ChIP, ChIP-seq, and ChIP-reChIP.
For each ChIP experiment, chromatin was prepared from snap-frozen livers, homogenized in phosphate-buffered saline (PBS), and cross-linked using 37% formaldehyde at a final concentration of 1%. After incubation (for 10 min while rotating at room temperature [RT]), 1M glycine was added to the cross-linked material to a final concentration of 0.125 M and the material was incubated for 10 min while rotating at RT, pelleted by centrifugation at 400 × g for 2 min at 4°C, washed two times in cold PBS, and resuspended in lysis buffer (1% Triton, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, 1 mM EGTA, 20 mM Tris [pH 8.0]) (200 μl/10 mg chromatin) before sonication according to the manufacturer's protocol using the Diagenode Bioruptor twin (twice for 20 cycles, 30 s on/off, maximum level). The samples were centrifuged for 2 min at 10,000 × g, and the supernatant was used for subsequent chromatin IP using antibodies against RXR and PPARα (sc553 and sc9000, respectively; Santa Cruz), RNA polymerase II (AC-0555-100; Diagenode), or LXR (28).
For each IP, 200 μl chromatin was diluted one time in IP wash buffer 1 (1% Triton, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, 1 mM EGTA, 20 mM Tris [pH 8.0], 2 μg/μl bovine serum albumin [BSA], and complete protease inhibitor) and incubated with antibody for 3 h while rotating at 4°C before addition of protein A beads and a further incubation overnight at 4°C while rotating. The beads were washed at 4°C twice with IP wash buffer 1, once with IP wash buffer 2 (1% Triton, 0.1% SDS, 500 mM NaCl, 2 mM EDTA, 1 mM EGTA, 20 mM Tris [pH 8.0]), once with IP wash buffer 3 (0.25 M LiCl, 1% NP-40, 1% deoxycholic acid, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris [pH 8.0]), and finally twice with Tris-EDTA buffer, all at 4°C. DNA-protein complexes were eluted with 400 μl of elution buffer (1% SDS and 0.1 M NaHCO3) and decross-linked by adding NaCl to a final concentration of 0.2 M and shaking overnight at 65°C. DNA was purified using phenol-chloroform and analyzed by quantitative PCR or ChIP-seq, as described previously (52). ChIP primers denoted “No gene” (forward primer, 5′-TGGTAGCCTCAGGAGCTTGC; reverse primer, 5′-ATCCAAGATGGGACCAAGCTG) aligning to a genomic region on chromosome 15 with no binding of the NRs investigated were used as a negative control.
For ChIP-reChIP, chromatin from the first IP was eluted after the final wash in 30 μl of 1% SDS and 10 mM fresh dithiothreitol (DTT) at 37°C while shaking for 20 to 30 min and then diluted 60 times in IP wash buffer 1 and subjected to a second round of immunoprecipitation. DNA was subsequently eluted and purified as described above.
Gene expression.
RNA was prepared from frozen liver samples, and relative mRNA levels were determined by quantitative reverse transcription-PCR (RT-PCR) and normalized to cyclophilin A (peptidylprolyl isomerase A [Ppia]) as described previously (39). Primer sequences used for quantitative PCR (qPCR) are available upon request.
Electrophoretic mobility shift assay.
In vitro translations were performed using a TNT kit according to the recommendations of the manufacturer (Promega). Double-stranded oligonucleotides were labeled using [γ-32P]ATP and polynucleotide kinase (Roche Molecular Biochemicals). _In vitro_-translated proteins were incubated for 20 min on ice in binding buffer [10 mM Tris (pH 8.0), 40 mM KCl, 1 mM dithioerythritol, 4% glycerol, 0.05% Nonidet P-40, and 2.4 μg of poly(dI-dC)]. Subsequently, 50 fmol (∼2 × 105 cpm) of 32P-labeled oligonucleotide was added, and the mixture was incubated for 20 min at room temperature. Free DNA and DNA-protein complexes were resolved by electrophoresis in 0.5× Tris-borate-EDTA (TBE) on 5% polyacrylamide gels.
Bioinformatic analyses.
Wiggle files for visualization of ChIP-seq data were generated by determining the number of overlapping sequence reads per base pair, averaged over a 10-bp window. Peaks were called using FindPeaks 4 (18) with the following parameters: 0.1 for subpeaks and 0.3 for trim-peaks. All peaks with less than 10 overlapping reads were discarded. De novo motifs were generated with GimmeMotifs version 0.60 (83) with analysis size xl and the tools Mdmodule, MEME, MotifSampler, trawler, Improbizer, BioProspector, and GADEM. All other parameters were set at the default.
For all motif analyses, randomly selected genomic sequences with a similar distribution relative to the transcription start sites (TSS) of genes with peaks were used as background. The de novo motif analysis was performed using the pwmscan.py utility included with GimmeMotifs. An individual motif-scanning threshold for both DR1 and DR4 was chosen to result in the same fraction of hits in the background sequences (1 every 100 sequences, or a false discovery rate [FDR] of 1%). The direct, inverted, and everted repeat motif analysis was carried out using pwmscan.py in combination with a custom Python script. All peaks were scanned for low-similarity matches to the half-site RGKTCA (converted to a position frequency matrix for use with pwmscan.py). The minimum match quality was defined as a pwmscan.py cutoff score of 0.8. Subsequently, all putative half-sites were combined and scored as direct, everted, or inverted repeats with a minimum spacer of 0 nucleotides and a maximum spacer of 5. All possible combinations per sequence with at least one half-site with a near perfect score (cutoff of 0.99) were reported.
Putative LXR-regulated genes were defined on the basis of RNA polymerase II ChIP-seq data as LXR-induced or -repressed genes. Putative genes were considered induced if the following condition was met: no. of tags for T09-treated WT mice/no. of tags for Veh-treated WT mice > mean plus 1.5SD, where no. is number, WT is wild type, Veh is vehicle, and 1.5SD is the standard deviation multiplied by 1.5. Putative genes were considered repressed if the following condition was met: no. of tags for Veh-treated WT mice/no. of tags for T09-treated WT mice > mean plus 1.5SD. T09-regulated genes were considered LXR dependent if following condition was met: [(no. of tags for Veh-treated WT mice/no. of tags for T09-treated WT mice)/(no. of tags for Veh-treated LXRdKO mice/no. of tags for T09-treated LXRdKO mice)] > mean plus 1.5SD.
RESULTS
Genome-wide mapping of hepatic LXR and RXR binding sites.
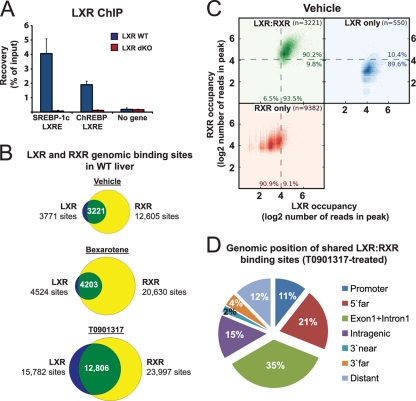
In order to generate genome-wide maps of LXR and RXR binding to chromatin in mouse liver, we performed ChIP-seq for each NR using cross-linked liver chromatin from mice treated with vehicle, the RXR agonist bexarotene (LGD1069, Targretin) (20) or the LXR agonist T0901317 (70), respectively. The RXR antibody has been tested extensively and used previously (51, 52, 74), and careful testing of the LXR antibody using chromatin samples from wild-type (WT) and LXRα/β double knockout (LXRdKO) mice confirmed the specificity of this antibody (Fig. 1A). From the ChIP-seq data, we generated genome-wide high-resolution maps of LXR and RXR binding sites using FindPeaks (FDR level of <0.001) (18). The number of mapped sequence tags obtained (see Table S1 in the supplemental material) was equalized for the different ChIP-seq reads to obtain the same number of total tags in all RXR reads and, similarly, the same number of tags in all LXR reads. We detected 12,605, 20,630, and 23,997 RXR binding sites in the livers from WT mice treated with vehicle, bexarotene, and T0901317, respectively (Fig. 1B). These binding profiles are rather distinct from RXR binding in 3T3-L1 cells (52) with only 12% of the liver RXR binding sites (vehicle treated) overlapping with RXR binding in 3T3-L1 cells on day 6 (see Fig. S1 in the supplemental material). For the LXR profiles, we identified 3,771, 4,524, and 15,782 LXR binding sites in vehicle-, bexarotene-, and T0901317-treated animals, respectively, and found that 81 to 93% of all LXR binding sites are shared with RXR (Fig. 1B), consistent with the notion that LXR binds to DNA as an obligate heterodimer with RXR. In contrast, shared LXR-RXR binding sites constitute only 26% and 20% of the RXR binding sites during vehicle or bexarotene treatment, respectively, in agreement with the ability of RXR to form heterodimers with several other NRs. However, following T0901317 treatment, the number of LXR sites increases, and LXR becomes associated with 53% of all RXR binding sites. The number of reads per peak for both receptors in the vehicle-treated condition showed that the group of shared binding sites is clearly distinct from the LXR- and RXR-specific groups (Fig. 1C), indicating that true “LXR-only” and “RXR-only” sites exist. Similar results were obtained for the bexarotene- and T0901317-treated animals (see Fig. S2 in the supplemental material).
Fig 1.
Genome-wide mapping of LXR and RXR binding sites in mouse liver. (A) LXR ChIP-qPCR on the livers from wild-type (WT) and LXRα/β-deficient (LXR double knockout [LXRdKO]) mice gavaged with the LXR agonist T0901317 (30 mg/kg body weight [mpk]) once daily for 14 days. Sterol regulatory element-binding protein 1c (SREBP-1c) (61) and carbohydrate response element-binding protein (ChREBP) (8) LXREs were used as positive controls for LXR binding, whereas “No gene” is a negative control (see Materials and Methods for details). Bars represent the means plus standard deviations (SD) (error bars) (n = 3). (B) Venn diagrams representing the number of “LXR only” (blue), “RXR only” (yellow), and shared LXR-RXR binding sites (green) in mouse liver detected by ChIP-seq. Mice were gavaged with vehicle (1% carboxymethyl cellulose), the RXR agonist bexarotene (100 mpk), or the LXR agonist T0901317 (30 mpk) once daily for 14 days. Peaks were called using FindPeaks (FDR > 0.001). (C) Scatterplots illustrating the intensity of LXR and RXR binding in vehicle-treated mouse liver (log2 number of reads per peak) at sites that are defined as LXR specific (blue), RXR specific (red), and shared (green). The percentages indicate the fractions of sites that are above or below the broken line. (D) Genomic positions of shared LXR-RXR binding sites in T0901317-treated mouse liver relative to the nearest gene (PinkThing) are shown as follows: Distant, distance to the TSS > 25 kb; 5′far, 25 to 5 kb upstream of the TSS; Promoter, <5 kb upstream of the TSS; Intragenic, intragenic peaks downstream of intron 1; 3′near, <5 kb downstream of the 3′ end; 3′far, 5 to 25 kb downstream of the 3′ end.
Using PinkThing (http://pinkthing.cmbi.ru.nl), we assigned hepatic LXR-RXR binding sites (12,806 in total) by proximity to a total of 5,524 genes (Fig. 1D). This analysis showed that LXR-RXR binding sites are distributed throughout the genome like other NRs analyzed by global ChIP experiments (10, 52, 75, 91) with 50% of all sites in introns and only 11% in the proximal 5′ region of the nearest gene. Notably, 35% of all binding sites and 70% of the intragenic binding sites are located in exon 1 and intron 1 (Fig. 1D).
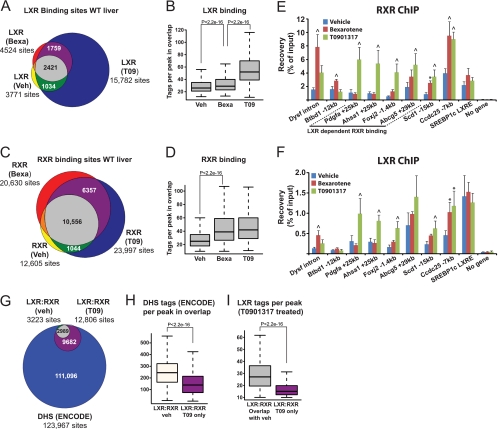
Comparison of genomic LXR binding in livers exposed to vehicle and to LXR and RXR agonists showed that the number of LXR binding sites increases dramatically by chronic treatment with LXR agonist, whereas treatment with RXR ligand results in only a modest increase in the number of LXR binding sites (Fig. 1B and 2A). Similarly, LXR peak intensities (number of sequence tags recovered in peak-detected areas) are markedly increased by treatment with T0901317 but only slightly increased by treatment with bexarotene (Fig. 2B). In contrast, both the number of RXR binding sites and the intensity of RXR binding are strongly increased by bexarotene as well as T0901317 treatment (Fig. 2C and D). The ligand-induced binding of RXR and LXR was validated by ChIP-qPCR at selected sites using livers from 4 different animals per group (Fig. 2E and F). The results demonstrate that the ChIP-qPCR faithfully mimics the ChIP-seq binding profiles at these sites (see Fig. S3 in the supplemental material; also data not shown). Thus, LXR and RXR binding can be induced specifically by bexarotene (Dysf intron), specifically by T0901317 (Pdgfa +25kb, Ahsa1 +25kb, Foxj2 −1.4kb, and Abcg5 +29kb) or by both ligands (Ccdc25 −7kb) (Fig. 2E and F). In contrast, binding to the SREBP-1c (Srebf1 product) LXRE, which is already highly occupied by LXR in vehicle-treated liver, is not significantly induced by ligand treatment. Interestingly, for sites with LXR-dependent RXR binding, i.e., sites where RXR binding is lost in livers from LXRdKO mice (see Fig. S3A to S3D), RXR occupancy is highly dependent on the LXR agonist in WT mice. This shows that at least for some sites the occupancy of the LXR-RXR heterodimer is primarily responsive to the LXR agonist.
Fig 2.
Assessment of genomic LXR and RXR binding in mouse liver during treatment with LXR and RXR agonists. Mice were gavaged with vehicle (Veh) (1% carboxymethyl cellulose), the RXR agonist bexarotene (Bexa) (100 mpk), or the LXR agonist T0901317 (T09) (30 mpk) once daily for 14 days. (A and C) Venn diagrams representing the number of genome-wide binding sites of LXR (A) and RXR (C) in livers from mice treated with vehicle (yellow), bexarotene (red), and T0901317 (blue). (B and D) Box plots illustrating the number of tags per LXR peak (B) and RXR peak (D) at sites that are conserved between the different treatments (gray area in panels A and C). (E and F) ChIP-qPCR validation of ligand-dependent RXR binding (E) and LXR binding (F) to selected loci identified by ChIP-seq (see Fig. S3 in the supplemental material) (n = 4). Values that are significantly different from the vehicle by one-way analysis of variance (ANOVA) are indicated as follows: ∧, P < 0.01; ∗, P < 0.05. (G) Venn diagram representing the number of LXR-RXR binding sites in vehicle- and T0901317-treated mice and overlap with ENCODE DNase I-hypersensitive (DHS) sites. (H and I) Box plots illustrating the number of DHS (H) and LXR (I) tags per peak in overlap with T0901317-independent and T0901317-dependent LXR binding sites. For all box plots, the rectangles show the interquartile ranges (IQR) from the first quartile to the third quartile and the lines in the middle of the boxes represent the medians. The whiskers are drawn to the nearest value not exceeding 1.5 times the IQR, and outliers are not shown. Wilcoxon test with Bonferroni's correction was used.
We also compared the identified LXR-RXR binding profiles with ENCODE DNase I hypersensitivity (DHS) whole-genome data from mouse liver (59) and found that 98.9% of the LXR-RXR binding sites were located within open chromatin regions (Fig. 2G, DHS FDR of <0.01). Notably, the average DNase I hypersensitivity was higher at T0901317-independent LXR-RXR binding sites present in vehicle-treated mice compared to the T0901317-dependent LXR-RXR binding sites that arise following LXR activation with T0901317 (Fig. 2H). Correlating with this, the average LXR peak intensity following T0901317 treatment was higher at the T0901317-independent binding sites compared to the T0901317-dependent binding sites (Fig. 2I). These findings suggest that chromatin accessibility may predetermine the ability of LXR to bind to DNA in mouse liver and that LXR agonism enables LXR to bind to sites that are less accessible.
In summary, the ChIP-seq data show that LXR binding overlaps with RXR binding sites in mouse liver and that exposure to an LXR agonist enhances occupancy of both LXR and RXR to their target sites, whereas exposure to bexarotene primarily increases occupancy of RXR, presumably through other RXR heterodimers than LXR-RXR. These changes in NR occupancy are mainly attributed to specific activation of RXR and LXR, since the expression levels of the predominant RXR and LXR isoforms in the liver, RXRα and LXRα (6, 47, 62) are only minimally affected by ligand treatment as measured by quantitative real-time PCR (see Fig. S4 in the supplemental material).
Identification of putative direct LXR target genes.
To assess the effects of long-term agonist treatment on hepatic gene transcription, we performed RNA polymerase II (RNAPII) ChIP-seq and counted the number of recovered tags within gene bodies (position plus 250 to the end of gene) as previously reported (52). Gene transcription was analyzed in WT and LXRdKO mice treated with vehicle or T0901317, respectively. This analysis identified a total of 161 genes (59 induced genes and 102 repressed genes) regulated by T0901317 in an LXR-dependent manner (see the gene list in Table S2 in the supplemental material). Functional annotation of the LXR-dependent T0901317-induced genes using DAVID (25) revealed that this group of genes is significantly associated with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways classically connected to LXR regulation, including pathways involved in lipid and bile acid metabolism (Table 1). Likewise, genes involved in complement and coagulation cascades were significantly downregulated during T0901317 treatment in an LXR-dependent manner. Intriguingly, two distinct sets of genes associated with PPAR signaling were also significantly induced or repressed, respectively, by LXR-mediated T0901317 action (see Table S2). The PPAR-associated group of genes induced by T0901317 in an LXR-dependent manner was mainly involved in lipogenesis and fatty acid oxidation, whereas the PPAR-associated group of genes repressed by T0901317 was mainly involved in lipoprotein metabolism (data not shown). This suggests that cross talk exists between hepatic LXR- and PPAR-regulated pathways.
Table 1.
Putative direct LXR target genesa
| Gene type and KEGG pathway (KEGG pathway identifier) | No. of genes | P value | % of genes with the adjacent LXR-RXR peak |
|---|---|---|---|
| LXR-induced genes (59 genes, 63% with peaks) | |||
| PPAR signaling pathway (mmu03320) | 7 | 3.32E−05 | 100 |
| Polyunsaturated fatty acid biosynthesis (mmu01040) | 4 | 8.82E−05 | 100 |
| Limonene and pinene degradation (mmu00903) | 3 | 0.008 | 100 |
| Benzoate degradation via CoA ligation (mmu00632) | 3 | 0.010 | 100 |
| Bile acid biosynthesis (mmu00120) | 3 | 0.013 | 100 |
| Glycerolipid metabolism (mmu00561) | 3 | 0.019 | 100 |
| Valine, leucine, and isoleucine degradation (mmu00280) | 3 | 0.021 | 100 |
| Fatty acid metabolism (mmu00071) | 3 | 0.022 | 100 |
| Fatty acid biosynthesis (mmu00061) | 2 | 0.032 | 100 |
| Glycerophospholipid metabolism (mmu00564) | 3 | 0.042 | 100 |
| LXR-repressed genes (102 genes, 70% with peaks) | |||
| Complement and coagulation cascades (mmu04610) | 8 | 3.21E−06 | 100 |
| PPAR signaling pathway (mmu03320) | 5 | 0.005 | 100 |
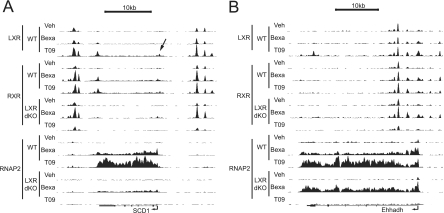
Regulated genes with associated shared LXR-RXR genomic binding sites were identified using PinkThing, which uses the nearest gene for peak assignment. This revealed a total of 104 putative direct LXR target genes (36 induced and 68 repressed). Binding of LXR to regions near these genes is in all cases except one (Foxq1) higher in T0901317-treated liver compared to vehicle-treated liver as determined by the LXR association strength (54) (see Table S2 in the supplemental material). The putative direct LXR targets include the genes encoding the stearoyl-CoA desaturase 1 (SCD1) and the enoyl-coenzyme A, hydratase/3-hydroxyacyl coenzyme A dehydrogenase (Ehhadh). We identified several novel LXR-RXR binding sites in the Scd1 locus that display strong association with LXR-RXR compared to the previously characterized LXRE in the proximal promoter (11) (Fig. 3A). Similarly, we identified several LXR-RXR binding sites in the Ehhadh locus (Fig. 3B), a locus previously shown to be regulated by both LXR and PPARα (24, 40). Importantly, all T0901317-regulated genes that were assigned to significantly enriched KEGG pathways (P value < 0.05) in our data set, have nearby shared LXR-RXR binding sites (Table 1), indicating that these genes are indeed direct LXR targets.
Fig 3.
LXR, RXR, and RNA polymerase II (RNAP2) binding to the Scd1 (A) and Ehhadh (B) gene loci in mouse liver. UCSC Genome Browser tracks derived from LXR, RXR, and RNAPII ChIP-seq data are shown. The black arrow shows the position of a previously reported LXRE (11).
LXR is not required for RXR association with the majority of LXR-RXR binding sites.
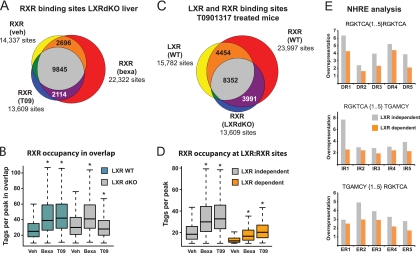
To further examine RXR binding to shared RXR-LXR sites, we analyzed the genome-wide RXR binding in LXRdKO mice treated with vehicle, the LXR agonist T0901317, or the RXR agonist bexarotene. As expected, the T0901317-induced increase in the number of RXR binding sites in WT liver (23,997 sites in T0901317-treated mice versus 12,605 in vehicle-treated mice [Fig. 2C]) is absent in the LXRdKO liver (13,609 sites in T0901317-treated mice versus 14,337 in vehicle-treated mice [Fig. 4A]). Similarly, the increased intensity of RXR binding in WT liver following T0901317 treatment is also compromised in the LXRdKO liver (Fig. 4B), confirming that the effects of the LXR agonist are almost entirely dependent on LXR. In contrast, the bexarotene-induced RXR binding to target sites is maintained and even enhanced in the LXRdKO liver (see Fig. S5 in the supplemental material). Moreover and quite surprisingly, we observed that RXR in LXRdKO mice is still binding to the majority of the shared LXR-RXR binding sites identified in WT mice (Fig. 4C), indicating that RXR also binds to these sites either as a homodimer or by forming heterodimers with other NRs. An example of this is the LXR-RXR site at the Ccdc25 −7kb region, where LXR and RXR occupancy is induced by both RXR and LXR agonism in the WT liver (Fig. 2E and F; see Fig. S6 in the supplemental material). RXR occupancy at this site is maintained in the LXRdKO liver and induced by bexarotene (see Fig. S6), indicating that this site is occupied by LXR-RXR as well as other RXR-containing dimers in WT mice.
Fig 4.
LXR is not required for binding of RXR to the majority of LXR-RXR binding sites. (A) Venn diagram showing the number of RXR binding sites in the livers of LXR dKO mice treated with vehicle (veh) (yellow), bexarotene (bexa) (red), and T0901317 (T09) (blue). (B) Boxplot illustrating the number of tags per RXR peak at sites that are conserved between the different treatments in panel A. Values that are significantly different (P < 2.2e−16 by Wilcoxon test with Bonferroni's correction) from those of vehicle-treated mice are indicated by an asterisk. (C) Venn diagram showing the number of genome-wide RXR binding sites in the livers of T0901317-treated WT (red) and LXR dKO mice (blue) and overlap with LXR binding sites in T0901317-treated WT mice (yellow). (D) RXR occupancy at LXR-RXR sites in WT mice (shared between vehicle-, bexarotene-, and T0901317-treated animals) that are present (LXR independent) or absent (LXR dependent) in LXRdKO mice. (E) Relative overrepresentation (fold enrichment compared to a random background) of direct, inverted, and everted repeats with the core sequence RGKTCA under LXR-RXR peaks where RXR binding is either LXR independent (gray) or LXR dependent (orange). For all box plots, the rectangles show the IQR from the first quartile to the third quartile and the lines in the middle of the boxes represent the medians. The whiskers are drawn to the nearest value not exceeding 1.5 times the IQR, and outliers are not shown.
Presumably, RXR binding sites that are lost in the LXRdKO mice (LXR-dependent RXR occupancy) represent sites where RXR binds only as a heterodimer with LXR and not with other receptors, whereas sites that are maintained in the LXRdKO mice (LXR-independent RXR occupancy) represent sites where RXR can bind with other receptors as well. To investigate how agonists affect RXR occupancy at these two different types of sites, we determined the RXR occupancy at 212 LXR1-dependent and 7,815 LXR-independent LXR-RXR binding sites in WT mice that show an absence of RXR binding in LXRdKO mice (Fig. 4D). This showed that LXR-independent binding sites on average have a significantly higher RXR occupancy, probably reflecting that more RXR heterodimers can bind to these sites. Notably however, RXR occupancy at both types of sites is similarly responsive to RXR and LXR agonist. This shows that, unlike what the few examples of LXR-dependent binding sites in Fig. 2E indicated, RXR occupancy at LXR-dependent sites is induced by bexarotene as well as T09. Thus, although LXR-RXR binding at some sites may be particularly responsive to T09, overall the occupancy of the LXR-RXR heterodimer is increased by both RXR and LXR agonism.
The finding that RXR binding to LXR target sites is maintained in the absence of LXR prompted us to investigate the nature of potential nuclear hormone response elements (NHREs) under the LXR-RXR peaks. We determined the enrichment of direct, inverted, and everted repeats of the core sequence RGKTCA separated by 1 to 5 nucleotides (DR1 to DR5, IR1 to IR5, and ER1 to ER5) under the shared LXR-RXR binding sites relative to random genomic regions (Fig. 4E). We found that all NHREs are enriched at the shared LXR-RXR binding sites and that the DR4 motif, as expected, is particularly enriched. Notably, the DR1, IR1, and ER2 motifs are also highly enriched, especially in the fraction of LXR-RXR binding sites where RXR binding is maintained in the LXRdKO liver. These data indicate that at the majority of LXR-RXR binding sites, RXR can also bind with dimerization partners other than RXR, in particular some that bind to either DR1, IR1, or ER2 elements.
Extensive overlap between LXR and PPARα binding and gene programs.
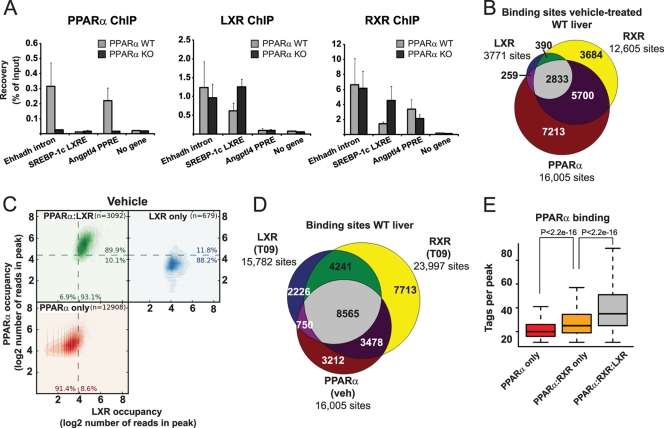
The overrepresentation of DR1 elements at shared LXR-RXR binding sites combined with the significant association of putative direct hepatic LXR target genes with PPAR-regulated pathways suggested the existence of cross talk between hepatic PPAR and LXR signaling. In the liver, PPARα is the dominant PPAR subtype, and in order to determine whether LXR and PPARα bind to common genomic sites, we performed PPARα, RXR, and LXR ChIP-qPCR on a number of LXR-RXR binding sites in WT and PPARα KO mice, respectively. Interestingly, PPARα occupies the LXR-RXR binding site of the putative enhancer in intron 1 of the Ehhadh gene (Fig. 5A) as well as several other LXR binding sites (data not shown). The fact that this binding is dependent on functional PPARα confirms the specificity of the antibody. We also found LXR binding sites that are not significantly occupied by PPARα, such as the SREBP-1c LXRE (61), and PPARα target sites that are not occupied by LXR, such as the peroxisome proliferator response element (PPRE) in intron 1 of the angiopoietin-like 4 gene (Angptl4) (46) (Fig. 5A). Notably, LXR binding to the common binding site in the Ehhadh locus is not affected by lack of functional PPARα, indicating that the binding of the two factors is not interdependent. In contrast, there is increased binding of both LXR and RXR to the SREBP-1c LXRE in PPARα KO mice.
Fig 5.
Genome-wide overlap between LXR and PPARα binding sites in mouse liver. (A) PPARα, LXR, and RXR ChIP-qPCR on selected loci in the livers of wild-type (WT) and PPARα knockout (KO) mice. Bars represent the mean plus range of two independent experiments. (B) Venn diagram representing the overlap between PPARα, LXR, and RXR binding sites in the livers of vehicle-treated WT mice. (C) Scatterplots illustrating the intensity of LXR and PPARα binding in vehicle-treated mouse liver (log2 number of reads per peak) at sites that are defined as LXR specific (blue), PPARα specific (red), and shared (green). The percentages indicate the fractions of sites that are above or below the broken line. (D) Venn diagram representing the overlap between PPARα binding sites in vehicle-treated WT liver with LXR and RXR binding sites in T0901317-treated (T09) WT liver. (E) Boxplot representing the number of PPARα ChIP-seq tags per peak in clusters that do not overlap with RXR and LXR binding (red), overlap with only RXR (orange) and overlap with both RXR and LXR (gray). Wilcoxon test with Bonferroni's correction was used. For all box plots, the rectangles show the IQR from the first quartile to the third quartile and the lines in the middle of the boxes represent the medians. The whiskers are drawn to the nearest value not exceeding 1.5 times the IQR, and outliers are not shown.
To determine the extent of the overlap between LXR and PPARα binding sites at a genome-wide level and to identify novel putative coregulated genes, we performed PPARα ChIP-seq on chromatin isolated from both WT and LXRdKO livers. We identified 16,005 PPARα binding sites in vehicle-treated WT liver, and as expected, a major part of these (53%) are shared with RXR (Fig. 5B). Interestingly, we observed that 88% of all shared LXR-RXR binding sites in vehicle-treated animals are also associated with PPARα, suggesting that the majority of LXR target genes may be coregulated by PPARα. In vehicle-treated liver, LXR cooccupies 19% of the PPARα binding sites (Fig. 5B and C), whereas after T0901317 treatment, LXR becomes associated with 58% of the PPARα binding sites identified in vehicle-treated liver (Fig. 5D). No nonspecific binding of LXR and PPARα antibodies was found at shared binding sites investigated by ChIP-qPCR in LXRdKO and PPARαKO mice, respectively (data not shown). Notably, the average intensity of PPARα genomic binding is significantly higher on shared LXR/RXR/PPARα target sites compared to “PPARα-only” and PPARα-RXR sites, indicating that in mouse liver LXR occupies most strong, high-confidence PPARα binding sites (Fig. 5E). Indeed, within the Ehhadh locus, the PPARα binding site with the highest intensity is the shared LXR/RXR/PPARα binding site in the first intron (Fig. 6A). Importantly, PPARα occupancy at this site as well as at most other shared binding sites (Fig. 6A to D and data not shown) is not dependent on LXR, although the total number of PPARα binding sites is reduced from 16,005 to 9,561 in LXRdKO mice compared to the WT mice (see Fig. S7 in the supplemental material). This excludes the possibility that PPARα is primarily binding via tethering to LXR.
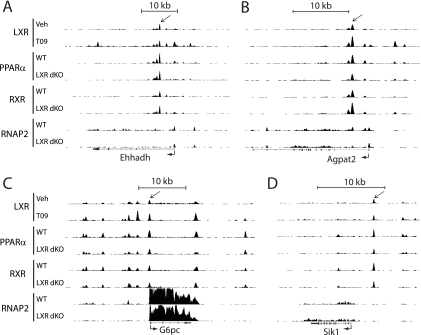
Fig 6.
UCSC Genome Browser tracks derived from ChIP-seq data showing LXR, PPARα, RXR, and RNAP2 binding to the Ehhadh (A), Agpat2 (B), Gbpc (C), and Sik1 (D) loci. The black arrows indicate overlapping binding of LXR, PPARα, and RXR.
Other examples of genomic regions cooccupied by LXR/RXR/PPARα include intron 1 of the 1-acylglycerol-3-phosphate-_O_-acyltransferase 2 (Agpat2) gene (Fig. 6B), the PPRE in the proximal glucose-6-phosphatase (G6pc) promoter (27) (Fig. 6C) and the proximal promoter of the AMP-activated protein kinase (AMPK) family member salt-inducible kinase 1 gene (Sik1) (Fig. 6D), which has been shown to repress hepatic gluconeogenesis via phosphorylation of TORC2 (36) and induce hepatic lipogenesis by modulating SREBP-1c activity (93). However, the ChIP-seq analyses confirmed that there are also a large number of PPARα-RXR binding sites that do not display any significant overlap with LXR binding, e.g., the proximal promoter of Hmcs2 (see Fig. S8B in the supplemental material), which has previously been shown to contain a PPRE in the rat genome (65). Similarly, there are several LXR binding sites that are not cooccupied by PPARα such as the previously mentioned SREBP-1c LXRE (61) (see Fig. S8A). Notably, when comparing our data with ENCODE DHS data from mouse liver, we find that the shared LXR/RXR/PPARα binding sites are located in more open chromatin regions compared to “PPARα-RXR-only” and “LXR-RXR-only” binding sites (see Fig. S9A and S9B). Whether these sites are more open due to the binding of multiple nuclear receptor complexes or whether more receptors bind to the sites because they are more open remains to be determined.
LXR and PPARα bind to overlapping sites with degenerate DR elements.
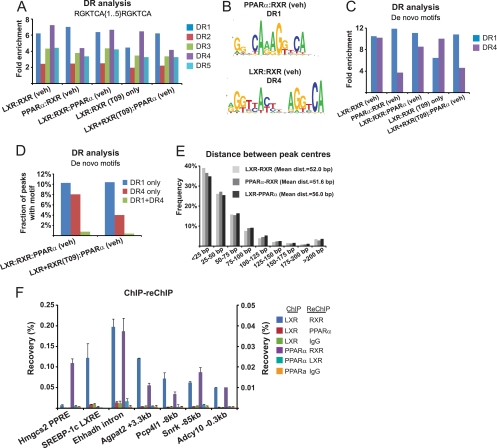
LXRs and PPARs are known to mediate their responses by binding with high specificity to DR4 (76, 79, 92) and DR1 (34, 81) elements, respectively. The extensive overlap between LXR and PPARα binding in mouse liver may arise by proximity of DR1 and DR4 elements at a high percentage of the binding sites. Alternatively, LXR and PPARα may bind to overlapping sequences, either through tethering or through alternating direct binding. The latter would require a promiscuity in binding that is far greater than what is observed in in vitro binding assays (34, 76, 79, 92). To address these possibilities, we first determined the relative enrichment of DR1 to DR5 elements with the core sequence RGKTCA at the shared LXR/RXR/PPARα binding sites and compared this to LXR-RXR and PPARα-RXR binding sites. While our analysis showed that all DR elements are enriched at the sites investigated, we found, as expected, that the DR4 element is particularly enriched at LXR-RXR binding sites in vehicle-treated liver (7.3-fold) and in “LXR-RXR-only” binding sites in T0901317-treated liver (6.5-fold), whereas the DR1 element is particularly enriched at PPARα-RXR binding sites (7-fold) (Fig. 7A). At the shared LXR/RXR/PPARα binding sites, the DR4 element is slightly more enriched than the DR1 element in vehicle-treated liver (6.7-fold versus 6.4-fold), whereas the DR1 element is most enriched in the group of binding sites that are shared between LXR and RXR in T0901317-treated liver and PPARα in vehicle-treated liver (6.2-fold). A similar result was reached when using DR4 and DR1 de novo motifs (GimmeMotifs, FDR of 1%) (83) identified in the LXR-RXR and PPARα-RXR groups, respectively (Fig. 7B and C).
Fig 7.
LXR and PPARα bind to common binding sites in a mutually exclusive manner. (A) Direct repeat motif search (RGKTCA[1.…5]RGKTCA) on LXR and PPARα genomic binding regions in mouse liver treated with vehicle (veh) and T0901317 (T09). LXR-RXR (T09) refers to LXR-RXR binding sites in T0901317-treated liver that are not overlapping with PPARα binding sites. (B) Web logos of DR1 and DR4 sequence motifs identified by de novo motif search on PPARα-RXR (vehicle) and LXR-RXR (vehicle) binding sites (ROC_AUC = 0.632/0.744). (C) Fold enrichment over background of DR1 and DR4 de novo motifs (FDR of 1%). (D) Fraction of peaks with either a single de novo DR1 or DR4 motif or both. (E) Distance between peak centers of overlapping LXR, RXR, and PPARα peaks determined by FindPeaks. Distances were calculated for each site and are shown as 25-bp bins. (F) ChIP-reChIP on liver chromatin from wild-type mice treated with 30 mpk T0901317 24 h and again 8 h before euthanasia. Bars indicate reChIP recoveries when using an antibody against LXR in the first ChIP and against either RXR, PPARα, or IgG in the second ChIP (left y axis), as well as when using an antibody against PPARα in the first ChIP and against either RXR, LXR, or IgG in the second ChIP (right y axis). Bars represent the mean plus range of data from two mice.
Since both DR4 and DR1 elements are enriched at the shared LXR and PPARα binding sites, we asked whether the overlapping binding of LXR and PPARα is the result of receptor binding to adjacent DR1 and DR4 elements or whether LXR and PPARα could bind to the same response element. We therefore determined the number of shared binding sites that contain only a DR1 element or only a DR4 element or both elements. We found that 10.4% and 4.0 to 8.0% of the sites contained only a DR1 element or a DR4 element, respectively, while just 0.4 to 0.8% of the sites contained two separate DR1 and DR4 elements (Fig. 7D). The low percentage of sites with DR1 or DR4 elements indicates that the LXR-RXR as well as the PPARα-RXR heterodimer binds to fairly degenerate DR elements in the liver. In these analyses, the stringency needs to be relatively high in order to distinguish between the different types of DR elements. When less stringent criteria that do not differentiate between the different DRs to the same extent are used in the motif search, 31% of the shared LXR/RXR/PPARα sites (vehicle) have a DR4 element, whereas 67% have a DR1 element.
The fact that many sites have no (80.8 to 85.2%) or only one (14.4 to 18.4%) DR1 or DR4 element per binding site that is recognized by the high-stringency motif search indicates that at the shared binding sites, LXR-RXR and PPARα-RXR are quite promiscuous and are recruited to rather degenerate DR motifs. It is possible that these heterodimers are being recruited to the same degenerate DR elements, or they could be recruited to separate degenerate DRs. To further investigate this, we determined the distance between peak centers of LXR and PPARα at the shared binding sites and compared this to the distance between LXR and RXR peak centers and PPARα and RXR peak centers. Since LXR and PPARα bind to DNA as obligate heterodimers with RXR, we reasoned that the distance to the RXR peak center would represent the distance that one can expect when comparing the binding of two transcription factors that bind to the same response element. We found that the average distance between LXR and PPARα peak centers (56 bp) is very close to the average distances between peak centers of LXR-RXR and PPARα-RXR (52 and 51.6 bp, respectively) and that the distribution pattern is highly similar (Fig. 7E). This suggests that LXR-RXR and PPARα-RXR are binding to the same or overlapping degenerate response elements at these sites.
Because our data indicated that LXR and PPARα bind to overlapping sites in vivo and that these sites show enrichment of the DR1 element, we reinvestigated whether LXR in the presence of RXR could bind directly to DR1 elements in vitro using gel mobility shift assays. For probes we used the acyl-coenzyme A oxidase (Aco) DR1, which is known to bind the PPARα-RXR heterodimer in vitro (81), as well as two DR1 elements identified by nuclear hormone receptor (NHR) scan (66) as the only DR elements present in sequences under shared LXR/RXR/PPARα peaks at the proximal promoters of the Rrbp1 and Adcy10 genes, respectively (see Fig. S11A and S11B in the supplemental material). As expected, LXR-RXR binds to the SREBP-1c DR4 LXRE (61), whereas PPARα-RXR binds to the Aco, Rrbp1, and Adcy10 DR1 elements. However, although LXR binds to sequences containing these DR1 elements in vivo (see Fig. S11), we failed to detect binding of LXR-RXR to these same sequences in vitro (see Fig. S12). This indicates that the chromatin context as well as cobinding proteins, including other transcription factors, associated with chromatin, plays a major role in determining whether a response element is recognized or not. The fact that the shared LXR/RXR/PPARα binding sites align with highly open chromatin regions (ENCODE DHS data [see Fig. S9A and Fig. S9B]) suggests that chromatin accessibility may be contributing to DR promiscuity.
LXR and PPARα bind to overlapping sites in a mutually exclusive manner.
As our results indicated that LXR and PPARα bind to adjacent or overlapping response elements, we wanted to investigate whether they do that simultaneously through a common protein complex or sequentially through different protein complexes. ChIP-reChIP experiments with the livers from WT mice treated with T0901317 for 24 h (Fig. 7F) showed that as expected, LXR and RXR cooccupy the SREBP-1c LXRE (61), whereas PPARα and RXR specifically cooccupy the Hmgcs2 PPRE (65). At the shared LXR/RXR/PPARα sites within the Ehhadh and Agpat2 gene loci (Fig. 6A and B), both LXR and PPARα show cooccupancy with RXR. Similar results were reached when analyzing the shared LXR/RXR/PPARα binding sites in the Pcp4l1 and Snrk gene loci, which are rare examples of sites that display adjacent DR1 and DR4 elements as well as the shared binding site in the Adcy10 locus which harbors a single consensus DR1 that is not bound by LXR in vitro (see Fig. S12 in the supplemental material). However, we were unable to detect cooccupancy of LXR and PPARα at any investigated sites using an LXR antibody in the first ChIP and a PPARα antibody in the second ChIP or vice versa (Fig. 7F and results not shown). Taken together, these data indicate that LXR-RXR and PPARα-RXR heterodimers are recruited to shared binding sites in a mutually exclusive manner.
Extensive overlap between nuclear receptor binding sites in mouse liver.
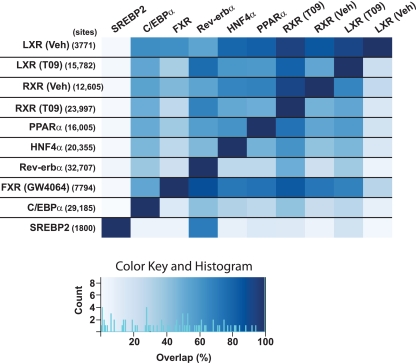
To determine whether the sharing of binding sites is a specific feature of LXR and PPARα, we compared our data with published mouse liver ChIP-seq data for the nuclear receptors HNF4α (68), Rev-erbα (19), and FXR (80) as well as with two other transcription factors that play important roles in hepatic metabolic gene regulation, the CCAAT enhancer-binding protein α (C/EBPα) (68) and sterol-responsive element-binding protein 2 (SREBP2) (71) (Fig. 8). The alignment shows that both LXR and PPARα binding sites overlap significantly with binding sites of the other NRs investigated, as well as with C/EBPα. In fact, in vehicle-treated liver, there are 2,000 LXR/RXR/PPARα binding sites that are also occupied by HNF4α, Rev-erbα (GW4064 treated), and FXR. In contrast, there are very few binding sites of any of the other transcription factors (TFs), except Rev-erbα, that overlap with SREBP2 binding.
Fig 8.
Genome-wide cooccurrence of mouse hepatic transcription factors (TFs). Comparison of LXR, RXR, and PPARα genome-wide binding with published binding profiles of HNF4α and C/EBPα (68), Rev-erbα (19), FXR (80), and SREBP2 (71). The percentage of binding sites of the transcription factors on the y axis that have any overlap (≥1 bp) with binding sites for the TFs on the x axis was calculated for each pair and visualized as a heat map. The total number of binding sites for each factor is indicated in parentheses after the TF name on the y axis.
Functional cross talk between LXR and PPARα agonism in mouse liver.
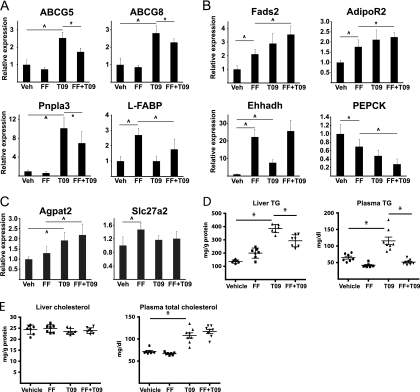
The extensive overlap between LXR and PPARα binding sites prompted us to investigate the possible cross talk between LXR and PPARα agonism. Mice were treated for 5 days with vehicle, the LXR agonist T0901317, the PPARα agonist fenofibrate, or both. Gene expression and physiological parameters were compared between the different groups of mice. As expected, LXR agonism resulted in induction of known hepatic LXR target genes involved in cholesterol transport (e.g., Abcg5 and Abcg8) (Fig. 9A). Likewise, PPARα agonist treatment resulted in induction of known PPARα target genes such as the liver-specific fatty acid-binding protein (Fabp1) and Ehhadh (Fig. 9A and B). There was no change in the expression of either LXR or PPARα following the different ligand treatments (data not shown). Interestingly, when investigating genes with nearby overlapping PPARα and LXR binding sites, we did observe interference between PPARα and LXR ligand treatments for genes that are mainly responsive to one agonist. Thus, PPARα agonism reduced LXR agonist activation of LXR-responsive genes (Abcg5, Abcg8, and Pnpla3), whereas LXR agonism reduced PPARα agonist activation of the PPARα agonist-responsive gene (L-FABP) (Fig. 9A). This suggests that binding of an activated PPARα to a site where this receptor is not engaged in activation of gene expression may interfere with the function of activated LXR binding to the same site and vice versa. Whether this interference is due to coregulator recruitment and/or competition for limiting amounts of RXR (26, 94) remains to be determined. Notably, however, other genes with nearby shared LXR and PPARα binding sites are activated (fatty acid desaturase 2 [_Fads2_], adiponectin receptor 2 [_Adipor2_], and Ehhadh) or repressed (phosphoenolpyruvate carboxykinase 1 [_Pck1_]) by both ligands, and in this case there is no interference but rather an additive tendency of the agonists (Fig. 9B). Finally, there are genes with nearby shared LXR and PPARα binding sites that are regulated only by either LXR agonism (Agpat2) or PPARα agonism (very long chain acyl-CoA synthetase, Slc27a2) without interference from the other agonist (Fig. 9C). Thus, the functional outcome of the LXR and PPARα overlap appears to be determined by the context of the genomic binding site(s) and the activity of the individual receptors at the particular sites. This indicates that cross talk is context dependent and less likely to arise from competition for RXR.
Fig 9.
Functional cross talk between LXR and PPARα in mouse liver. Wild-type C57BL6 mice were treated with PPARα agonist fenofibrate (FF) (200 mpk) (n = 7), LXR agonist T0901317 (T09) (30 mpk) (n = 8), both FF and T09 (200 and 30 mpk, respectively) (n = 8), or vehicle (n = 7) once daily for 5 days. On the fifth day of treatment, the animals were sacrificed, and liver and plasma samples were taken. (A to C) mRNA levels of selected genes in liver samples as determined by qPCR relative to cyclophilin A. PEPCK, phosphoenolpyruvate carboxykinase. (D) Liver and plasma triglyceride (TG); (E) liver and plasma total cholesterol. Values that are significantly different by one-way ANOVA are indicated by black lines and the following symbols: ∧, P < 0.01; ∗, P < 0.05.
In further support of functional cross talk between LXR and PPARα pathways, we found that fibrate treatment neutralized the increased liver TG as well as hypertriglyceridemia displayed in mice treated with LXR agonist (Fig. 9D), whereas there was no effect of fibrate treatment on the elevated liver cholesterol levels and hypercholesterolemia (Fig. 9E). Cumulatively, these data indicate that there is significant cross talk between the LXR and PPARα gene programs in mouse liver, which may be due in part to the sharing of genomic binding sites.
DISCUSSION
The critical role of LXR in liver cholesterol and fatty acid metabolism is well-known, but the genome-wide binding of this NR and its obligate heterodimerization partner RXR has hitherto not been investigated in the liver. Here we identify 12,605 RXR binding sites and 3,771 LXR binding sites in the livers of vehicle-treated mice. The number of RXR binding sites and the degree of binding are dramatically increased by the LXR agonist T0901317, indicating that LXR is a major heterodimerization partner for RXR at these sites. Intriguingly, we find a substantial overlap between LXR and PPARα binding sites, indicating that LXR-RXR and PPARα-RXR heterodimers bind to overlapping regions in chromatin. Since we did not detect any simultaneous binding of PPARα and LXR to the shared sites investigated, LXR does not appear to be recruited indirectly via association with PPARα and vice versa. Consistent with this, the majority of PPARα binding sites that overlap with LXR are maintained in LXRdKO mice, i.e., PPARα binding is not dependent on LXR. Thus, LXR and PPARα are most likely recruited directly to most of the overlapping binding sites.
Notably, only a few percent of the LXR-RXR or PPARα-RXR binding sites contain a well-defined DR4 or DR1 element, respectively, indicating that these heterodimers bind to rather degenerate DR motif sequences. Our finding that LXR binds to a large number of sites with no DR4 resemblance is consistent with the results of Heinz and colleagues showing that a rather low percentage of the binding sites of LXR in macrophages have a sequence that shows resemblance to the DR4 consensus sequence (22). Similarly, PPARα binds to a large number of sites with no DR1 resemblance. Since the binding of PPARα and LXR receptors does not appear to be interdependent, we consider it likely that the receptors bind directly to overlapping or adjacent degenerate DR response elements. This indicates that binding in a chromatin context is significantly more promiscuous in terms of DNA sequence than the in vitro binding properties that have been used to define the consensus sequences. Furthermore, we suggest that this promiscuity involves stabilizing protein-protein interactions with other transcription factors binding to the enhancers, and/or the impact of chromatin structure and epigenetic marks that in the present case leads to clustering of LXR and PPARα at transcription factor “hot spots” (74) at open chromatin regions. An alternative explanation is that the vast majority of LXR-RXR and PPARα-RXR binding in the genome is indirect through tethering; however, we consider this possibility unlikely, given the lack of major interdependence between LXR and PPARα binding.
We cannot formally show whether LXR-RXR and PPARα-RXR bind to the same or adjacent DR motif; however, the lack of detectable cooccupancy of LXR-RXR and PPARα-RXR to shared sites and the overlap of the peak centers between LXR and PPARα tracks suggest that the receptors may bind to the same degenerate element in a mutually exclusive manner. Interestingly, we identify approximately 2,000 LXR/RXR/PPARα binding sites that overlap with previously reported hepatic binding sites for HNF4α, FXR, and Rev-erbα. This indicates the existence of NR “hot spots” in the liver to which multiple NRs bind. It appears unlikely that each of these receptors would have their separate response elements under the shared peaks, and we suggest that there is considerable sharing of degenerate DR elements by these NRs. Traditionally, this would have been considered to give rise to competition for binding; however, recent data from Voss and colleagues using ectopic expression of wild-type glucocorticoid receptor (GR) and an estrogen receptor (ER) with a GR DNA binding domain (DBD) indicate that the occupancy time for GR is so low (<10% of the time) that there is no competition between the two types of receptors binding to the same element (86). In fact, the receptors appear to cooperate in gene activation although they use the same response element. Our finding of extensive overlap between the binding of multiple NRs is fully consistent with this work and suggests that many response elements may serve multipurpose functions in the liver.
It should be noted that although hepatocytes are by far the most abundant cell type in the liver, comprising more than 60% of all liver cells, the liver is a heterogeneous tissue consisting of at least 15 different cell types (reviewed in reference 45). Since we did not detect any simultaneous recruitment of LXR and PPARα to any of the shared LXR and PPARα binding sites examined, we cannot rule out the possibility that a certain part of the overlap between LXR and PPARα genomic binding could be due to contributions from different cell types, i.e., that LXR binds to a particular site in one cell type, and PPARα binds to the same site in another cell type. Kupffer cells and the sinusoidal endothelial cells are the most abundant nonhepatocyte cells, representing up to 20% and 15% of all liver cells, respectively (45). PPARα is not detected in isolated rat Kupffer cells (58), whereas LXRα is highly expressed in these cells (90). It is therefore possible that a minor portion of the detected LXR binding sites originate from Kupffer cells and these sites would not have overlapping LXR and PPARα binding in hepatocytes.
At a functional level, it is intriguing that LXR and PPARα seem to bind to the same genomic sites, given that LXR and PPARα regulate opposing pathways in the liver. Thus, LXR has been shown to activate the lipogenic gene program (8, 30, 61, 70), whereas PPARα activates fatty acid catabolism (32, 41). Consistent with this, we show here that administration of the PPARα agonist fenofibrate alleviates the lipogenic effects of LXR activation and blunts LXR activation of specific LXR target genes. Notably however, there are also target genes where the two receptors appear to cooperate in the activation of gene expression. This indicates that the cross talk between LXR and PPARα is context dependent and not due to competition for limiting amounts of RXR. Such context-dependent cross talk may arise through sharing of direct target genes in a manner in which the two receptors bind to different sites and either cooperate or antagonize the action of each other. However, our data demonstrating an extensive overlap between the binding sites of LXR and PPARα in mouse liver indicate that a novel level of cross talk exists.
In conclusion, we provide the first in vivo genome-wide map of hepatic binding of the NRs LXR, PPARα, and their common dimerization partner RXR and demonstrate a surprising overlap between the binding sites of LXR and PPARα. Our data indicate that this represents a new level of potential cross talk between LXR and PPARα that can be positive as well as negative. Future studies should be directed toward unraveling the molecular mechanisms of this cross talk.
Supplementary Material
Supplemental material
ACKNOWLEDGMENTS
This study was supported by grants to the EU FP6 STREP project X-TRA-NET and grants from NordForsk given to the Nordic Center of Excellence MitoHealth and grants from the Danish Natural Science Research Council and the Danish Health Science Research Council. Jan-Åke Gustafsson was supported by a grant from the Swedish Science Council. Thomas Åskov Petersen is employed at Novo Nordisk A/S and owns shares in the company.
We thank Corine Rommens and Emmanuel Bouchaert for technical assistance.
Footnotes
Published ahead of print 12 December 2011
REFERENCES
- 1.Alberti S, et al. 2001. Hepatic cholesterol metabolism and resistance to dietary cholesterol in LXRbeta-deficient mice. J. Clin. Invest. 107:565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apfel R, et al. 1994. A novel orphan receptor specific for a subset of thyroid hormone-responsive elements and its interaction with the retinoid/thyroid hormone receptor subfamily. Mol. Cell. Biol. 14:7025–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baes M, et al. 1994. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol. Cell. Biol. 14:1544–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedoucha M, Atzpodien E, Boelsterli UA. 2001. Diabetic KKAy mice exhibit increased hepatic PPARgamma1 gene expression and develop hepatic steatosis upon chronic treatment with antidiabetic thiazolidinediones. J. Hepatol. 35:17–23 [DOI] [PubMed] [Google Scholar]
- 5.Blumberg B, et al. 1998. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 12:3195–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bookout AL, et al. 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126:789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugge A, Mandrup S. 2010. Molecular mechanisms and genome-wide aspects of PPAR subtype specific transactivation. PPAR Res. 2010:pii:169506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cha JY, Repa JJ. 2007. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 282:743–751 [DOI] [PubMed] [Google Scholar]
- 9.Choi HS, et al. 1997. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J. Biol. Chem. 272:23565–23571 [DOI] [PubMed] [Google Scholar]
- 10.Chong HK, et al. 2010. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res. 38:6007–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu K, Miyazaki M, Man WC, Ntambi JM. 2006. Stearoyl-coenzyme A desaturase 1 deficiency protects against hypertriglyceridemia and increases plasma high-density lipoprotein cholesterol induced by liver X receptor activation. Mol. Cell. Biol. 26:6786–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costet P, Luo Y, Wang N, Tall AR. 2000. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 275:28240–28245 [DOI] [PubMed] [Google Scholar]
- 13.Denson LA, et al. 2000. Interleukin-1beta suppresses retinoid transactivation of two hepatic transporter genes involved in bile formation. J. Biol. Chem. 275:8835–8843 [DOI] [PubMed] [Google Scholar]
- 14.Desvergne B, Wahli W. 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 20:649–688 [DOI] [PubMed] [Google Scholar]
- 15.Duvic M, et al. 2001. Bexarotene is effective and safe for treatment of refractory advanced-stage cutaneous T-cell lymphoma: multinational phase II-III trial results. J. Clin. Oncol. 19:2456–2471 [DOI] [PubMed] [Google Scholar]
- 16.Duvic M, et al. 2001. Phase 2 and 3 clinical trial of oral bexarotene (Targretin capsules) for the treatment of refractory or persistent early-stage cutaneous T-cell lymphoma. Arch. Dermatol. 137:581–593 [PubMed] [Google Scholar]
- 17.Farol LT, Hymes KB. 2004. Bexarotene: a clinical review. Expert Rev. Anticancer Ther. 4:180–188 [DOI] [PubMed] [Google Scholar]
- 18.Fejes AP, et al. 2008. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24:1729–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng D, et al. 2011. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331:1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamann LG. 2000. An efficient, stereospecific synthesis of the dimer-selective retinoid X receptor modulator (2E,4E,6Z)-7-[5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-(n-propyloxy)naphthalen-3-yl]-3-methyl octa-2,4,6-trienoic acid. J. Org. Chem. 65:3233–3235 [DOI] [PubMed] [Google Scholar]
- 21.He A, Kong SW, Ma Q, Pu WT. 2011. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. U. S. A. 108:5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S, et al. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38:576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. 1998. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol. Cell. Biol. 18:5652–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu T, et al. 2005. Hepatic peroxisomal fatty acid beta-oxidation is regulated by liver X receptor alpha. Endocrinology 146:5380–5387 [DOI] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 26.Ide T, et al. 2003. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. II. LXRs suppress lipid degradation gene promoters through inhibition of PPAR signaling. Mol. Endocrinol. 17:1255–1267 [DOI] [PubMed] [Google Scholar]
- 27.Im SS, et al. 2011. Peroxisome proliferator-activated receptor alpha is responsible for the up-regulation of hepatic glucose-6-phosphatase gene expression in fasting and db/db mice. J. Biol. Chem. 286:1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsson T, et al. 2009. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Mol. Cell 34:510–518 [DOI] [PubMed] [Google Scholar]
- 29.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 383:728–731 [DOI] [PubMed] [Google Scholar]
- 30.Joseph SB, et al. 2002. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277:11019–11025 [DOI] [PubMed] [Google Scholar]
- 31.Kennedy MA, et al. 2001. Characterization of the human ABCG1 gene: liver X receptor activates an internal promoter that produces a novel transcript encoding an alternative form of the protein. J. Biol. Chem. 276:39438–39447 [DOI] [PubMed] [Google Scholar]
- 32.Kersten S, et al. 1999. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 103:1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kliewer SA, et al. 1998. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82 [DOI] [PubMed] [Google Scholar]
- 34.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. 1992. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358:771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight BL, et al. 2005. A role for PPARalpha in the control of SREBP activity and lipid synthesis in the liver. Biochem. J. 389:413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koo SH, et al. 2005. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437:1109–1111 [DOI] [PubMed] [Google Scholar]
- 37.Laffitte BA, et al. 2001. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. U. S. A. 98:507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lalloyer F, et al. 2009. Rexinoid bexarotene modulates triglyceride but not cholesterol metabolism via gene-specific permissivity of the RXR/LXR heterodimer in the liver. Arterioscler. Thromb. Vasc. Biol. 29:1488–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lalloyer F, et al. 2011. Peroxisome proliferator-activated receptor-alpha gene level differently affects lipid metabolism and inflammation in apolipoprotein E2 knock-in mice. Arterioscler. Thromb. Vasc. Biol. 31:1573–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SS, et al. 1995. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15:3012–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leone TC, Weinheimer CJ, Kelly DP. 1999. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. U. S. A. 96:7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang G, et al. 2002. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 277:9520–9528 [DOI] [PubMed] [Google Scholar]
- 43.Makishima M, et al. 2002. Vitamin D receptor as an intestinal bile acid sensor. Science 296:1313–1316 [DOI] [PubMed] [Google Scholar]
- 44.Makishima M, et al. 1999. Identification of a nuclear receptor for bile acids. Science 284:1362–1365 [DOI] [PubMed] [Google Scholar]
- 45.Malarkey DE, Johnson K, Ryan L, Boorman G, Maronpot RR. 2005. New insights into functional aspects of liver morphology. Toxicol. Pathol. 33:27–34 [DOI] [PubMed] [Google Scholar]
- 46.Mandard S, et al. 2004. The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment. J. Biol. Chem. 279:34411–34420 [DOI] [PubMed] [Google Scholar]
- 47.Mangelsdorf DJ, et al. 1992. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 6:329–344 [DOI] [PubMed] [Google Scholar]
- 48.Miller VA, et al. 1997. Initial clinical trial of a selective retinoid X receptor ligand, LGD1069. J. Clin. Oncol. 15:790–795 [DOI] [PubMed] [Google Scholar]
- 49.Moran-Salvador E, et al. 2011. Role for PPARγ in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. FASEB J. 25:2538–2550 [DOI] [PubMed] [Google Scholar]
- 50.Naik SU, et al. 2006. Pharmacological activation of liver X receptors promotes reverse cholesterol transport in vivo. Circulation 113:90–97 [DOI] [PubMed] [Google Scholar]
- 51.Nielsen R, Grontved L, Stunnenberg HG, Mandrup S. 2006. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Mol. Cell. Biol. 26:5698–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen R, et al. 2008. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22:2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oosterveer MH, et al. 2009. Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. J. Biol. Chem. 284:34036–34044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouyang Z, Zhou Q, Wong WH. 2009. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 106:21521–21526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parks DJ, et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science 284:1365–1368 [DOI] [PubMed] [Google Scholar]
- 56.Patsouris D, Reddy JK, Muller M, Kersten S. 2006. Peroxisome proliferator-activated receptor alpha mediates the effects of high-fat diet on hepatic gene expression. Endocrinology 147:1508–1516 [DOI] [PubMed] [Google Scholar]
- 57.Peet DJ, et al. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93:693–704 [DOI] [PubMed] [Google Scholar]
- 58.Peters JM, Rusyn I, Rose ML, Gonzalez FJ, Thurman RG. 2000. Peroxisome proliferator-activated receptor alpha is restricted to hepatic parenchymal cells, not Kupffer cells: implications for the mechanism of action of peroxisome proliferators in hepatocarcinogenesis. Carcinogenesis 21:823–826 [DOI] [PubMed] [Google Scholar]
- 59.Raney BJ, et al. 2011. ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res. 39:D871–D875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Repa JJ, et al. 2002. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 277:18793–18800 [DOI] [PubMed] [Google Scholar]
- 61.Repa JJ, et al. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 14:2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Repa JJ, Mangelsdorf DJ. 2000. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu. Rev. Cell Dev. Biol. 16:459–481 [DOI] [PubMed] [Google Scholar]
- 63.Repa JJ, et al. 2000. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science 289:1524–1529 [DOI] [PubMed] [Google Scholar]
- 64.Rizvi NA, et al. 1999. A phase I study of LGD1069 in adults with advanced cancer. Clin. Cancer Res. 5:1658–1664 [PubMed] [Google Scholar]
- 65.Rodriguez JC, Gil-Gomez G, Hegardt FG, Haro D. 1994. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. J. Biol. Chem. 269:18767–18772 [PubMed] [Google Scholar]
- 66.Sandelin A, Wasserman WW. 2005. Prediction of nuclear hormone receptor response elements. Mol. Endocrinol. 19:595–606 [DOI] [PubMed] [Google Scholar]
- 67.Sanderson LM, Boekschoten MV, Desvergne B, Muller M, Kersten S. 2010. Transcriptional profiling reveals divergent roles of PPARalpha and PPARbeta/delta in regulation of gene expression in mouse liver. Physiol. Genomics 41:42–52 [DOI] [PubMed] [Google Scholar]
- 68.Schmidt D, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328:1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schulman IG, Shao G, Heyman RA. 1998. Transactivation by retinoid X receptor-peroxisome proliferator-activated receptor gamma (PPARgamma) heterodimers: intermolecular synergy requires only the PPARgamma hormone-dependent activation function. Mol. Cell. Biol. 18:3483–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schultz JR, et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14:2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seo YK, et al. 2011. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 13:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seol W, Choi HS, Moore DD. 1995. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol. Endocrinol. 9:72–85 [DOI] [PubMed] [Google Scholar]
- 73.Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. 2004. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell 116:417–429 [DOI] [PubMed] [Google Scholar]
- 74.Siersbaek R, et al. 2011. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 30:1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. 2007. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song C, Kokontis JM, Hiipakka RA, Liao S. 1994. Ubiquitous receptor: a receptor that modulates gene activation by retinoic acid and thyroid hormone receptors. Proc. Natl. Acad. Sci. U. S. A. 91:10809–10813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Staels B. 2001. Regulation of lipid and lipoprotein metabolism by retinoids. J. Am. Acad. Dermatol. 45:S158–S167 [DOI] [PubMed] [Google Scholar]
- 78.Talukdar S, Hillgartner FB. 2006. The mechanism mediating the activation of acetyl-coenzyme A carboxylase-alpha gene transcription by the liver X receptor agonist T0-901317. J. Lipid Res. 47:2451–2461 [DOI] [PubMed] [Google Scholar]
- 79.Teboul M, et al. 1995. OR-1, a member of the nuclear receptor superfamily that interacts with the 9-cis-retinoic acid receptor. Proc. Natl. Acad. Sci. U. S. A. 92:2096–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas AM, et al. 2010. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology 51:1410–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tugwood JD, et al. 1992. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 11:433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Meer DL, et al. 2010. Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 38:2839–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Heeringen SJ, Veenstra GJ. 2011. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics 27:270–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Venkateswaran A, et al. 2000. Human white/murine ABC8 mRNA levels are highly induced in lipid-loaded macrophages. A transcriptional role for specific oxysterols. J. Biol. Chem. 275:14700–14707 [DOI] [PubMed] [Google Scholar]
- 85.Venteclef N, et al. 2010. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes Dev. 24:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Voss TC, et al. 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146:544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wallerman O, et al. 2009. Molecular interactions between HNF4a, FOXA2 and GABP identified at regulatory DNA elements through ChIP-sequencing. Nucleic Acids Res. 37:7498–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3:543–553 [DOI] [PubMed] [Google Scholar]
- 89.Wang Y, et al. 2008. Regulation of cholesterologenesis by the oxysterol receptor, LXRalpha. J. Biol. Chem. 283:26332–26339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang YY, et al. 2006. Activation of the liver X receptor protects against hepatic injury in endotoxemia by suppressing Kupffer cell activation. Shock 25:141–146 [DOI] [PubMed] [Google Scholar]
- 91.Welboren WJ, et al. 2009. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28:1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Willy PJ, et al. 1995. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 9:1033–1045 [DOI] [PubMed] [Google Scholar]
- 93.Yoon YS, Seo WY, Lee MW, Kim ST, Koo SH. 2009. Salt-inducible kinase regulates hepatic lipogenesis by controlling SREBP-1c phosphorylation. J. Biol. Chem. 284:10446–10452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoshikawa T, et al. 2003. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARs suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. Mol. Endocrinol. 17:1240–1254 [DOI] [PubMed] [Google Scholar]
- 95.Yoshikawa T, et al. 2001. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 21:2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material