Chondrocytic Atf4 regulates osteoblast differentiation and function via Ihh (original) (raw)
Abstract
Atf4 is a leucine zipper-containing transcription factor that activates osteocalcin (Ocn) in osteoblasts and indian hedgehog (Ihh) in chondrocytes. The relative contribution of Atf4 in chondrocytes and osteoblasts to the regulation of skeletal development and bone formation is poorly understood. Investigations of the Atf4–/–;Col2a1-Atf4 mouse model, in which Atf4 is selectively overexpressed in chondrocytes in an _Atf4_-null background, demonstrate that chondrocyte-derived Atf4 regulates osteogenesis during development and bone remodeling postnatally. Atf4 overexpression in chondrocytes of the Atf4–/–;Col2a1-Atf4 double mutants corrects the reduction in stature and limb in _Atf4_–/– embryos and rectifies the decrease in Ihh expression, Hh signaling, proliferation and accelerated hypertrophy that characterize the _Atf4_–/– developing growth plate cartilages. Unexpectedly, this genetic manipulation also restores the expression of osteoblastic marker genes, namely Ocn and bone sialoprotein, in _Atf4_–/– developing bones. In Atf4–/–;Col2a1-Atf4 adult mice, all the defective bone parameters found in Atf4–/– mice, including bone volume, trabecular number and thickness, and bone formation rate, are rescued. In addition, the conditioned media of ex vivo cultures from wild-type or _Atf4_–/–;Col2a1-Atf4, but not _Atf4_–/– cartilage, corrects the differentiation defects of _Atf4_–/– bone marrow stromal cells and Ihh-blocking antibody eliminates this effect. Together, these data indicate that Atf4 in chondrocytes is required for normal Ihh expression and for its paracrine effect on osteoblast differentiation. Therefore, the cell-autonomous role of Atf4 in chondrocytes dominates the role of Atf4 in osteoblasts during development for the control of early osteogenesis and skeletal growth.
Keywords: Atf4, Chondrocytes, Osteoblasts, Mouse
INTRODUCTION
In vertebrate embryos at early stages of development, the skeleton consists of avascular cartilage that is gradually replaced by bone later during development, with the exception of areas that require resilient but flexible stiffening. These areas include the walls of larger respiratory passageways and the articular surfaces of joints. Bone, as a mineralized connective tissue, forms by one of the following two processes: replacement of pre-existing mesenchyme, which is defined as intramembranous ossification, or replacement of a pre-existing cartilage mold, which is defined as endochondral ossification. The latter process forms the majority of the bony elements in the body. In mice, ossification centers in the middle of diaphysis starts to form at embryonic day (E) 13 or 13 days post-coitum. Blood vessels invade in the midshaft of the cartilaginous mold of limb and vertebra (Hall, 1988; Reddi, 1994), which brings in precursors of osteoblasts and osteoclasts and transforms it into marrow (Maes et al., 2010; Karsenty and Wagner, 2002; Karsenty et al., 2009). The distal ends of the cartilage template are retained so that the outer surfaces become the articular joints. The inner areas of the cartilage at each of the two distal ends develop into the epiphyseal growth plate that contains four types of chondrocytes: resting, proliferative, prehypertrophic and hypertrophic chondrocytes (Karsenty and Wagner, 2002; Kronenberg, 2003; Karsenty et al., 2009).
Cartilage is not only an ontological precursor of bone, but is also an active participant in bone formation because it secretes local cytokines that regulate both the elongation of the cartilaginous rudiment and the formation of the bony skeleton. Indian hedgehog (Ihh) is one of the local cytokines that plays an indispensable role in controlling skeletal growth. In the developing cartilage, Ihh is expressed by prehypertrophic chondrocytes that have just exited the cell cycle and signals proliferative chondrocytes to divide and perichondrial mesenchymal cells to differentiate into osteoblasts. Thus, Ihh directs both longitudinal growth and bone collar formation. Ihh–/– mice exhibit a reduction in chondrocyte proliferation, impaired chondrocyte maturation, a delay in hypertrophic vascularization, and absence of osteoblasts (Vortkamp et al., 1996; St-Jacques et al., 1999; Chung et al., 2001; Long et al., 2001), establishing the role of Ihh in coupling chondrogenesis and osteogenesis during skeletal development. Importantly, deletion of Ihh specifically in chondrocytes leads to perinatal lethality and skeletal defects recapitulating _Ihh_–/– animals in which Ihh is globally eliminated (Razzaque et al., 2005), thus confirming that the function of Ihh is not restricted to the cartilage. Postnatally, Ihh is essential for maintaining growth plate and trabecular bone as conditional deletion of Ihh in chondrocytes after birth results in destruction of the articular surfaces in long bones and premature fusion of growth plates, as well as loss of trabecular bone over time (Maeda et al., 2007). Two osteoblast differentiation factors, Runx2 and Atf4, have been shown to activate Ihh expression and regulate chondrocyte proliferation and differentiation by binding directly to the Ihh proximal promoter (Yoshida et al., 2004; Wang et al., 2009). However, the role of these transcription factors in chondrocytes in controlling osteoblast differentiation and function has not been established.
Atf4 is a leucine zipper-containing transcription factor and a member of the cAMP response element-binding protein (CREB) family. It was originally identified as a nuclear binding activity enriched in osteoblasts (Ducy and Karsenty, 1995; Schinke and Karsenty, 1999). Global deletion of Atf4 in mice led to severe osteopenia, impaired osteoblast terminal differentiation, reduced Ocn expression and decreased type I collagen synthesis (Yang et al., 2004). Subsequent studies revealed that Atf4 plays an indispensable role in the regulation of chondrocyte proliferation and differentiation during skeletal development (Wang et al., 2009). Atf4–/– embryos and pups exhibit cartilage defects, characterized by a reduced and disorganized proliferative zone, decreased proliferation, expanded hypertrophic zone and reduced Ihh transcription and Hh signaling in chondrocytes (Wang et al., 2009). The latter two genetic studies established the role of Atf4 in osteoblasts and chondrocytes, respectively. However, the global nature of gene deletion in the Atf4–/– mouse model precluded the analysis of a putative role of Atf4 in chondrocytes on chondrocyte-osteoblast coupling during osteogenesis and skeletal development in vivo.
In this study, we created and analyzed _Atf4_–/–;Col2a1-Atf4 mice, in which Atf4 was overexpressed specifically in chondrocytes under the control of the promoter/enhancer of the mouse type II collagen α1 chain (Col2a1) gene (Metsaranta et al., 1991; Metsaranta et al., 1995) in an _Atf4_–/– genetic background. _Atf4_–/–;Col2a1-Atf4 double mutants lack Atf4 in every cell type except for chondrocytes, providing a unique model to study the specific contribution of dysfunctional osteoblasts, chondrocytes or the combination of the two cell types to the skeletal abnormalities of _Atf4_–/– mutants. Analysis of this model provides evidence that chondrocyte-derived Atf4, via its transcriptional target Ihh, is required for proper osteoblast differentiation, function and bone mass acquisition.
MATERIALS AND METHODS
Animals
Animals were maintained in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and were studied according to Institutional Animal Care and Use Committee approved protocols. Col2a1-Atf4 transgene was constructed by inserting an entire Atf4 locus in between a 3 kb fragment of the Col2a1 chain gene promoter and its 3 kb chondrocyte-specific enhancer within the first intron (Metsaranta et al., 1995). Southern blot was performed using _Bam_HI/_Xho_I digested tail genomic DNA and an Atf4 cDNA probe to select transgenic founders. Atf4 cDNA probe for Southern blot was described previously (Wang et al., 2009). Two lines of transgenic mice containing high copy numbers of the Col2a1-Atf4 were used for subsequent matings with wild type (WT) to maintain the transgenic lines or Atf4+/– mice to generate experimental animals. PCR was also performed for genotyping of transgenic mice using the primer pairs 5′-GCCTCGCTGCGCTTCGC-3′ and 5′-GCTTAGGCCGGTGGGGGTTG-3′ and Atf4 mutant mice as described (Yang et al., 2004).
Quantitative real time RT-PCR (qRT-PCR)
Long bone cartilage and bone were isolated from P0 newborn pups by separating white articular cartilages and red midshaft marrow-containing bones under a dissecting microscope. Total RNA from tail, limb cartilage and long bones of indicated genotypes at indicated developmental stages was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocols. DNase I-treated RNA (2 μg) was reverse-transcribed with 100 units of Superscript II plus RNase H– Reverse Transcriptase (Invitrogen) using 100 μM random hexamer primers. qRT-PCR was performed using a standard TaqMan PCR kit protocol on an Applied Biosystems 7300 Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA). Specific forward and reverse oligonucleotide primers used for Atf4 (Mm00515325_m1), Ihh (Mm00439613_m1), patched (Mm00436029_m1), Gli1 (Mm00494645_m1), PTH/PTHrP receptor (PPR; Pth1r – Mouse Genome Informatics) (Mm00441046_m1) were from Applied Biosystems. 18S rRNA was used as an internal control (QuantumRNA Classic 18S Internal Standard). For each genotype, at least three pups were analyzed and standard errors were calculated based on qRT-PCR results from at least three sample repetitions.
Skeletal preparation and histology
Embryos and pups were isolated and skeletal preparation was performed according to standard protocols as described (Wang et al., 2009). For histology, embryos and postnatal day (P) 0 mice were fixed in 4% paraformaldehyde, embedded in paraplast and sectioned at 5 μm. Sections were stained with Hematoxylin and Eosin (H&E). For each genotype, at least three embryos or mice were analyzed.
In vivo proliferation assay
BrdU (5-bromo-2′-deoxyuridine) in vivo labeling of proliferative chondrocytes of embryos and newborn pups (0.1 mg/g of body weight) were performed. Embryos and pups were fixed, sectioned and stained by immunohistochemistry using a BrdU staining kit (Zymed Laboratories, San Francisco, CA, USA) as described previously (Wang et al., 2009).
In situ hybridization
Alternate sections of forelimb were used for histological analysis were hybridized with indicated probes as described previously (Wang et al., 2009). The probe for patched was from Dr Chin Chiang’s laboratory (Vanderbilt University, Nashville, TN, USA).
Microcomputed tomography (μCT) analysis
Femurs from 1-month-old and 3-month-old mice were collected and fixed overnight in 4% paraformaldehyde (pH 7.4) and then 70% ethanol. Distal femurs were scanned using a μCT imaging system (Scanco μCT 40; Scanco Medical, Bassersdorf, Switzerland). Tomographic cross-sectional images were acquired at 55 kV, medium resolution, and 100 slides. Contours were fitted to the outer perimeter of the distal femurs using the auto-contouring feature in the Scanco Software with the threshold of 220 mg hydroxyapatite/cm3.
Cartilage and bone marrow stromal cells culture
Cartilages isolated under a dissecting microscope from P0 to P2 newborn pups with the same genotype were pooled and cultured in αMEM (6 mg cartilage/ml) containing 10% serum and 1% antibiotics. To harvest cartilage-conditioned media, 50% of the cartilage media filtered through cell culture inserts (pore size 1 microns, BD Falcon, Franklin Lakes, NJ, USA) was collected every two days and used as cartilage-conditioned medium (CM). An equal amount of fresh medium was added back to the organ culture. Bone marrow stromal cells (BMSCs) from 2-month-old mice were isolated from long bones by a centrifugation method as described (Dobson et al., 1999). BMSCs were plated in 6-well tissue culture plate with 2×105 cells/well and 4×105 cells/well for alkaline phosphatase-positive colony forming units (CFU-ap) and mineralized nodules (CFU-ob), respectively, following a standard protocol except that cartilage CM (100 μl) was supplemented. For blocking Hh signaling, anti-Hh antibody (5E1, 2 μg/ml; Developmental Studies Hybridoma Bank), cyclopamine (10 μM) or anti-PTHrP antibody as a control (2 μg/ml, a gift from Dr T. J. Martin, University of Melbourne, Australia) was added together with the CM into BMSC cultures. In a similar manner, purmorphamine (10 μM) was added to stimulate Hh signaling. Alkaline phosphatase (ap) and calcified tissues were stained by ap activity staining and von Kossa staining as described previously (Yang et al., 2000; Lian et al., 2009). These experiments were repeated at least three times.
RESULTS
Generation of _Atf4_–/–;Col2a1-Atf4 mice
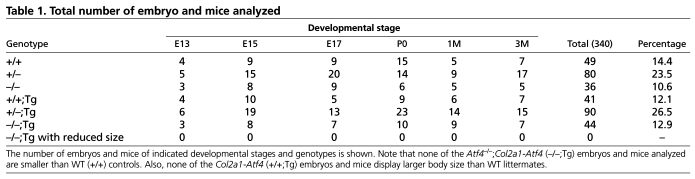
A 2.5 kb sequence containing all three exons and two introns of the Atf4 gene was inserted between the 3 kb promoter and 3 kb enhancer of the mouse type II collagen α1 chain gene (Col2a1, Fig. 1A) (Metsaranta et al., 1991; Metsaranta et al., 1995). Among 28 mice screened by Southern hybridization of genomic DNA, five expressed the Col2a1-Atf4 transgene (Fig. 1B). Male and female transgenic mice reproduced and transmitted the Col2a1-Atf4 transgene to their progenies normally. The founders and their offspring showed no difference compared with their wild-type (WT) littermates in gross body size, weight and activity for up to 1 year and at least three generations of observation, suggesting that Atf4 overexpression in chondrocytes does not affect skeletal development or metabolism. We selected and expanded two transgenic lines (2 and 9) that contained higher copy numbers of the transgene and thus expressed higher levels of Atf4 in chondrocytes. Quantitative real time PCR (qRT-PCR), using total RNA from 6-week-old mouse tails, revealed that Atf4 mRNA level was 4 and 5.5 times higher in lines 2 and 9, respectively, than the endogenous level in WT littermates (Fig. 1C). Both of these lines were used interchangeably to mate with Atf4+/– mice to produce Atf4+/–;Col2a1-Atf4 mice that were then mated with Atf4+/– mice to obtain four types of experimental mouse littermates, WT, _Atf4_–/–, Col2a1-Atf4 and _Atf4_–/–;_Col2a1-Atf4 m_ice at the expected percentages (12.5% each genotype). These results indicate that the Col2a1-Atf4 transgene is not toxic, and is thus providing a useful tool for the study of the function of Atf4 in chondrocytes.
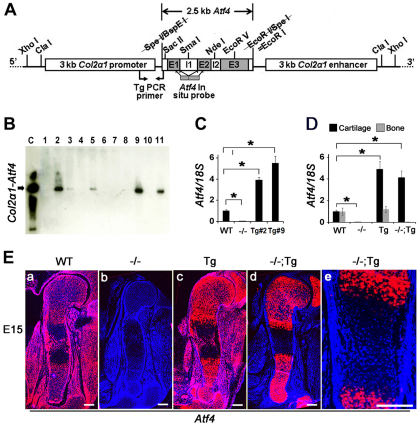
Fig. 1.
Generation of Col2a1-Atf4 transgenic (Tg) mice. (A) Schematic of the Col2a1-Atf4 transgenic construct. Restriction sites, primers used for genotyping, and cDNA probe covering the coding region of the Atf4 gene are indicated. E, exon; I, intron. (B) Southern hybridization of tail genomic DNA identified five founders (2, 3, 5, 9 and 11). (C) qRT-PCR of tail RNA revealed two transgenic lines (Tg#2 and Tg#9) overexpressing Atf4. Data were normalized to endogenous Atf4 level in WT mice and 18S rRNA (_n_=3). Error bars represent s.e.m. *P<0.05 by paired Student’s _t_-test. (D) qRT-PCR of cartilage and bone RNA demonstrated that Atf4 is specifically overexpressed in cartilage but not in bone in Col2a1-Atf4 (Tg) and _Atf4_–/–;Col2a1-Atf4 (–/–;Tg, #9) mice. (E) In situ hybridization of E15 sections of humeri showing Atf4 expression pattern. Note that Atf4 expression is ubiquitous in WT (a) but absent in _Atf4_–/– (b) limb. The Col2a1-Atf4 expression in Col2a1-Atf4 (Tg, c) and _Atf4_–/–;Col2a1-Atf4 (–/–;Tg, d) limbs is restricted to chondrocytes and at a higher level than that in WT limbs. (e) Higher magnification of the ossification center region of E15 _Atf4_–/–;Col2a1-Atf4 humeral sections showing the absence of Col11-Atf4 expression in osteoblasts. _n_=3. Scale bars: 0.2 mm.
Selective chondrocytic Atf4 expression in _Atf4_–/– mice
qRT-PCR using RNA samples isolated from cartilage and bone from P0 newborn pups confirmed that Atf4 mRNA levels were significantly (P<0.05) higher in Col2a1-Atf4 and _Atf4_–/–;_Col2a1_-Atf4 cartilage compared with the WT endogenous level in the cartilage and bone, with a 4.9-fold increase in Col2a1-Atf4 and 4.1-fold increase in _Atf4_–/–;Col2a1-Atf4 cartilage, respectively (Fig. 1D). To pinpoint the exact cell populations in which Atf4 was overexpressed within the cartilage in the transgenic embryos, we performed in situ hybridization using E15, E17 and P0 limb sections and an Atf4 cDNA probe that hybridizes with both endogenous and transgenic Atf4 mRNA (Fig. 1A). As expected, Atf4 mRNA was detected in WT humeri and surrounding tissues and was absent in all cell types in _Atf4_–/– humeri at E15 (Fig. 1Ea,b), E17 and P0 (supplementary material Fig. S1a,b,e,f). The expression pattern of Atf4 in Col2a1-Atf4 transgenic humeri was similar to that of WT humeri; however, the level of Atf4 mRNA was visibly higher in epiphyseal growth plate chondrocytes where the endogenous Col2a1 mRNA is highly expressed at E15 (Fig. 1Ea,c), E17 and P0 (supplementary material Fig. S1a,c,e,g). Cell specificity was also shown by an identical expression pattern of the Col2a1-Atf4 transgene to endogenous Col2a1 (Fig. 4Am) but not to endogenous Atf4 in _Atf4_–/–;Col2a1-Atf4 humeri at E15 (Fig. 1Ea,d), E17 and P0 (supplementary material Fig. S1a,d,e,h). No ectopic expression in hypertrophic chondrocytes, osteoblasts in the midshaft ossification centers, or cells within the perichondrium and periosteum areas was noted in E15 (Fig. 1Ee), E17 and P0 humeri (supplementary material Fig. S1). Together, these data demonstrate that the mouse Col2a1 promoter/enhancer drives overexpression of Atf4 exclusively in _Col2a1_-expressing chondrocytes.
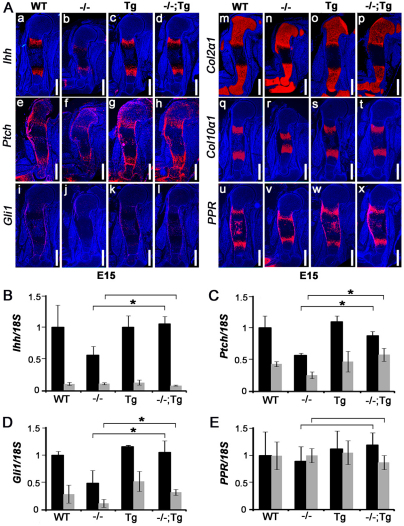
Fig. 4.
Overexpression of Atf4 in chondrocytes corrects the reduced expression of Ihh and its target genes in _Atf4_–/– mice. (Aa-x) In situ hybridization of E15 humeral sections. Note that the decrease in Ihh, Ptch1 and Gli1 expression in _Atf4_–/– humeri was completely rescued in _Atf4_–/–;Col2a1-Atf4 humeri, whereas the expression levels of Col2a1, Col10a1 and PPR were not affected. _n_=3. Scale bars: 0.5 mm. (B-E) qRT-PCR analysis showing expression levels of Ihh, Ptch1 Gli1 and PPR in P0 cartilages (black bars) and bones (gray bars). Error bars represent s.e.m. _N_=3. *P<0.05 by paired Student’s _t_-test. Tg, Col2a1-Atf4; –/–;Tg, _Atf4_–/–;Col2a1-Atf4.
Overexpression of Atf4 in chondrocytes rescues the short stature of _Atf4_–/– mice
Atf4–/– embryos and mice have shorter long bones compared with their WT littermates, which is caused by defects in both cartilage and bone development (Yang et al., 2004; Wang et al., 2009). To examine the role of the Col2a1-Atf4 transgene in chondrocytes and growth plate development, E13, E15 and E17 embryos as well as newborn pups (P0) were analyzed by skeletal preparation followed by Alcian Blue and Alizarin Red staining of cartilage and bone, respectively. At E13, the skeleton of embryos of all four genotypes was cartilaginous as expected, indicated by lack of Alizarin Red-stained ossification centers (Fig. 2A). The visibly smaller skeleton of _Atf4_–/– embryos was not observed in the _Atf4_–/–;Col2a1-Atf4 skeleton, which instead was comparable to WT and Col2a1-Atf4 transgenic skeletons, suggesting that overexpression of Atf4 in chondrocytes corrected the reduced cartilage growth in _Atf4_–/– embryos. At E15, the delayed formation of ossification centers in _Atf4_–/– skeletons was not observed in _Atf4_–/–;Col2a1-Atf4 skeletons, which were still larger than _Atf4_–/– skeletons and the same size as WT and Col2a1-Atf4 littermates. This held true for every later stage examined (Fig. 2A).
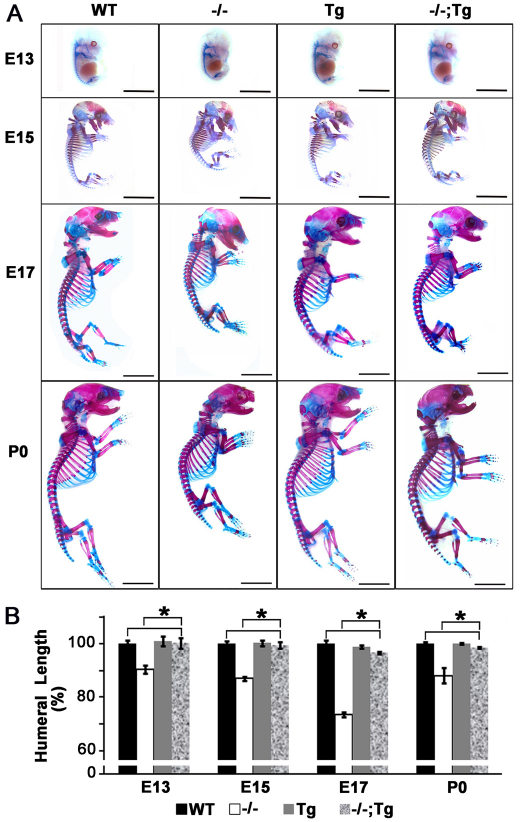
Fig. 2.
Overexpression of Atf4 in chondrocytes rescues shortened statures in _Atf4_–/– mutant mice. (A) Alizarin Red and Alcian Blue staining of skeletons of embryos and newborn pups at the indicated developmental stages. Scale bars: 0.5 mm. _n_=3. (B) Quantification of humeral length in embryos and pups of indicated genotypes at indicated developmental stages. Error bars represent s.e.m. _n_=3. *P<0.05 by paired Student’s _t_-test. Tg, Col2a1-Atf4; –/–;Tg, _Atf4_–/–;Col2a1-Atf4.
Quantitative measurement showed that the 8-30% reduction in E13, E15, E17 and P0 humeral length of _Atf4_–/– embryos and pups was completely rescued in _Atf4_–/–;Col2a1-Atf4 mutants (Fig. 2B). None of the Col2a1-Atf4 transgenic mutants was found to be larger than WT littermates at every stage examined (Fig. 2, Table 1), suggesting that forced expression of Atf4 in chondrocytes to four to five times higher than the endogenous level does not cause excessive growth of _Atf4_–/– skeletons embryonically and postnatally. Collectively, these data support the notion that Atf4 in chondrocytes is a crucial determinant of skeletal growth, and confirm that the dwarfism phenotype of _Atf4_–/– mice is caused by a developmental defect that is chondrocyte autonomous.
Table 1.
Total number of embryo and mice analyzed
Overexpression of Atf4 in chondrocytes rescues the proliferation and differentiation defects of _Atf4_–/– growth plate chondrocytes
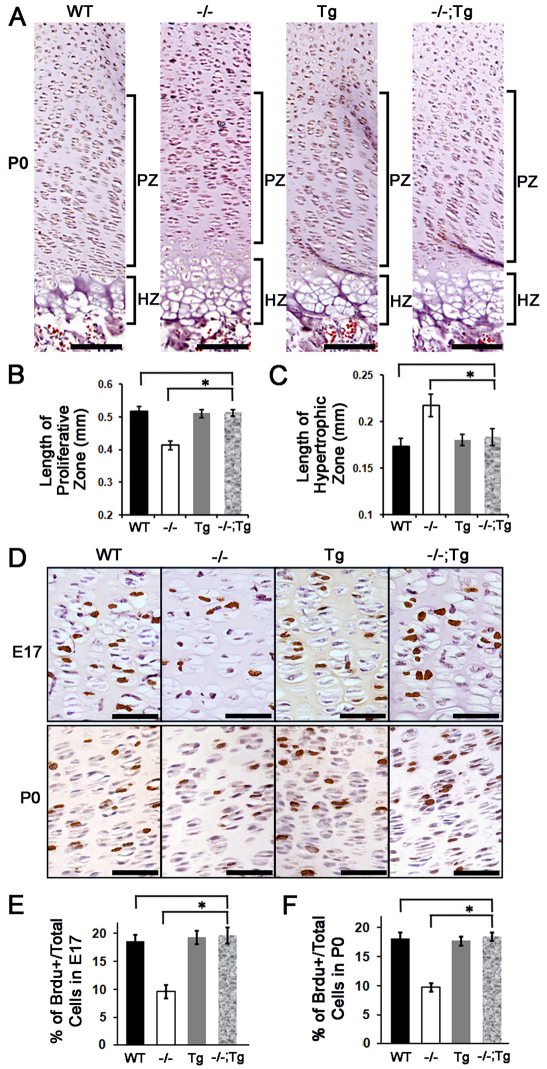
In developing cartilage, growth plate proliferative chondrocytes divide rapidly to form a stack of flattened cells, creating characteristic columns that are readily visible in E17 sections. In P0 growth plates, the stacks of chondrocytes appeared as long columns containing 20-30 cells, whereas in _Atf4_–/– growth plates these columns were short, disorganized and contained only 10-15 cells (Fig. 3A) (Wang et al., 2009). The 30% reduction in the length and the disorganization of the proliferative zone characteristic of _Atf4_–/– growth plates was not found in the _Atf4_–/–;Col2a1-Atf4 growth plate (Fig. 3A,B). Furthermore, the 30% expansion in the hypertrophic zone of Atf4–/– humeral growth plate was also restored to normal in _Atf4_–/–;Col2a1-Atf4 humeri (Fig. 3A,C). No lengthening of proliferative zone and shortening in hypertrophic zone of Col2a1-Atf4 growth plates was observed compared with the WT growth plates (Fig. 3A-C). Accordingly, the decreased proliferation rate (10%) in _Atf4_–/– developing cartilage (Wang et al., 2009), as calculated by the percentage of BrdU-positive cells over total cells in proliferative zones, was corrected in _Atf4_–/–;_Col2a1_-Atf4 limbs (20%), which was the same as that of Col2a1-Atf4 (19%) and WT (19%) growth plates at E17 and P0 (Fig. 3D-F). Therefore, we conclude that overexpression of Atf4 in _Col2a1_-expressing chondrocytes in _Atf4_–/– mutants rescues the proliferation and expanded hypertrophy defects but failed to cause overgrowth of properly formed WT growth plates.
Fig. 3.
Overexpression of Atf4 in chondrocytes restores growth plate chondrocyte defects in _Atf4_–/– mice. (A) H&E staining of humeral sections of indicated genotypes with proliferative chondrocyte zones (PZ) and hypertrophic chondrocyte zones (HZ) of growth plate chondrocytes are indicated. Scale bars: 0.1 mm. (B,C) Quantification of the length of the PZ and HZ in growth plates of indicated genotypes. Error bars represent s.e.m. _n_=3. *P<0.05 by paired Student’s _t_-test. (D) BrdU immunohistochemistry of humeral sections showing BrdU-positive (brown) proliferating growth plate chondrocytes at indicated developmental stages. Scale bars: 0.05 mm. (E,F) Quantification of the BrdU immunohistochemistry results shown in D. Error bars represent s.e.m. _n_=3. *P<0.05 by paired Student’s _t_-test. Tg, Col2a1-Atf4; –/–;Tg, _Atf4_–/–;Col2a1-Atf4.
Overexpression of Atf4 in chondrocytes restores Hh signaling in _Atf4_–/– cartilage and bone
We have previously shown that the expression of Ihh and Hh signaling target genes is decreased in _Atf4_–/– growth plates (Wang et al., 2009). To determine whether the Col2a1-Atf4 transgene could restore Ihh expression in Atf4–/– growth plates, we performed in situ hybridization. Our data showed a comparable level of Ihh expression in prehypertrophic chondrocytes of WT, Col2a1-Atf4 and _Atf4_–/–;Col2a1-Atf4 humeri, which was stronger than in _Atf4_–/– humeri at every developmental stage examined, including E15 (Fig. 4Aa-d), E17 and P0 (supplementary material Fig. S2). Ihh expression was not detected in resting and proliferating chondrocytes in the Col2a1-Atf4 or _Atf4_–/–;Col2a1-Atf4 growth plate, suggesting that overexpression of Atf4 in _Col2a1_-expressing chondrocytes is sufficient to rescue Ihh transcription in prehypertrophic cells where Ihh is expressed endogenously, but is not sufficient to induce Ihh ectopic expression in other chondrocyte populations in vivo.
The expression of patched1 (Ptch1) and Gli1, two direct downstream targets of Hh signaling during skeletal development (Alcedo and Noll, 1997; Mak et al., 2008), was also examined by in situ hybridization to determine whether Hh signaling was also rescued in _Atf4_–/–;Col2a1-Atf4 embryos and pups. As expected, strong Ptch1 expression was found in prehypertrophic chondrocytes, in cells of the perichondrium and periosteum, and in midshaft osteoblasts in WT growth plates at E15 (Fig. 4Ae), E17 and P0 (supplementary material Fig. S2). The expression level of Ptch1 was noticeably decreased in _Atf4_–/– growth plates (Fig. 4Af), but rescued to normal levels by the Col2a1-Atf4 transgene in _Atf4_–/–;Col2a1-Atf4 humeri at every developmental stage examined (Fig. 4Ae-h; supplementary material Fig. S2). Similarly, the decreased Gli1 expression seen in _Atf4_–/– humeri was not observed in _Atf4_–/–;Col2a1-Atf4 humeri (Fig. 4Ai-l; supplementary material Fig. S2). As controls, chondrocyte markers, such as Col2a1 in resting, proliferative and prehypertrophic chondrocytes, Col10a1 in hypertrophic chondrocytes, and PTHrP receptor (PPR) in prehypertrophic chondrocytes, periosteum and osteoblasts (Wang et al., 2009), remained unchanged in Col2a1-Atf4 and _Atf4_–/–;Col2a1-Atf4 humeri compared with WT and _Atf4_–/– controls (Fig. 4Am-x; supplementary material Fig. S2). Quantitative measurements by qRT-PCR demonstrated that the 50% decrease in the expression of Ihh, Ptch1 and Gli1 in _Atf4_–/– cartilage was restored in _Atf4_–/–;_Col2a1_-Atf4 cartilages to a level comparable to that of WT and _Col2a1_-Atf4 cartilages (Fig. 4B-D), thus confirming the in situ hybridization results. Based on these results, we conclude that overexpression of Atf4 in chondrocytes rescues Hh signaling in _Atf4_–/– mice.
As Ptch1, Gli1 and PPR are also expressed in osteoblasts, we then examined the expression of these genes in bones by qRT-PCR. In agreement with the absence of Ihh mRNA signal in humeri at all the developmental stages examined by in situ hybridization (Fig. 4Aa-d; supplementary material Fig. S2), Ihh mRNA level in bone was 90% lower than that in cartilage of all four genotype pups examined (Fig. 4B). The expression of the Hh downstream genes Ptch1 and Gli1 was 50% and 70% lower, respectively, in bone than in cartilage of WT newborns. Ptch1 and Gli1 expression decreased about 50% in _Atf4_–/– bones compared with the WT and _Col2a1_-Atf4 bones, suggesting defective Hh signaling in _Atf4_–/– bones. Interestingly, this Hh signaling defect was corrected in _Atf4_–/–;_Col2a1_-Atf4 bones (Fig. 4B-D), showing that the _Col2a1_-Atf4 transgene in chondrocytes rescued impaired Hh signaling in Atf4–/– bones. The expression of PPR in cartilages and bones was similar in all four genotypes examined (Fig. 4E), suggesting a specific effect of the _Col2a1_-Atf4 transgene in rescuing the Hh signaling defects present in _Atf4_–/– skeletons. Collectively, these results indicate that selective overexpression of Atf4 in chondrocytes restores Ihh expression in cartilage and Hh signaling in both cartilage and bone of the _Atf4_–/–;_Col2a1_-Atf4 developing skeletons. Thus, these results confirm that Atf4 plays a chondrocyte-autonomous role in growth plate development, and suggest that Atf4 in chondrocytes might positively and indirectly regulate bone development and osteogenesis.
Overexpression of Atf4 in chondrocytes rescues defective osteogenesis in _Atf4_–/– embryos
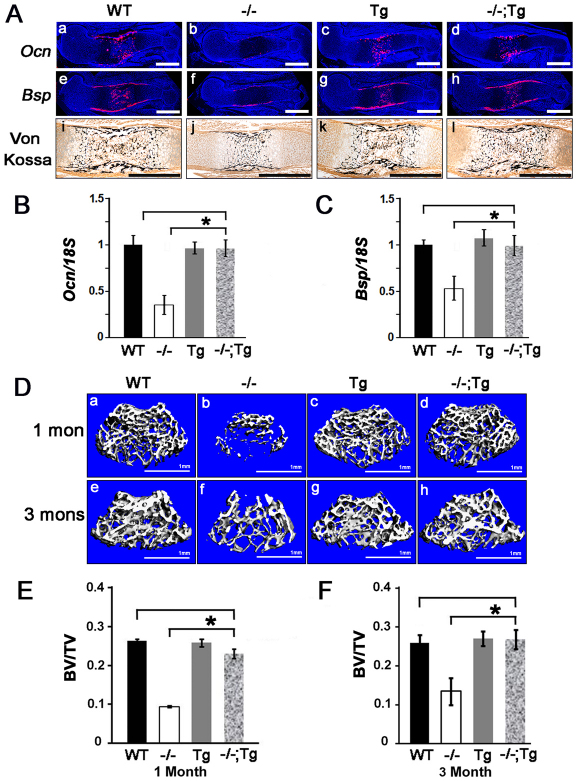
The growth of the skeleton after E14 relies on the proliferation and differentiation of both chondrocytes and osteoblasts as primary ossification centers start to form in the midshaft of long bones and vertebrae. The observation of a complete skeletal size rescue in _Atf4_–/–;_Col2a1_-Atf4 embryos at E15 and E17 and pups at P0 (Fig. 2) alluded to a possible beneficial effect of the _Col2a1_-Atf4 transgene in both chondrocytes and osteoblasts. To test this hypothesis, we first examined the expression of two genetic markers of mature osteoblasts, osteocalcin (Ocn; Bglap – Mouse Genome Informatics) and bone sialoprotein (Bsp) by in situ hybridization. As expected, the expression of both Ocn and Bsp was almost absent in the periosteum and trabecular bone in E15 _Atf4_–/– humeri but was strong in WT controls (Fig. 5Aa,b,e,f), E17 and P0 (supplementary material Fig. S3). The level and pattern of Ocn and Bsp expression in the _Col2a1_-Atf4 transgenic humeri were similar to those in WT controls at every developmental stage examined (Fig. 5Aa,c,e,g; supplementary material Fig. S3), indicating that Atf4 overexpression in chondrocytes does not affect osteoblast differentiation in a WT background. Unexpectedly, however, Ocn and Bsp expression levels in _Atf4_–/–;_Col2a1_-Atf4 humeri were much higher than those in _Atf4_–/– humeri and were comparable with those in WT and _Col2a1_-Atf4 transgenic humeri (Fig. 5Ad,h; supplementary material Fig. S3), indicating that overexpression of Atf4 in chondrocytes rescues the osteoblast differentiation defect in _Atf4_–/– humeri. Furthermore, in contrast to the severely delayed formation of primary ossification centers in the midshafts in E15 _Atf4_–/– humeri, well-developed ossification centers with two times larger mineralized areas were observed in _Atf4_–/–;_Col2a1_-Atf4 humeri, similar to those observed in WT and _Col2a1_-Atf4 humeri (Fig. 5Ai-l; supplementary material Fig. S3). These results provide the first line of evidence that Atf4 in chondrocytes controls osteogenesis during skeletal development.
Fig. 5.
Overexpression of Atf4 in chondrocytes restores osteoblast differentiation and bone formation in _Atf4_–/– mice. (Aa-l) In situ hybridization of sections through E15 humeri showing that the decreased level of Ocn and Bsp in _Atf4_–/– bones was restored to normal level in _Atf4_–/–;Col2a1-Atf4 bones. von Kossa staining of sections through E15 humeri showing rescued mineralization in _Atf4_–/–;Col2a1-Atf4 primary ossification centers. Scale bars: 0.5 mm. (B,C) qRT-PCR analysis of P0 bones quantifying Ocn (B) and Bsp (C) expression. Error bars represent s.e.m. _n_=3. *P<0.05 by paired Student’s _t_-test. (D) Microtomographic image showing rescued bone formation in 1- (1 mon) and 3-month (3 mons) _Atf4_–/–;Col2a1-Atf4 femur heads. _n_=6. Scale bars: 1 mm. (E,F) Quantification of 1- and 3-month femur bone volume/tissue volume (BV/TV) measured by μCT. Error bars represent s.e.m. _N_=6. *P<0.05 by paired Student’s _t_-test. Tg, Col2a1-Atf4; –/–;Tg, _Atf4_–/–;Col2a1-Atf4.
Overexpression of Atf4 in chondrocytes rescues the defects in bone formation in _Atf4_–/– adult mice
To investigate whether overexpression of Atf4 in _Atf4_–/– chondrocytes could rescue bone defects beyond birth in Atf4–/– mice, we examined adult mice from 1- to 6-months-old. Among all the mice examined, none of the _Atf4_–/–;_Col2a1_-Atf4 mutants was smaller than the WT littermates, as measured by nose-to-tail base length from 1- to 3-months-old. The penetrance of this genetic rescue in adult mice was also 100% (Table 1). Consistent with what was observed in embryos and pups, _Col2a1_-Atf4 transgenic mice displayed the same body size as WT littermates, indicating that overexpression of Atf4 in chondrocytes is not sufficient to cause the overgrowth of bone. To determine whether the osteoblast differentiation and bone mass defects in _Atf4_–/– mice were corrected by overexpression of Atf4 in chondrocytes in vivo, we measured trabecular bone mass in distal femurs by 3D microcomputed tomography (μCT). As expected, and in agreement with the low bone mass in _Atf4_–/– vertebrae at all ages examined (Yang et al., 2004), bone mass, as measured by a ratio of bone volume over total tissue volume (BV/TV), was 0.09±0.003% in _Atf4_–/– femurs of 1-month-old mice, which was significantly (P<0.05) lower than the ratio of 0.23±0.009% in _Atf4_–/–;_Col2a1_-Atf4, 0.26±0.005% in WT, and 0.27±0.008% in _Col2a1_-Atf4 littermate controls (Fig. 5D,E). The rescue in BV/TV by the Col2a1-Atf4 transgene was also observed at 3 months of age with a BV/TV of 0.26±0.020% in WT, 0.27±0.020% in _Col2a1_-Atf4, 0.27±0.024% in _Atf4_–/–;_Col2a1_-Atf4 and 0.13±0.034% in _Atf4_–/– mice (Fig. 5D,F).
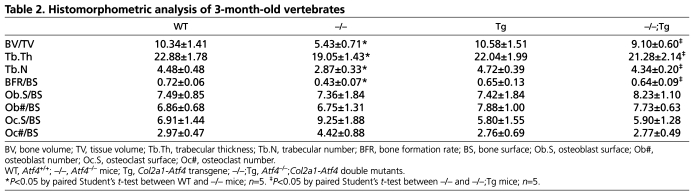
Histomorphometric analysis of vertebrae in 3-month-old mice confirmed that the decreased BV/TV in _Atf4_–/– mice was restored to normal levels in _Atf4_–/–;_Col2a1_-Atf4 femurs (Table 2). Importantly, the markedly reduced bone formation rate over bone surface (BFR/BS) in _Atf4_–/– mice (0.43±0.07 in _Atf4_–/– vs 0.72±0.06 in WT, P<0.05) was normalized in _Atf4_–/–;_Col2a1_-_Atf4_ mutants (0.64±0.09). BFR/BS in _Col2a1_-_Atf4_ transgenic mice (0.65±0.13) was similar to that of WT mice. These data indicate that overexpression of _Atf4_ in chondrocytes rescues the suboptimal osteoblast terminal differentiation and activity in _Atf4_–/– mice (Yang et al., 2004). The ratio of osteoblast number over bone surface (Ob#/BS) was normal in _Atf4_–/– mutants compared with WT littermates, which is consistent with our previous finding (Yang et al., 2004). Ob#/BS was also normal in _Col2a1_-_Atf4_ and _Atf4_–/–;_Col2a1_-_Atf4_ vertebrae, indicating that _Atf4_ overexpression in chondrocytes is not sufficient to induce osteoblast proliferation. Lastly, the osteoclast number (Oc#/BS) and the osteoclast-covered bone surface over total bone surface (Oc.S/BS) in _Atf4_–/– vertebrae were higher than that of WT, _Col2a1_-_Atf4_ and _Atf4_–/–;_Col2a1_-_Atf4_ vertebrae, but these differences were not statistically significant (_P_>0.5, Table 2). Therefore, our data indicate that Atf4 overexpression in chondrocytes promotes osteoblast differentiation and bone formation but does not significantly affect osteoclastogenesis. This finding is important as it suggests that the loss of Atf4 in osteoblasts can be compensated for by overexpression of Atf4 in chondrocytes.
Table 2.
Histomorphometric analysis of 3-month-old vertebrates
Overexpression of Atf4 in chondrocytes corrects defective Hh signaling in _Atf4_–/– bone marrow stromal cells
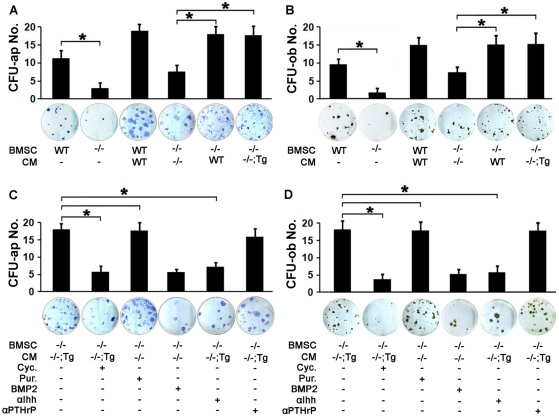
That chondrocytic Atf4 rescued the bone defects in _Atf4_–/– mice suggested that diffusible factors in cartilage of _Atf4_–/–;_Col2a1_-Atf4 double mutants could make up for the loss of Atf4 in osteoblasts to support proper osteoblast differentiation and bone formation. To address this hypothesis, we utilized an ex vivo system in which the conditioned media (CM) of WT, _Atf4_–/– and _Atf4_–/–;_Col2a1_-Atf4 cartilage explants was collected and supplemented to the osteogenic media of WT, _Atf4_–/ and _Col2a1_-Atf4 bone marrow stromal cell (BMSC) cultures. Without supplementation with cartilage CM, an 80% decrease in the number of alkaline phosphatase-positive colony forming units (CFU-ap) and mineralized nodules (CFU-ob) in _Atf4_–/– BMSC cultures compared with WT BMSC cultures was observed (Fig. 6A,B). This indicates that there is an intrinsic osteoblast differentiation defect in _Atf4_–/– BMSCs. Supplementation of WT cartilage CM led to a 45% increase in CFU-ap and CFU-ob formation in WT BMSC cultures. Interestingly, supplementation of _Atf4_–/– cartilage CM caused also a 62% increase in CFU-ap and CFU-ob formation in _Atf4_–/– BMSC cultures. These results reveal that cells in cartilage can exert a potent paracrine and stimulatory effect on osteoblast differentiation of BMSC progenitors. Importantly, supplementation of WT or _Atf4_–/–;_Col2a1_-Atf4 cartilage CM resulted in an 86% increase in CFU-ap and CFU-ob formation in _Atf4_–/– BMSC cultures (Fig. 6A,B). This demonstrates that WT or _Atf4_–/–;_Col2a1_-Atf4 CM contains an osteogenic diffusible factor(s), which is not present in _Atf4_–/– cartilage CM and which is responsible for the complete rescue of the osteoblast differentiation defect of _Atf4_–/– BMSCs.
Fig. 6.
Cartilage conditioned media contain osteogenic activities. (A,B) Quantification of CFU-ap (A) and CFU-ob (B) formed in BMSC cultures under osteoblast differentiation condition. Note that _Atf4_–/– BMSC failed to properly differentiate into mature osteoblasts, which can be partially rescued by Atf4_–/– cartilage CM and completely rescued by CM of WT and Atf4_–/–;Col2a1-Atf4 cartilages. Error bars represent s.e.m. _n_=3. *P<0.05 by paired Student’s _t_-test. (C,D) Quantification of CFU-ap (C) and CFU-ob (D) formed in BMSC cultures in osteoblast differentiation assays. Note that purmorphamine (Pur.) increases CFU-ap and CFU-ob formed by _Atf4_–/– BMSCs supplemented with _Atf4_–/– cartilage CM and cyclopamine (Cyc.) and 5E1, but not anti-PTHrP, decrease the CFU-ap and CFU-ob formed by _Atf4_–/– BMSCs supplemented with _Atf4_–/–;Col2a1-Atf4 cartilage CM. Error bars represent s.e.m. _N_=3. *P<0.05 by paired Student’s t_-test. Tg, Col2a1-Atf4; –/–;Tg, Atf4_–/–;Col2a1-Atf4.
Based on the knowledge that Ihh is a transcriptional target gene of Atf4 in chondrocytes and a crucial determinant of osteogenesis, as well as the observation that defective Hh signaling present in _Atf4_–/– bones was corrected in _Atf4_–/–;_Col2a1_-Atf4 bones (Fig. 4), we tested whether Ihh was one of the osteogenic factors in WT or _Atf4_–/–;_Col2a1_-Atf4 cartilage CM that promoted osteoblast differentiation of _Atf4_–/– BMSCs. We first modulated Hh signaling in the ex vivo system described above pharmacologically with cyclopamine, a Hh signaling blocker, and purmorphamine, a Hh signaling activator. Cyclopamine caused a 63% decrease in CFU-ap and CFU-ob formation in _Atf4_–/– BMSC cultures supplemented with _Atf4_–/–;_Col2a1_-Atf4 cartilage CM. Conversely, and importantly, purmorphamine, but not Bmp2, completely restored CFU-ap and CFU-ob formation defect in _Atf4_–/– BMSC cultures supplemented with _Atf4_–/– cartilage CM (Fig. 6C,D). These data demonstrate that Hh signaling is required for _Atf4_–/– BMSCs to differentiate into mature osteoblasts. Next, direct blocking Ihh with the neutralizing antibody 5E1 but not an anti-PTHrP, reversed the rescue of CFU-ap and CFU-ob formation in _Atf4_–/– BMSCs by supplementation of _Atf4_–/–;_Col2a1_-Atf4 cartilage CM (Fig. 6C,D). Given that Ihh is not expressed in bones (Fig. 4A,B) and that Ihh is the only member of the Hh protein family expressed in chondrocytes (Bitgood and McMahon, 1995; Kronenberg, 2003), we conclude that Ihh is one of the major osteogenic factors differentially secreted by _Atf4_–/–;_Col2a1_-Atf4 cartilage. Taken together, these data demonstrate that Atf4 overexpression in _Atf4_–/–;_Col2a1_-Atf4 cartilage, by increasing the production of Ihh, rescues the differentiation defect of _Atf4_–/– BMSC cultures.
DISCUSSION
In this study we provide evidence that restoration of Atf4 expression specifically in _Col2a1_-expressing chondrocytes in _Atf4_–/– mice rescues not only the growth defects, but also the osteogenesis defects and reduced bone mass observed in _Atf4_–/– mice. Therefore, this study identifies Atf4 in chondrocytes as a crucial transcriptional regulator acting in a chondrocyte-autonomous fashion to regulate longitudinal growth of the skeleton and bone formation.
Atf4 was originally identified as a nuclear binding activity specific to osteoblasts (Ducy and Karsenty, 1995; Schinke and Karsenty, 1999), and its global deletion in mice was shown to lead to severe osteopenia, impaired osteoblast terminal differentiation, reduced osteocalcin expression and decreased type I collagen synthesis (Yang et al., 2004). Subsequent studies revealed that Atf4 is also expressed in chondrocytes, in which it controls chondrocyte proliferation and differentiation during skeletal development by transcriptionally activating Ihh (Wang et al., 2009). These two studies have established the indispensable role of Atf4 in osteoblasts and chondrocytes, respectively. However, the relative contribution of dysfunctional osteoblasts, chondrocytes or the combination of defects in these two cell types to the skeletal abnormalities observed in _Atf4_–/– mutants could not be defined in this global _Atf4_–/– mouse model because Atf4 is expressed in both cell types in equal abundance (Wang et al., 2009). By utilizing the _Atf4_–/–;_Col2a1_-Atf4 model, we were able to restore the expression of Atf4 specifically in chondrocytes, with osteoblasts remain deficient for Atf4. The bone elongation defects caused by dysfunctional growth plate chondrocytes in _Atf4_–/– cartilage were completely rescued by this genetic manipulation. These defects include shortened long bones and stature, disorganization of proliferating chondrocyte columns, shortening of the proliferating zone, a decrease in BrdU-positive chondrocyte number, expansion of the hypertrophic chondrocyte zone and decreased Ihh expression and Hh signaling (Figs 1, 2) (Wang et al., 2009). The complete correction of chondrogenesis in _Atf4_–/– mutants by the _Col2a1_-Atf4 transgene thus demonstrates that the reduced stature of _Atf4_–/– mice is not caused by an endocrine imbalance, and that Atf4 regulates growth plate development, chondrocyte proliferation and differentiation in vivo in a chondrocyte-autonomous manner.
A novel finding in this study is that the transcriptional activity of Atf4 in chondrocytes controls osteogenesis during development and postnatal bone mass accrual. This conclusion is supported by the observation that Atf4–/–;Col2a1-Atf4 mice have normal bone mass (Fig. 5), bone formation rate, trabecular bone thickness and trabecular numbers (Table 2) in contrast to _Atf4_–/– mice, and despite complete absence of endogenous or ectopic Atf4 expression in osteoblasts (Fig. 1; supplementary material Fig. S1). It is important to note that, unlike the rat Col2a1 promoter and enhancer (Nakamura et al., 2006; Chen et al., 2007; Hilton et al., 2007), the mouse Col2a1 promoter and enhancer used to produce the Col2a1-Atf4 transgenic construct in this study is chondrocyte-specific and does not show leakiness in any other tissues (Mukhopadhyay et al., 1995; Zhou et al., 1995; Ovchinnikov et al., 2000). This specificity is further confirmed by our in situ hybridization data showing that expression of the Col2a1-Atf4 transgene was never detected in the periosteum, where osteoblasts progenitors are present, or in any cells adjacent to _Col2a1_-expressing chondrocytes, at all the developmental stages analyzed (Fig. 1; supplementary material Figs S1, S2). These observations support the crucial role of Atf4 in chondrocytes for the regulation of osteoblast differentiation and osteogenesis.
Based on our previous study (Wang et al., 2009) and on the ex vivo assays (Fig. 6) reported here, Ihh appears to be one of the main mediators of the effect of chondrocytes on osteogenesis that is missing in _Atf4_–/– cartilage. The role of Hh signaling in the regulation of osteoblast differentiation and function is supported by the observations that Hh signaling components, including smoothened, patched and Gli genes, are expressed in osteoblasts (Ohba et al., 2008; Joeng and Long, 2009) and deletion of Ihh in mice leads to bone defects in embryos and pups (St-Jacques et al., 1999). However, osteoblasts might not be a source of Ihh in vivo. Some in vitro studies suggested that Ihh is expressed in osteoblastic cell lines (Murakami et al., 1997; Jemtland et al., 2003), yet Ihh expression level is minimal, if not absent, in bone marrow osteoblasts in vivo (Fig. 2) (Chung et al., 2001; Wang et al., 2009). This strongly suggests that Ihh expression by osteoblastic cell lines was characteristic of immortalized transformed cells because Ihh is known to be upregulated in many tumor cells (Kayed et al., 2004; Xuan and Lin, 2009). Three main observations strongly suggest that the transcriptional activity of Atf4 in chondrocytes is required for proper osteogenesis, via its control of Ihh production. First, Ihh is specifically expressed by prehypertrophic chondrocytes but not in osteoblasts in vivo (Fig. 4) (Razzaque et al., 2005; Maeda et al., 2010). Second, the differentiation of _Atf4_–/– osteoblasts induced by the conditioned media of WT or Atf4–/–;Col2a1-Atf4 cartilage can be blocked by neutralization of Ihh (Fig. 6). Lastly, deletion of Ihh specifically in _Col2a1_-expressing chondrocytes impairs osteoblast differentiation leading to loss of trabecular bone over time (Maeda et al., 2007). Additionally, our ex vivo data indicates that Ihh is not the only factor secreted by cartilage to promote osteogenesis, given that the WT and _Atf4_–/– cartilage CM stimulates the osteoblast differentiation of WT and _Atf4_–/– BMSC cultures, respectively (Fig. 6A,B). Additional factors, such as fibroblast growth factors, Bmps and Wnts, which are not direct transcriptional targets of Atf4 and are produced by the cartilage, might contribute to the osteogenic activity observed in _Atf4_–/– cartilage CM.
The exact population of cells that chondrocyte-derived Atf4 and Ihh target and the time point(s) during skeletal development at which this Atf4-Ihh effect occurs remain unclear and warrant further investigation. Mesenchymal progenitors before bone marrow formation might be the earliest target cells of chondrocyte-derived Ihh. Later during development, perichondrial and periosteal mesenchymal osteoblast progenitors might be relevant targets and they could contribute, at least partly, to the mechanism whereby Atf4 and Ihh from chondrocytes indirectly control osteogenesis. It is also unclear whether chondrocyte-derived Ihh can act as a paracrine factor to control osteoblast differentiation in the primary spongiosa at later developmental stages once the marrow has formed. If it is biologically relevant, the regulation of Ihh by Atf4 should be restricted to the bone growth period and cease or diminish once the growth plate has closed, because the source of Ihh then disappears. These speculative mechanisms will be best addressed by the use of conditional and inducible Atf4 mutant mice.
The observation that overexpression of Atf4 solely in chondrocytes rescues low bone mass in _Atf4_–/– mice (Figs 2, 3, 4, 5) challenges the original notion that dysfunctional osteoblasts, characterized by impaired differentiation and collagen production (Yang et al., 2004), underlie this phenotype. This study supports the concept that bone mass accrual, which relies on osteoblast differentiation and activity, might be controlled, at least in part, by the transcriptional activity of Atf4 in chondrocytes at early stages of development. The cell-autonomous role of Atf4 in osteoblasts (Yang et al., 2004) might be secondary at these developmental stages, but might become more prominent in adults once the growth plate has closed, and contribute to the processes of bone remodeling, bone repair or glucose homeostasis (Hinoi et al., 2009; Yoshizawa et al., 2009) via its effects on Ocn production, amino acids import and stress responses (Harding et al., 2003; Sowa and Karsenty, 2007). Future studies comparing chondrocyte- and osteoblast-specific Atf4 mutant mice will be instrumental to finding the answers to these questions.
Supplementary Material
Supplementary Material
Acknowledgments
We thank Drs Chin Chiang and T. Jack Matin for cDNA probe and anti-PTHrP antibody. The monoclonal anti-Hh antibody 5E1 developed by Dr. Thomas M. Jessell and Susan Brenner-Morton was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Science, Iowa, IA 52242.
Footnotes
Funding
This work was supported by the National Institutes of Health [AR055972 to X.Y.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
W.W. performed the majority of the research and drafted the paper. N.L. and Y.M. performed co-culture assays and part of the bone histology. L.L. generated and maintained the Col2-Atf4 transgenic mice. R.C.G. helped genotyping the mice. F.E. participated in the design of experimental approaches and in the redaction of the manuscript. X.Y. supervised the research and wrote the manuscript.
References
- Alcedo J., Noll M. (1997). Hedgehog and its patched-smoothened receptor complex: a novel signalling mechanism at the cell surface. _Biol. Chem._378, 583–590 [DOI] [PubMed] [Google Scholar]
- Bitgood M. J., McMahon A. P. (1995). Hedgehog and BMP genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. _Dev. Biol._172, 126–138 [DOI] [PubMed] [Google Scholar]
- Chen M., Lichtler A. C., Sheu T. J., Xie C., Zhang X., O’Keefe R. J., Chen D. (2007). Generation of a transgenic mouse model with chondrocyte-specific and tamoxifen-inducible expression of Cre recombinase. _Genesis_45, 44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung U. I., Schipani E., McMahon A. P., Kronenberg H. M. (2001). Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. _J. Clin. Invest._107, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson K. R., Reading L., Haberey M., Marine X., Scutt A. (1999). Centrifugal isolation of bone marrow from bone: an improved method for the recovery and quantitation of bone marrow osteoprogenitor cells from rat tibiae and femurae. _Calcif. Tissue Int._65, 411–413 [DOI] [PubMed] [Google Scholar]
- Ducy P., Karsenty G. (1995). Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. _Mol. Cell. Biol._15, 1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. K. (1988). The embryonic development of bone. _Am. Sci._76, 174–181 [Google Scholar]
- Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. (2003). An integrated stress response regulates amino Acid metabolism and resistance to oxidative stress. _Mol. Cell_11, 619–633 [DOI] [PubMed] [Google Scholar]
- Hilton M. J., Tu X., Long F. (2007). Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. _Dev. Biol._308, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi E., Gao N., Jung D. Y., Yadav V., Yoshizawa T., Kajimura D., Myers M. G., Jr, Chua S. C., Jr, Wang Q., Kim J. K., et al. (2009). An Osteoblast-dependent mechanism contributes to the leptin regulation of insulin secretion. _Ann. New York Acad. Sci._1173 Suppl. 1, E20–E30 [DOI] [PubMed] [Google Scholar]
- Jemtland R., Divieti P., Lee K., Segre G. V. (2003). Hedgehog promotes primary osteoblast differentiation and increases PTHrP mRNA expression and iPTHrP secretion. _Bone_32, 611–620 [DOI] [PubMed] [Google Scholar]
- Joeng K. S., Long F. (2009). The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. _Development_136, 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G., Wagner E. F. (2002). Reaching a genetic and molecular understanding of skeletal development. _Dev. Cell_2, 389–406 [DOI] [PubMed] [Google Scholar]
- Karsenty G., Kronenberg H. M., Settembre C. (2009). Genetic control of bone formation. _Annu. Rev. Cell Dev. Biol._25, 629–648 [DOI] [PubMed] [Google Scholar]
- Kayed H., Kleeff J., Keleg S., Guo J., Ketterer K., Berberat P. O., Giese N., Esposito I., Giese T., Buchler M. W., et al. (2004). Indian hedgehog signaling pathway: expression and regulation in pancreatic cancer. _Int. J. Cancer_110, 668–676 [DOI] [PubMed] [Google Scholar]
- Kronenberg H. M. (2003). Developmental regulation of the growth plate. _Nature_423, 332–336 [DOI] [PubMed] [Google Scholar]
- Lian N., Wang W., Li L., Elefteriou F., Yang X. (2009). Vimentin inhibits ATF4-mediated osteocalcin transcription and osteoblast differentiation. _J. Biol. Chem._284, 30518–30525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F., Zhang X. M., Karp S., Yang Y., McMahon A. P. (2001). Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. _Development_128, 5099–5108 [DOI] [PubMed] [Google Scholar]
- Maeda Y., Nakamura E., Nguyen M. T., Suva L. J., Swain F. L., Razzaque M. S., Mackem S., Lanske B. (2007). Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. _Proc. Natl. Acad. Sci. USA_104, 6382–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Schipani E., Densmore M. J., Lanske B. (2010). Partial rescue of postnatal growth plate abnormalities in Ihh mutants by expression of a constitutively active PTH/PTHrP receptor. _Bone_46, 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C., Kobayashi T., Selig M. K., Torrekens S., Roth S. I., Mackem S., Carmeliet G., Kronenberg H. M. (2010). Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. _Dev. Cell_19, 329–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak K. K., Kronenberg H. M., Chuang P. T., Mackem S., Yang Y. (2008). Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. _Development_135, 1947–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsaranta M., Toman D., de Crombrugghe B., Vuorio E. (1991). Mouse type II collagen gene. Complete nucleotide sequence, exon structure, and alternative splicing. _J. Biol. Chem._266, 16862–16869 [PubMed] [Google Scholar]
- Metsaranta M., Garofalo S., Smith C., Niederreither K., de Crombrugghe B., Vuorio E. (1995). Developmental expression of a type II collagen/beta-galactosidase fusion gene in transgenic mice. _Dev. Dyn._204, 202–210 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay K., Lefebvre V., Zhou G., Garofalo S., Kimura J. H., de Crombrugghe B. (1995). Use of a new rat chondrosarcoma cell line to delineate a 119-base pair chondrocyte-specific enhancer element and to define active promoter segments in the mouse pro-alpha 1(II) collagen gene. _J. Biol. Chem._270, 27711–27719 [DOI] [PubMed] [Google Scholar]
- Murakami S., Nifuji A., Noda M. (1997). Expression of Indian hedgehog in osteoblasts and its posttranscriptional regulation by transforming growth factor-beta. _Endocrinology_138, 1972–1978 [DOI] [PubMed] [Google Scholar]
- Nakamura E., Nguyen M. T., Mackem S. (2006). Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. _Dev. Dyn._235, 2603–2612 [DOI] [PubMed] [Google Scholar]
- Ohba S., Kawaguchi H., Kugimiya F., Ogasawara T., Kawamura N., Saito T., Ikeda T., Fujii K., Miyajima T., Kuramochi A., et al. (2008). Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. _Dev. Cell_14, 689–699 [DOI] [PubMed] [Google Scholar]
- Ovchinnikov D. A., Deng J. M., Ogunrinu G., Behringer R. R. (2000). Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. _Genesis_26, 145–146 [PubMed] [Google Scholar]
- Razzaque M. S., Soegiarto D. W., Chang D., Long F., Lanske B. (2005). Conditional deletion of Indian hedgehog from collagen type 2alpha1-expressing cells results in abnormal endochondral bone formation. _J. Pathol._207, 453–461 [DOI] [PubMed] [Google Scholar]
- Reddi A. H. (1994). ‘Bone and cartilage differentiation. _Curr. Opin. Genet. Dev._4, 737–744 [DOI] [PubMed] [Google Scholar]
- Schinke T., Karsenty G. (1999). Characterization of Osf1, an osteoblast-specific transcription factor binding to a critical cis-acting element in the mouse osteocalcin promoters. _J. Biol. Chem._274, 30182–30189 [DOI] [PubMed] [Google Scholar]
- Sowa H., Karsenty G. (2007). ATF4 is a key molecule linking food intake and skeletal development. _J. Musculoskelet. Neuronal Interact._7, 326–327 [PubMed] [Google Scholar]
- St-Jacques B., Hammerschmidt M., McMahon A. P. (1999). Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. _Genes Dev._13, 2072–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vortkamp A., Lee K., Lanske B., Segre G. V., Kronenberg H. M., Tabin C. J. (1996). Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. _Science_273, 613–622 [DOI] [PubMed] [Google Scholar]
- Wang W., Lian N., Li L., Moss H. E., Wang W., Perrien D. S., Elefteriou F., Yang X. (2009). Atf4 regulates chondrocyte proliferation and differentiation during endochondral ossification by activating Ihh transcription. _Development_136, 4143–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan Y., Lin Z. (2009). Expression of Indian Hedgehog signaling molecules in breast cancer. _J. Cancer Res. Clin. Oncol._135, 235–240 [DOI] [PubMed] [Google Scholar]
- Yang X., Ji X., Shi X., Cao X. (2000). Smad1 domains interacting with Hoxc-8 induce osteoblast differentiation. _J. Biol. Chem._275, 1065–1072 [DOI] [PubMed] [Google Scholar]
- Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H. C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T. M., et al. (2004). ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. _Cell_117, 387–398 [DOI] [PubMed] [Google Scholar]
- Yoshida C. A., Yamamoto H., Fujita T., Furuichi T., Ito K., Inoue K., Yamana K., Zanma A., Takada K., Ito Y., et al. (2004). Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. _Genes Dev._18, 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T., Hinoi E., Jung D. Y., Kajimura D., Ferron M., Seo J., Graff J. M., Kim J. K., Karsenty G. (2009). The transcription factor ATF4 regulates glucose metabolism in mice through its expression in osteoblasts. _J. Clin. Invest._119, 2807–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Garofalo S., Mukhopadhyay K., Lefebvre V., Smith C. N., Eberspaecher H., de Crombrugghe B. (1995). A 182 bp fragment of the mouse pro alpha 1(II) collagen gene is sufficient to direct chondrocyte expression in transgenic mice. _J. Cell Sci._108, 3677–3684 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material