Ubiquitination, localization, and stability of an anti-apoptotic BCL2-like protein, BCL2L10/BCLb, are regulated by Ubiquilin1 (original) (raw)
Abstract
We have previously shown that all six members of the anti-apoptotic BCL2 gene family can cooperate with (myelocytomatosis oncogene) MYC in a mouse model of leukemia, but three of them are significantly less potent contributors to leukemogenicity than the other three. The protein encoded by one of these less potent genes, BCL2L10/BCLb, was recently shown to vary dramatically in many primary human cancers by immunohistochemistry, and the protein levels were inversely correlated with survival in patients with several cancer types. We examined BCLb mRNA in a panel of human cancer cell lines and did not observe the extensive variation in mRNA that would be required to explain the vast differences in protein levels. We found that the levels of BCLb protein diminish quickly after inhibition of protein synthesis with cycloheximide, so we searched for interacting proteins that might affect posttranslational stability of BCLb. Using a variety of approaches, including immunoaffinity and mass spectrometry, we identified a protein, Ubiquilin1 (Ubqln), that specifically interacts with BCLb, and not with other anti-apoptotic BCL2-like proteins. Ubqln stabilizes BCLb protein, while also promoting monoubiquitination on multiple lysine residues and relocation to the cytosol. Furthermore, primary lung adencarcinomas have more Ubqln mRNA than normal adjacent lung tissue, and higher Ubqln mRNA levels are associated with shorter survival of lung cancer patients, suggesting that potentiation of the anti-apoptotic potential of BCLb through regulation of its stability by Ubqln may be an important factor in tumor progression.
Keywords: apoptosis, PLIC, BH3
Disruption of biological pathways that regulate a cell’s normal “life versus death” decisions is a hallmark of cancer cells (1). Whether this disruption is caused by aberrant oncogene activation, loss of tumor suppressors, or mutation in genes directly involved in apoptotic decisions, the outcome is increased cell number. According to most models, the proapoptotic members of the BCL2 family of proteins are required for nearly all forms of cellular death, including apoptosis caused by intrinsic signals within all cells and extrinsic signals that affect some cells (2–4). Conversely, the anti-apoptotic BCL2 proteins are required to sustain cell viability during normal homeostasis. Loss of proapoptotic or gain of anti-apoptotic BCL2 proteins probably contributes to survival of most, if not all, human cancer cells (5–7).
The six anti-apoptotic members of the BCL2 family are BCL2, BCLxl (BCL2L1), BCLw (BCL2L2), BFL1 (BCL2A1), MCL1, and BCLb (BCL2L10) (8). Until recently it was assumed that aberrant expression of genes that encode any of these six proteins could participate in tumorigenesis with equivalent potency. Using a (myelocytomatosis oncogene) MYC-driven mouse model of myeloid leukemogenesis, we demonstrated directly that expression of cDNA’s encoding any of the six anti-apoptotic BCL2-like proteins can potentiate leukemogenesis (9). However, we observed significant differences in the potency of individual BCL2 family cDNA’s in cooperation with MYC in this model: the genes fall into two distinct classes, based on cooperative potential, with BCL2, BCLxL, and BCLw more potent than BCLb, BFL1, and MCL1. These findings suggested that there may be unrecognized factors that influence how effectively an anti-apoptotic gene can contribute to tumorigenesis.
Because anti-apoptotic BCL2 gene products are believed to be important in human cancers, multiple inhibitors of BCL2 proteins have been developed and are being tested in clinical trials (10–12). Many of these inhibitors have demonstrated impressive therapeutic efficacy in mouse models and human cell lines, but they have limited specificity. One small molecule, ABT-737, inhibits the three most potent anti-apoptotic BCL2 proteins, but its effect is limited by the frequent appearance of primary or acquired drug resistance (13). In many instances, resistance is due to high level expression of the one of the three BCL2 proteins not inhibited by the drug (13). A better understanding of the biological and biochemical regulation of each of the anti-apoptotic BCL2 proteins is likely to improve our ability to target this class of proteins and the pathways that regulate their function with additional drugs.
There is intense interest in understanding the mechanisms that lead to aberrant expression of proteins that can contribute to oncogenic transformation. By examining the stability, abundance, and interacting partners of anti-apoptotic BCL2 proteins, we have begun to define mechanisms that might regulate these proteins. Here we have identified Ubiquilin1 (Ubqln) as a specific interaction partner of BCLb. Following interaction with Ubqln, BCLb is apparently monoubiquitinated on multiple lysine residues and subsequently stabilized. Concurrent with Ubqln-stimulated ubiquitination, BCLb is relocalized from membranes to the cytoplasm. We have also observed a significant increase in Ubqln levels in primary human lung cancer samples compared to adjacent normal tissue, and this increase is associated with decreased survival of patients with lung adenocarcinoma.
Results
We recently demonstrated that cDNA’s encoding all six anti-apoptotic BCL2-family proteins cooperate with MYC in a mouse model of leukemogenesis, but with significant differences in the oncogenic potency of individual family members (9). Mice receiving cells that expressed the MYC oncogene and BCL2, BCLxl,, or BCLw cDNA’s developed aggressive myeloid leukemia with a much shorter latency than observed in mice receiving cells that express MYC and cDNA’s for BCLb, MCL1, or BFL1. A number of possibilities could explain this observation, including inherent differences in protein function, protein-protein interactions, mRNA translation rates, or differences in mRNA or protein stability. For example, a more labile protein would be less abundant and potentially less able to contribute to oncogenic transformation.
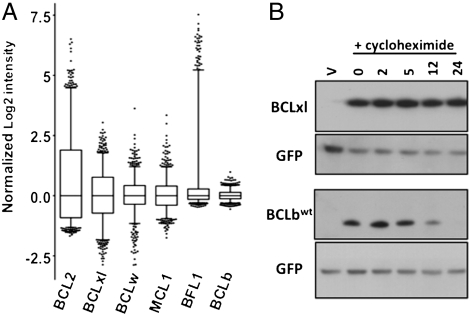
The Reed laboratory has demonstrated that high levels of BCLb protein are detected in many primary human cancers (14). In fact, in many cancer types, the levels of BCLb protein correlated inversely with patient survival, with higher BCLb levels associated with more advanced disease or a poorer prognosis. A common mechanism that leads to increased protein level is increased production of mRNA. However, upon examination of the Sanger Institute’s Cancer Cell Line Project microarray database of 732 cancer cell lines (http://www.broadinstitute.org/cgi-bin/cancer/datasets.cgi), we noted that BCLb was the only BCL2-family gene for which the mRNA concentration was basically invariant (Fig. 1A). These data suggest that there are alternative mechanisms that lead to alterations in BCLb protein levels in human cancers.
Fig. 1.
BCLb is a labile protein that is not regulated by alterations in mRNA levels. (A) Invariant levels of BCLb mRNA in cell lines. The mRNA levels for all six anti-apoptotic BCL2 family genes were analyzed in a set of 732 cancer cell lines from diverse tissues of origin using microarray data from the Sanger Institute’s Cancer Cell Line Project (http://www.sanger.ac.uk/genetics/CGP/CellLines/). Normalized log2 intensity values for each gene were median-centered across all the cell lines and plotted to show the variation in expression. Box-plots depict the median group expression (mid line), the 25th and 75th percentiles (box), and the limits of 95% of samples for each group “whiskers” with values for all other samples falling outside of this range represented by black dots. (B) Varied response of BCL2 proteins following cycloheximide treatment. 293T cells were transfected with plasmids expressing GFP and BCLb or BCLxl. Forty-eight hours posttransfection cells were left untreated (0) or treated with cycloheximide (+cycloheximide) for the indicated amount of time (in hours). Western blot analysis of lysates was performed with antibodies specific to BCLb or BCLxl. GFP, expressed from the same mRNA as the respective BCL2-proteins, was detected as a loading control. V = empty vector containing only GFP.
We then examined if the stability of the individual BCL2 proteins could explain the differences in oncogenic potency. Levels of BCLxl (a potent MYC cooperator) and BCLb (a less potent MYC cooperator) were examined following inhibition of the translational machinery with cycloheximide (Fig. 1B). BCLxl and BCLb were produced in cells by introducing plasmids that each encode a BCL2 family member, as well as GFP following an internal ribosomal entry sequence. We performed a Western blot on all lysates, using an antibody against GFP as an internal control, because GFP is known to have a long half-life and is encoded by the same mRNA as the BCL2 family protein. Western blots of the BCL2 proteins from lysates of cells treated for 0, 2, 5, 12, and 24 h with cycloheximide demonstrated striking differences in the abundance of BCLxl and BCLb, with BCLxl showing no decrease and BCLb showing a dramatic decrease, when compared to the untreated cells (Fig. 1B), implying that BCLb protein is inherently less stable in this cell type.
To determine if this observation could be extended to the other BCL2 proteins, we expanded the analysis to include other less potent MYC collaborators, MCL1 and BFL1, as well as BCLw, another potent MYC cooperator (Fig. S1). BCLb, MCL1, and BFL1 levels fell dramatically in cells following cycloheximide treatment, whereas levels of BCL2 proteins that were more potent in cooperating with MYC (BCLw and BCLxl) were unaffected when protein synthesis was blocked with cycloheximide . These observations suggest that the relative stability of BCL2 proteins, as judged by the relative decay of the individual proteins following cycloheximide treatment, may reflect an underlying mechanism that determines oncogenic potential.
Stabilization of Bclb Protein Leads to More Potent Oncogenicity.
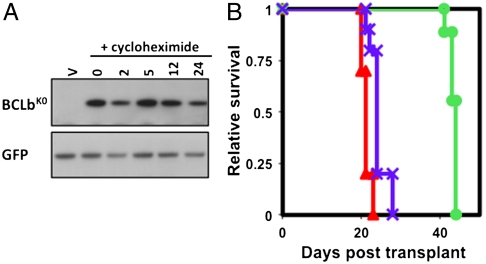
Protein degradation is frequently mediated by ubiquitination of proteins on target lysine residues, so we asked whether the instability of BCLb that we observed in cells is also mediated by a lysine-dependent, ubiquitin-mediated mechanism. To this end, we engineered a BCLb protein devoid of all four lysine residues (BCLbK0); this protein cannot be modified by the traditional ubiquitination machinery. The stability of BCLbK0 was compared to that of wild-type BCLb (BCLbwt) following treatment with cycloheximide (Fig. 2A). BCLbK0 protein levels did not change during 24 h of cycloheximide treatment. These findings reemphasize the idea that BCLb is turned over by a lysine- and ubiquitin-dependent mechanism that determines protein decay following translation inhibition.
Fig. 2.
Lysineless BCLb (BCLbK0) is more stable and a more potent oncogene than wild-type BCLb. (A) Stability of BCLbK0 protein following inhibition of protein synthesis. 293T cells were transfected with BCLbK0 and 48 h posttransfection cells were left untreated (0) or treated with cycloheximide (+cycloheximide) for the indicated amount of time (in hours). BCLb protein levels were determined with an anti-BCLb antibody. Bottom box, western blot analysis for GFP. V = empty vector containing only GFP (B) BCLbK0 is a more potent oncogene. Kaplan-Meier survival analysis of mice reconstituted with cells expressing MYC and BCLbwt (circles), BCLbK0 (crosses) or BCLxl (triangles) (plot is a combination of two independent experiments where nine animals were followed for each group; n = 9).
BCLbK0 is more stable than BCLbwt, so we asked whether BCLbK0 is more potent than BCLbwt in cooperating with MYC during leukemogenesis using the bone marrow infection and transplantation strategy, as previously described (9, 15). Briefly, we infected bone marrow cells from mice harboring a tetracycline operator-regulated MYC transgene (Tet-O-MYC) with a murine retroviral vector (MSCV) encoding BCLbwt, BCLbK0, or BCLxl, and a tetracycline transactivator (Fig. 2B). The survival times of mice receiving transplants showed that infection with vectors encoding BCLbK0 causes a more aggressive leukemia than does infection with vectors encoding BCLbwt. In fact, mice that received cells expressing MYC and BCLbK0 succumbed to leukemia just as quickly as mice that received cells expressing MYC and BCLxl. Thus, by removing the residues critical for degradation of BCLb, we created a more potent oncogene, implying that a mechanism that stablizes BCLb might potentiate its oncogenic effect.
Ubqln Interacts with Bclb.
The results in the preceding section indicate that proteins capable of affecting the stability of BCLb might have an important role in neoplasia. For example, proteins that normally promote the degradation of BCLb could be inactivated or sequestered, producing aberrantly high levels of BCLb that contribute to tumorigenesis. We therefore set out to identify proteins that interact with BCLb by performing immunoprecipitations of FLAG epitope-tagged BCL2 proteins, followed by mass spectrometry to identify interacting peptides. In initial experiments, we compared peptides from anti-FLAG immunoprecipitates from lysates of 293T cells containing FLAG-BCLb, FLAG-BCLw, FLAG-BFL1, or only the FLAG epitope, with the goal of identifying proteins that associate specifically with BCLb and regulate its stability.
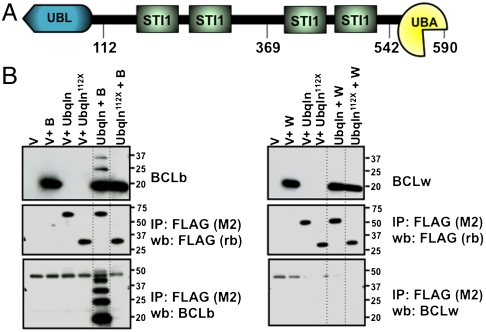
In triplicate biological experiments, the most abundant protein found by mass spectrometry in FLAG-BCLb immunoprecipitates, was a multidomain protein named Ubqln. No Ubqln peptides were identified in FLAG immunoprecipitates from FLAG-BCLw, FLAG-BFL1, or control lysates (Table S1). In previous reports, Ubqln has been shown to contain a ubiquitin-like (UBL) domain, a ubiquitin-associated (UBA) domain, and multiple heat-shock chaperonin (STI) domains (Fig. 3A) (16–18). The domain structure and previous literature on Ubqln suggest that it interacts with ubiquitinated proteins through its UBA domain and with the proteasome through its UBL domain (19, 20). Based on these interactions, Ubqln is thought to serve as a bridge between ubiquitinated proteins and the proteasome to facilitate degradation of the ubiquitinated targets.
Fig. 3.
Ubqln interacts specifically with BCLb. (A) Schematic of the Ubquilin1 protein. UBL domain; STI domain; UBA domain. The numbers below the diagram indicate amino acid residues. (B) Ubqln interacts specifically with BCLb. 293T cells were transfected with plasmids containing BCLb (B) and FLAG-tagged Ubqln (left box) or BCLw (W) and FLAG-tagged Ubqln (right box). Forty-eight hours posttransfection cell lysates were prepared and immunoprecipitations were performed with monoclonal anti-FLAG antibodies (M2) (to immunoprecipitate Ubqln-containing complexes). Western blots were performed to detect Ubqln (FLAG rb; poly-clonal FLAG antibody; middle box) and BCLb or BCLw (bottom box). Top box represents 10% of input used for immunoprecipitations. V = empty vector, Ubqln112_X_ = Ubqln encoding the first 112 amino acid of Ubqln containing only the UBL domain. The band migrating at approximately 40 kDa in both the right and left bottom boxes is a nonspecific band detected by the antibodies.
Recently, Ubqln has been shown to play a role in a variety of other cellular processes, including autophagy, ER-associated protein degradation (ERAD) and receptor trafficking, presumably by regulating the abundance of proteins implicated in these events (21–30). Furthermore, UBQLN is one member of a family of at least five proteins (UBQLN1, UBQLN2, UBQLN3, UBQLN4, and UBQLNL) that share a high degree of similarity at the level of both amino acid and domain structure. In fact, in multiple experiments, we also identified Ubiquilin2 as a specific BCLb-interacting protein, increasing our confidence in the specificity and robustness of the methods we have used to detect proteins bound to BCLb.
To validate the specificity of the mass spectrometry result indicating that BCLb, but not other BCL2 proteins, interact with Ubqln, we repeated the immunoprecipitation experiment in “reverse” by coexpressing untagged BCLb or BCLw with FLAG-tagged Ubqln in 293T cells (Fig. 3B). Following immunoprecipitation of Ubqln, we used antibodies specific for BCL2 family members to detect associated proteins by Western blotting. In agreement with our mass spectrometry data, we detected BCLb, but not BCLw, in the anti-FLAG-Ubqln immunoprecipitates. We then used the same strategy with the additional four anti-apoptotic BCL2 proteins and did not observe any interaction with Ubqln, indicating that Ubqln binds specifically to BCLb among this protein family (Fig. S2). In addition to a band migrating at the size corresponding to unmodified BCLb, multiple forms of BCLb were detected in both the immunoprecipitated sample and the unfractionated extract of cells coexpressing FLAG-Ubqln and BCLb. Because Ubqln is claimed to bind ubiquitinated proteins through the UBA domain and we have presented evidence that BCLb can be degraded by a lysine-dependent mechanism, we suspected that the slower migrating bands correspond to ubiquitinated forms of BCLb.
Ubqln Leads to Ubiquitination of Bclb.
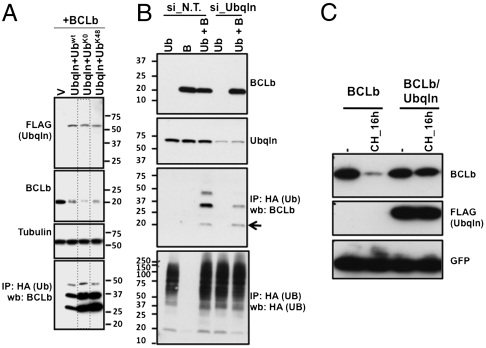
To demonstrate directly that the slowly migrating forms of BCLb seen in the presence of Ubqln are ubiquitinated forms of BCLb, we used an anti-HA antibody to immunoprecipitate ubiquitin-containing proteins from lysates after coexpressing BCLb, Ubqln, and different HA-tagged ubiquitin (Ub) constructs (wild-type Ub, lysineless Ub, or Ub containing only lysine 48) (Fig. 4A). We observed the same pattern of BCLb, consisting of multiple bands migrating at approximately 8 kDa intervals, following immunoprecipitation from cells expressing the three forms of ubiquitin. This finding is important because, when ubiquitin chains are conjugated with substrate proteins, the specific lysine residue of the ubiquitin protein used during conjugation can determine the functional outcome of ubiquitination (31–33). In some cases, ubiquitin can be added as a single conjugative event to the target protein, leading to changes in subcellular localization or other signaling events, but monoubiquitination is not recognized as a signal for proteasome-mediated destruction (34). Therefore, ubiquitin that does not contain any lysine residues (UbK0) can be conjugated to a target protein, but additional ubiquitin molecules cannot be added; in this way, single ubiquitin moieties are conjugated to lysine residues in the target protein (monoubiquitination) (35). Because the ubiquitination pattern of BCLb is the same with all ubiquitin mutants used in the experiment shown in Fig. 4 and because there are multiple forms of ubiquitinated BCLb observed, it is likely that the observable BCLb is monoubiquitinated on multiple lysine residues.
Fig. 4.
Ubqln promotes stabilization of BCLb and accumulation of monoubiquitinated BCLb. (A) BCLb is ubiquitinated on multiple lysine residues. Lysates were prepared from 293T cells expressing BCLb and FLAG-Ubqln, in addition to wild-type HA-ubiquitin (Ubwt), lysineless HA-ubiquitin (UbK0) or HA-ubiquitin-containing only lysine 48 (UbK48). Immunoprecipitations were performed with anti-HA antibodies (to immunoprecipitate Ubiquitin-containing proteins) and followed by Western blots with anti-BCLb antibodies (bottom box). Top three boxes represent 10% of the input used for immunoprecipitations, and anti-Tubulin antibodies were used to show equal amount of total protein extracts were used for immunoprecipitations. (B) Endogenous Ubqln can promote ubiquitination of BCLb. 293T cells were transfected with either nontargeting siRNA (si_N.T.) or siRNA against Ubqln (si_Ubqln). Twenty-four hours later these cells were transfected with vectors encoding BLCb (B) and/or HA-tagged ubiquitin (Ub). Twenty-four hours later cell lysates were prepared and immunoprecipitations were performed with anti-HA antibodies (to immunoprecipitate Ubiquitin-containing proteins) and Western blots to detect BCLb (middle lower box) and total ubiquitinated proteins (lower box) were performed. Top two boxes each represent 10% of the input used for immunoprecipitations. Endogenous Ubqln levels were detected with a poly-clonal antibody raised against Ubqln. The arrow in the middle lower box represents the nonubiquitinated form of BCLb. (C) Ubqln stabilizes BCLb following cycloheximide treatment. Cells expressing BCLb, or BCLb and FLAG-Ubqln were treated with cycloheximide (CH) for 16 h. Cell lysates were prepared and Western blots were performed using anti-BCLb antibodies (top box), anti-FLAG antibodies to detect Ubqln (middle box) or anti-GFP antibodies as controls (bottom box).
To exclude the possibility that ubiquitination of BCLb was an artifact of overexpression of Ubqln, we used previously characterized siRNA directed against Ubqln to examine the effect of reduced levels of endogenous Ubqln on ubiquitination of BCLb. These siRNAs reduced endogenous Ubqln levels, and a concomitant decrease in the levels of ubiquitinated BCLb was observed (Fig. 4B, 21).
We have demonstrated that Ubqln interacts with BCLb and that this interaction potentiates ubiquitination at multiple sites on BCLb. However, it is still not clear how Ubqln affects BCLb abundance and function. To determine if Ubqln can alter BCLb protein levels, we expressed BCLb alone or in combination with Ubqln and treated cells with cycloheximide (Fig. 4C). Coexpression of Ubqln stabilized BCLb in the presence of cycloheximide, in contrast to what is observed when BCLb is expressed alone (Fig. 1A). The stability of BCLb following cycloheximide in the presence of Ubqln was similar to that of BCLbK0 in the absence of exogenous Ubqln (Fig. 2A). These findings suggest that expression of Ubqln stabilizes BCLb.
Ubqln Affects Subcellular Localization of Bclb.
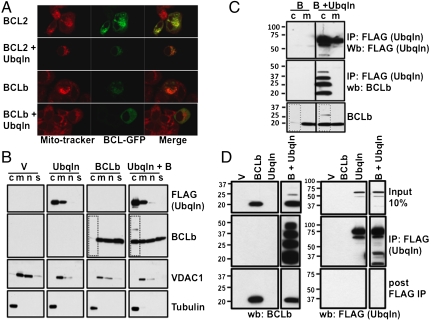
Ubqln stabilizes BCLb and apparently promotes monoubiquitination of BCLb. A known function of monoubiquitination is to alter the cellular location of proteins. We therefore wanted to determine if increased levels of Ubqln can alter localization of BCLb. We first compared cells programmed to express GFP-tagged BCLb alone or GFP-tagged BCLb and Ubqln. After 48 h of transfection, cells were stained with a mitochondrial dye, then viewed with confocal microscopy (Fig. 5A). In the absence of exogenous Ubqln, BCLb colocalizes almost exclusively with mitochondria, whereas, when Ubqln is coexpressed, BCLb protein appears in extramitochondrial cytoplasmic sites. In a parallel experiment with GFP-BCL2, we did not observe extramitochondrial BCL2 when it was coexpressed with Ubqln.
Fig. 5.
Ubqln alters subcellular localization of BCLb, and preferentially interacts with BCLb in the cytoplasm. (A) Ubqln alters the subcellular localization of BCLb, but not BCL2. H358 cells expressing GFP-tagged BCLb or GFP-tagged BCL2 and either empty vector or a vector containing Ubqln were treated with Mito-Tracker dye and subjected to confocal microscopy. (B) Ubqln promotes the accumulation of ubiquitinated BCLb in the cytosol. 293T cells expressing BCLb, Ubqln, or BCLb and Ubqln were subjected to subcellular fractionation by differential centrifugation, as described in Materials and Methods, followed by Western blots. Cells were fractionated into n-nuclear, m-membranes, c-cytosolic, and s-structural/cytoskeletal compartments. Western blots of VDAC1, a membrane bound protein, and tubulin, a cytoplasmic protein, anti-sera were used to document the validity of the fractionations. (C) Ubqln and BCLb interact in the cytosol. Fractionated lysates expressing either BCLb or BCLb and Ubqln were subjected to immunoprecipitation with anti-FLAG antibody to detect Ubqln interactions. Only the cytosolic (c) and membranous (m) fractions were used, as these were the only fractions previously determined to contain BCLb (B) or Ubqln. Following anti-FLAG immunoprecipitations, Western blots were performed to detect Ubqln and BCLb. BCLb was also detected using 10% of the total protein used for immunoprecipitations (bottom box). (D) Ubiquitinated BCLb interacts preferentially with Ubqln. Cell extracts containing BCLb, FLAG-Ubqln, or both BCLb and FLAG-Ubqln were used to immunoprecipitate Ubqln using anti-FLAG antibody. Following immunoprecipitation, Western blots were performed with both the precipitate (IP: FLAG) and the remaining supernatant (post FLAG IP). Antibodies against BCLb and against FLAG (to detect Ubqln) were used to probe the Western blots.
We then performed biochemical fractionation of cells to confirm and extend these observations. Western blots of fractionated lysates of cells expressing BCLb and/or Ubqln were performed for the two proteins (Fig. 5B). In accord with other groups, we found BCLb in membrane fractions, excluded from the cytosolic fractions (36). In cells expressing exogenous Ubqln, a large portion of BCLb was found in cytosolic fractions (see boxed area). Because BCLb in the cytosolic fraction appeared to be ubiquitinated, it may be a signal for relocalizing the BCLb protein from the membranes into the cytosol.
To determine where Ubqln and BCLb interact within the cell, we attempted to immunoprecipitate Ubqln from the cytosolic and membrane fractions (Fig. 5C). Although Ubqln and BCLb could be readily detected in both the cytosolic and membrane fractions, we were able to coprecipitate Ubqln and BCLb only from the cytosolic fraction. This result indicates that the presence of the two proteins within the same compartment, such as membranes, is not sufficient to elicit interaction, so specific signaling events are likely required for an interaction to occur.
To determine whether Ubqln preferentially interacts with ubiquitinated BCLb or facilitates ubiquitination of BCLb, we immunoprecipited Ubqln from whole cell lysates and sought ubiquitinated and nonubiquitinated forms of BCLb in the immunoprecipates and in the remaining supernatant (Fig. 5D). We observed nonubiquitinated BCLb in both the Ubqln immunoprecipitate and the remaining supernatant, but the ubiquitinated forms of BCLb were detected only in the fraction that was immunoprecipitated with Ubqln. All Ubqln was effectively removed by the immunoprecipitation, because we were not able to detect Ubqln in the depleted supernatant. These data demonstrate that the ubiquitinated fraction of BCLb is stably and preferentially bound to Ubqln in cells.
Ubqln in Human Cancer.
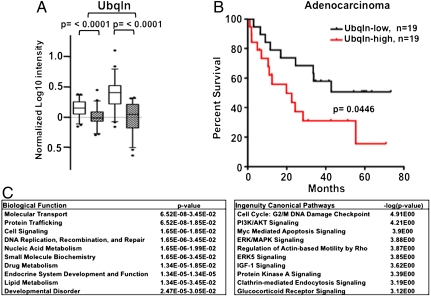
We wanted to determine if variation in Ubqln mRNA could be observed in human cancer samples. Previous work had shown that Ubqln mRNA and protein levels are increased in primary lung adenocarcinomas, a cancer type that also features high BCLb protein levels (14, 37). To build on these observations, we examined Ubqln mRNA in an independent set of 46 primary adenocarcinomas and matched normal lung samples (38). Both Ubqln probes demonstrated significantly increased mRNA levels in the primary adenocarcinoma samples compared to their corresponding normal lung samples (p < 0.0001 for both probes, shaded boxes correspond to “normal lung” samples, Fig. 6A).
Fig. 6.
Levels of Ubqln RNA in human lung adenocarcinoma predict survival outcome in patients. (A) Ubqln RNA was analyzed in a set of 46 tumor/normal pairs from the same patients. The microarray platform contained two individual probes for each gene that are depicted separately. Normalized log 10 expression values for each probe were compared between the normal and tumor samples using a two-tailed Wilcox matched-pairs sign rank test. The expression values for each data point were normalized to the average probe expression of the respective normal samples before plotting. Shaded boxes represent “normal” samples; open boxes represent “tumors.” (B) High Ubqln mRNA levels are associated with poor survival in lung adenocarcinoma. A set of 58 lung adenocarcinomas were separated into tertiles based on levels of Ubqln mRNA expression. Overall survival curves for the top and bottom tertiles were then plotted and compared using the Log-rank (Mantel-Cox) test. There is a significant association between the highest tertile of Ubqln RNA and poor prognosis (p = 0.0446). (C) IPA was performed on the 343 genes that were found to be significantly different between the Ubqln-high and Ubiqln-low human lung adenocarcinoma samples.
To investigate the potential clinical significance of expression of Ubqln in patients with lung adenocarcinoma, we asked whether there was any significant relationship between Ubqln mRNA levels and patient outcome. Although limited by the number of published studies that determined Ubqln RNA and included clinical information, we found one lung adenocarcinoma dataset that met these criteria (Duke GEO accession #GSE3141) (39). Patients in that dataset could be stratified into groups with significantly different overall survival based on their level of Ubqln mRNA: individuals with relatively high levels of Ubqln mRNA had a poorer outcome (Fig. 6B).
Because tumors with higher levels of Ubqln mRNA are associated with a shorter survival time, we asked whether other genes are differentially expressed in these two groups. The Significance Analysis of Microarrays program revealed 343 genes that were differentially expressed (fold-change greater than 2 or less than 0.5, _q_-value threshold = 0) between the “Ubqln high” and the “Ubqln low” expressing groups (40). Ingenuity Pathway Analysis (IPA) was then applied to this gene set to examine the genes in a systematic manner and determine their association with specific biological functions or canonical biological pathways (Fig. 6C). Molecular transport and protein trafficking, two processes in which Ubqln is thought to play a major role, were among the top ten most significant annotated biological functions represented by this gene set. Within the canonical pathways that are significantly represented in this gene set are many signaling pathways that are known to be involved in the process of neoplastic transformation, including MYC signaling, cell cycle regulation, and AKT signaling. Whether genes on this list represent functions and pathways related to, or regulated by, Ubqln or are simply representative of more aggressive tumors will require further analysis.
Discussion
We have sought to understand how the six anti-apoptotic BCL2 proteins can exhibit different activities in a cooperative mouse model of MYC-induced leukemia (9), and have shown that the strength of the cooperative oncogenic activity correlates with the abundance of the proteins (Fig. 1A and 2B). In an effort to explain these differences we performed an unbiased screen to identify proteins that interact with BCL2 proteins, using immunoprecipitation followed by mass spectrometry. We identified and characterized one such interacting protein, Ubqln that interacts specifically with BCLb. Interaction between Ubqln and BCLb results in a number of alterations to the BCLb protein, including accumulation of BCLb that is ubiquitinated on multiple lysines, stabilization following treatment of cells with cycloheximide, and subcellular relocalization of BCLb.
Under normal homeostatic conditions, BCLb protein is regulated by a lysine- and presumably proteasome-dependent mechanism. If ubiquitination of BCLb is blocked, by altering all of the lysine residues, for example, BCLb is stabilized, producing a substantial increase in the oncogenic potential of BCLb in our model of leukemia. Furthermore, we have extended previously published studies and shown that Ubqln is highly expressed in some human lung adenocarcinomas, and patients with higher Ubqln expression have shorter survival times. We suggest the possibility that the increase in BCLb protein levels, observed by others, could be due to high expression of Ubqln also found in these cancers. The exact role of Ubqln in human cancers requires further examination.
Ubqln contains a UBA domain and UBL domain and is thought to be involved in ERAD, among other processes. Ubqln binds ubiquitinated proteins through the UBA, at the same time binding the proteasome through the UBL; it appears to act as a scaffold to facilitate proteasomal degradation of ubiquitinated proteins that have been exported from the ER (24, 25). This function could explain most of the proposed functions of Ubqln, such as regulation of autophagy, unfolded protein response, and interaction with both poly-alanine and poly-glutamine expanded proteins (21, 22, 27–30, 41). Additional, nonERAD dependent functions for Ubqln, and related proteins, have also been proposed, including a role in the trafficking of amyloid precursor protein and endocytosis of G-protein coupled receptors (18, 21, 23, 26).
We have proposed a mechanism by which Ubqln could regulate apoptosis. Following expression of Ubqln there is an accumulation of both ubiquitinated and nonubiquitinated BCLb that is relocated to the cytosol. The mechanisms that lead to the ubiquitination and the function of this form of BCLb are not well understood. Stabilization of BCLb by Ubqln involves more than binding of Ubqln to ubiquitinated BCLb: Ubqln interacts with both nonubiquitinated BCLb and ubiquitinated BCLb, and the nonubiquitinated form appears to be stabilized as revealed after protein synthesis is arrested with cyclohexamide. We are attempting to determine the sites of interaction between BCLb and Ubqlin, any new function of BCLb located in the cyctosol after interaction with Ubqln, and the effects of the BCLb-Ubqln complex on ubiquitin ligases that might be responsible for the increase in ubiquitinated BCLb following interaction with Ubqln.
It is also unclear how BCLb translocates from membranes into the cytosol and is stabilized following interaction with Ubqln. At least two models could explain how expression of Ubqln might relocalize BCLb to the cytosol. The first posits that Ubqln binds to BCLb on membranes, leading to release into the cytosol; the second argues that BCLb is released from membranes by an unknown event and will interact with any available Ubqln. Once Ubqln and BCLb interact, BCLb might not be capable of reestablishing interaction with membranes. If the first model were true, we would expect to see interaction between Ubqln and BCLb in both the cytosolic and membrane fractions; in the second model, interaction would be observed only in the cytosol. In Fig. 5C we show that although BCLb and Ubqln are present in both the cytoplasmic and the membrane fractions, we were able to detect the interaction of BCLb and Ubqln only in the cytoplasm. This result suggests that Ubqln likely interacts with BCLb only after BCLb is released from membranes, although we cannot formally rule out the possibility that interaction between Ubqln and BCLb leads to an immediate release of both proteins from membranes.
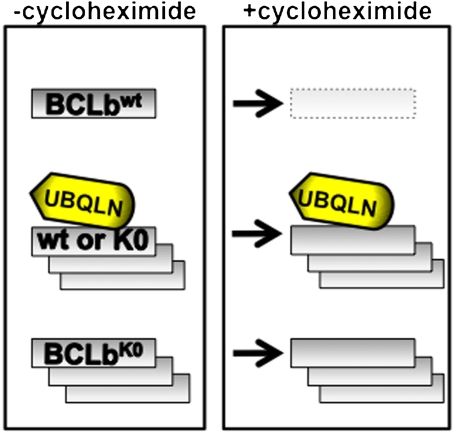
A particularly interesting aspect of this work is the observation that BCLb can be regulated by mechanisms that have not previously been reported (Fig. 7). One well established mechanism is illustrated following treatment with cycloheximide, when BCLbwt is apparently degraded by a lysine-dependent mechanism, whereas the levels of BCLbK0, which lacks lysine residues, are unaffected after 20 h of translational inhibition (Fig. 1B, Fig. 4C, and Fig. 7, +cycloheximide). We have presented data to indicate that this protection may arise from the ability of Ubqln to sequester BCLb in the cytoplasm. We currently favor a model whereby interaction of BCLb or BCLbK0 with Ubqln in the cytoplasm protects BCLb from signals that would lead to its degradation. Once the degradation signals have been attenuated, BCLb might be released from Ubqln and resume its normal function in the membrane-associated fraction. Such actions could help to explain the relationship between Ubqln levels and survival, responses of lung adenocarcinomas to therapy, or both.
Fig. 7.
The regulation of concentrations of BCLb by Ubiquillin and ubiquitination. Treatment of cells expressing BCLbwt or BCLbK0 with cycloheximide lead to varied outcomes of the BCLb proteins depending on whether BCLb contains lysine residues or if Ubiquilin is present. Treatment of cells expressing BCLbwt with cycloheximide leads to rapid degradation of wild-type BCLb proteins (top). In contrast, treatment of cells expressing BCLbK0 with cycloheximide has no affect on BCLbK0 levels (bottom). When Ubqln is expressed with either BCLbwt or BCLbK0, there is a stabilization of BCLb proteins that is maintained following prolonged treatment with cycloheximide (middle).
Materials and Methods
Antibodies Used for Analysis in This Study.
BCL2 #50E3, BCLxl #54H6, BFL1 #4647, BCLw, #31H4 and VDAC #4866 (Cell Signaling); MCL1 #559027 (Becton Dickenson); BCLb #ab45412 and GFP #ab6673 (Abcam); tubulin #B512, FLAG M2 conjugated agarose beads, FLAG poly-clonal #F7425 (Sigma); Ubqln poly-clonal was made by inoculating rabbits with a peptide specific to Ubqln1 (Yenzym Antibodies LLC); HA affinity matrix and HA #3F10 (Roche).
Bone Marrow Harvest, Infection, and Transplantation Assay.
Bone marrow was flushed from the femurs and tibias of 6 to 8 w old, previously untreated, donor mice that carried the Tet-O-MYC transgene. Red blood cells were lysed using Red Blood Cell Lysis buffer (Sigma). Infections were then carried out in the presence of poybrene (4 ug/mL) for 1 h during centrifugation (2,000 × g @ 30 °C) using MSCV based retroviral supernatants packaged in 293T cells. After infection cells were immediately transplanted into the tail vein of lethally irradiated FVB/n recipients acquired from Taconic.
Tissue Culture and Protein Analysis.
293T cells were cultured in DMEM supplemented with 10% FBS. DNA ransfections were done using Fugene6, as per the suppliers recommendations. siRNA transfections were done using Dharmafect1, as per the suppliers recommendations. The siRNA sequences used are as follows: nontargeting siRNA#1 catalogue #D-001810-01 (dharmacon), Ubqln1-GAAGAAAUCUCUAAACGUUUU. All cell extracts were prepared following scrape harvesting of 293T cells using CHAPS lysis buffer (1% CHAPS detergent, 150 mM NaCl, 50 mM Tris pH 7, 5 mM EDTA), except subcellular fractionation experiments, which were performed using the ProteoExtract Subcellular Proteome Extraction kit (Calbiochem/Merck), as per the manufacturer’s protocol. For immunoprecipitations, 200 ug of protein was incubated in 400 uL of total CHAPS buffer and incubated with indicated affinity matrix for 1 h at 4 °C. Following incubation, the matrix was washed three times in CHAPS buffer and then SDS loading buffer was added directly to washed matrix, boiled, and loaded directly into the wells of a PAGE gel. Drug treatments were performed as described in the text using 20 uM cycloheximide or 25 uM proteasome inhibitor MG132. For confocal microscopy, H358 cells were transfected with the indicated plasmids using Fugene6, as per the manufacturer’s protocol. Forty hours after transfection cells were treated with 100 nM MitoTracker Red CMXRos for 30 min. Cells were then washed one time with 1X PBS and replenished with fresh DMEM. Live cells were then subjected to confocal microscopy using a Zeiss LSM 510 NLO Meta System. Multiple images were acquired from multiple experiments and representative images are presented.
Plasmids.
All experiments using expression of BCL2 proteins were performed with vectors engineered for this study. All non-epitope-tagged BCL2-proteins were subcloned into the murine stem cell virus based retroviral vector MIGRX (MIGRX was created by altering the order of restriction enzyme sites in the multiple cloning site of the MIGR1 vector that was originally provided by W. Pear, University of Pennsylvania). The following accession numbers were used to design oligos for PCR based cloning from cDNA libraries generated from a “universal human RNA” library (Stratagene/Agilent Technologies): BCL2 #NM_000633, BCLxl #NM_138578, BCLw #NM_004050, MCL1 #NM_021960, BFL1 #NM_004049, BCLb #NM_020396. FLAG-tagged BCL2 constructs were generated by performing PCR of the MIG-BCL constructs using an oligo that fused the FLAG coding sequences to the amino terminus of each BCL2-protein. This product was then subcloned back into the MIGRX vector. Retroviral vectors used for the in vivo leukemia experiments were described previously (9). The following plasmids were obtained from Addgene; FLAG-Ubqln (Addgene plasmid 8663, deposited by Peter Howley), FLAG-Ubqln-112X (Addgene plasmid 8664, deposited by Peter Howley), pRK5-HA-Ubiquitin-WT (Addgene plasmid 17608, deposited by Ted Dawson), pRK5-HA-Ubiquitin-K0 (Addgene plasmid 17603, deposited by Ted Dawson), pRK5-HA-Ubiquitin-K48 (Addgene plasmid 17605, deposited by Ted Dawson), GFP-BCL2 (Addgene plasmid 17999, deposited by Clark Distelhorst). The GFP-tagged BCLb construct was purchased from OriGene catalogue #RG211604. Lysineless BCLb construct was engineered using the MIG- or MIT-BCLb constructs described above to do site directed mutagenesis with the Quick Change Multi site Mutagenesis kit (Stratagene/Ambion), as per the manufacturer’s protocol.
Mass Spectrometry.
FLAG-tag purified protein complexes were partially resolved using SDS-polyacrylamide gel electrophoresis; the mixtures were concentrated into a single, 3-mm wide “stack” by electrophoresing through an SDS “stacking gel” until entering the “separation gel,” followed by brief staining with Coomassie Blue and excision of the stacked protein band. In situ trypsin digestion of polypeptides in each gel slice was performed as described (42). The tryptic peptides were purified using a 2-μL bed volume of Poros 50 R2 (Applied Biosystems) reversed-phase beads packed in Eppendorf gel-loading tips (43). The purified peptides were diluted to 0.1% formic acid and then subjected to nano-liquid chromatography coupled to tandem mass spectrometry (nanoLC-MS/MS) analysis as follows. Peptide mixtures (in 20 μL) were loaded onto a trapping guard column (0.3 × 5 mm Acclaim PepMap 100 C18 cartridge from LC Packings) using an Eksigent nano MDLC system (Eksigent Technologies, Inc.) at a flow rate of 20 μL/ min. After washing, the flow was reversed through the guard column and the peptides eluted with a 5–45% acetonitrile gradient over 85 min at a flow rate of 200 nL/ min, onto and over a 75-micron × 15-cm fused silica capillary PepMap 100 C18 column (LC Packings). The eluant was directed to a 75-micron (with 10-micron orifice) fused silica nano-electrospray needle (New Objective). The electrospray ionization needle was set at 1,800 V. A linear ion quadrupole trap-Orbitrap hybrid analyzer (LTQ-Orbitrap, ThermoFisher) was operated in automatic, data-dependent MS/MS acquisition mode with one MS full scan (450–2,000 m/z) in the Orbitrap analyzer at 60,000 mass resolution and up to five concurrent MS/MS scans in the LTQ for the five most intense peaks selected from each survey scan. Survey scans were acquired in profile mode and MS/MS scans were acquired in centroid mode. The collision energy was automatically adjusted in accordance with the experimental mass (m/z) value of the precursor ions selected for MS/MS. Minimum ion intensity of 2,000 counts was required to trigger an MS/MS spectrum; dynamic exclusion duration was set at 60 s.
Initial protein/peptide identifications from the LC-MS/MS data were performed using the Mascot search engine (Matrix Science, version 2.3.02) with the human segment of International Protein Index protein database (89,952 sequences; European Bioinformatics Institute). The search parameters were as follows: (i) one missed cleavage tryptic site was allowed; (ii) precursor ion mass tolerance = 10 ppm; (iii) fragment ion mass tolerance = 0.8 Da; and (iv) variable protein modifications were allowed for methionine oxidation, cysteine acrylamide derivatization, and protein N-terminal acetylation. MudPit scoring was typically applied using significance threshold score p < 0.01. Decoy database search was always activated and, in general, for merged LS-MS/MS analysis of a gel lane with p < 0.01, false discovery rate averaged around 1%.
Scaffold (Proteome Software Inc.), version 3_1_2 was used to further validate and cross-tabulate the MS/MS-based peptide and protein identifications. Protein and peptide probability was set at 95% with a minimum peptide requirement of one.
Supplementary Material
Supporting Information
Acknowledgments.
We thank former members of the Varmus laboratory at Memorial Sloan-Kettering Cancer Center, especially G Sanchez, A Giannakou, R DeMatteo, and MA Melnick, for expert handling of the mouse colony, including genotyping. We thank current members of the Varmus laboratory at the National Institutes of Health (NIH), D O’Mard, J Idol, and S-Q Lee-Lin for technical support and A Unni for critical reading of manuscript and scientific discussions. We thank investigators that deposited plasmids at Addgene. We thank Laura Fabrizio for help with sample preparation for mass spectrometry. Funding for this project was provided, in part, by NIH P01 2P01CA094060-06A1 (to H.V.) and by intramural NCI funds. This work was also supported by NCI Cancer Center support Grant P30 CA08748 to Microchemistry and Proteomics Core Lab (H.E.-B.) and by funds from the James Graham Brown Cancer Center and the Kosair Pediatric Cancer Research Fund (L.J.B.).
Footnotes
The authors declare no conflict of interest .
See Author Summary on page 657.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 3.Horvitz HR. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 1999;59(7 Suppl):1701s–1706s. [PubMed] [Google Scholar]
- 4.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death . Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 5.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 6.Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103:839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 7.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 8.Lanave C, Santamaria M, Saccone C. Comparative genomics: the evolutionary history of the Bcl-2 family. Gene. 2004;333:71–79. doi: 10.1016/j.gene.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Beverly LJ, Varmus HE. MYC-induced myeloid leukemogenesis is accelerated by all six members of the antiapoptotic BCL family. Oncogene. 2009;28:1274–1279. doi: 10.1038/onc.2008.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen M, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci USA. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 12.Reed JC. Drug insight: cancer therapy strategies based on restoration of endogenous cell death mechanisms. Nat Clin Pract Oncol. 2006;3:388–398. doi: 10.1038/ncponc0538. [DOI] [PubMed] [Google Scholar]
- 13.Konopleva M, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Krajewska M, et al. Bcl-B expression in human epithelial and nonepithelial malignancies. Clin Cancer Res. 2008;14:3011–3021. doi: 10.1158/1078-0432.CCR-07-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beverly LJ, Capobianco AJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell. 2003;3:551–564. doi: 10.1016/s1535-6108(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 16.Kleijnen MF, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 17.Wu AL, Wang J, Zheleznyak A, Brown EJ. Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol Cell. 1999;4:619–625. doi: 10.1016/s1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- 18.Wu S, et al. Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim Biophys Acta. 2002;1542:41–56. doi: 10.1016/s0167-4889(01)00164-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 21.Hiltunen M, et al. Ubiquilin 1 modulates amyloid precursor protein trafficking and Abeta secretion. J Biol Chem. 2006;281:32240–32253. doi: 10.1074/jbc.M603106200. [DOI] [PubMed] [Google Scholar]
- 22.Kim SH, et al. Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J Biol Chem. 2009;284:8083–8092. doi: 10.1074/jbc.M808064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko HS, Uehara T, Nomura Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J Biol Chem. 2002;277:35386–35392. doi: 10.1074/jbc.M203412200. [DOI] [PubMed] [Google Scholar]
- 24.Lim PJ, et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol. 2009;187:201–217. doi: 10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu A, et al. Effects of ubiquilin 1 on the unfolded protein response. J Mol Neurosci. 2009;38:19–30. doi: 10.1007/s12031-008-9155-6. [DOI] [PubMed] [Google Scholar]
- 26.N’ Diaye E-N, et al. The ubiquitin-like protein PLIC-2 is a negative regulator of G protein-coupled receptor endocytosis. Mol Biol Cell. 2008;19:1252–1260. doi: 10.1091/mbc.E07-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.N’ Diaye E-N, et al. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothenberg C, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–3232. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, et al. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum Mol Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Monteiro MJ. Ubiquilin overexpression reduces GFP-polyalanine-induced protein aggregates and toxicity. Exp Cell Res. 2007;313:2810–2820. doi: 10.1016/j.yexcr.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 32.Galan JM, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buttner C, et al. Ubiquitination precedes internalization and proteolytic cleavage of plasma membrane-bound glycine receptors. J Biol Chem. 2001;276:42978–42985. doi: 10.1074/jbc.M102121200. [DOI] [PubMed] [Google Scholar]
- 34.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 35.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 36.Ke N, Godzik A, Reed JC. Bcl-B, a novel Bcl-2 family member that differentially binds and regulates Bax and Bak. J Biol Chem. 2001;276:12481–12484. doi: 10.1074/jbc.C000871200. [DOI] [PubMed] [Google Scholar]
- 37.Chen G, et al. Autoantibody profiles reveal ubiquilin 1 as a humoral immune response target in lung adenocarcinoma. Cancer Res. 2007;67:3461–3467. doi: 10.1158/0008-5472.CAN-06-4475. [DOI] [PubMed] [Google Scholar]
- 38.Lockwood WW, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med. 2010;7:e1000315. doi: 10.1371/journal.pmed.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 40.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Nat’l Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360:423–427. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winkler SG, et al. Isolation and mass spectrometry of transcription factor complexes. Methods. 2002;26:260–269. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- 43.Erdjument-Bromage H, et al. Micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
Proc Natl Acad Sci U S A. 2012 Jan 17;109(3):657-658.
AUTHOR SUMMARY
Alterations in the processes that affect homeostatic cell death mechanisms are found in nearly all malignancies (1). One of the most well studied families of proteins that regulate cell death is the BCL2 family of proteins. BCL2 proteins can be either the guardians of the stability of mitochondrial outer membrane permeability (anti-apoptotic BCL2 family members) or the executioners of cell death caused by induction of increased mitochondrial membrane permeability (proapoptotic BCL2 family members) (2). An imbalance in the levels of anti- vs. proapoptotic BCL2 family members is frequently observed during cancer progression, and the imbalance can determine the outcome of cancer therapy. The imbalance can be either quantitative, determined by the total amount of a particular anti- or proapoptotic signal, or qualitative, as determined by the particular signal that is disrupted (3).
We have been studying the activities of the six human anti-apoptotic BCL2 proteins. We previously reported that each of the six BCL2 genes could cooperate with MYC (myelocytomatosis oncogene) in a mouse model of leukemogenesis (4). However, we documented differences in the potency with which individual members could cooperate: BCL2, BCLxl, and BCLw were more potent oncogenes than BCLb, BFL1, and MCL1.
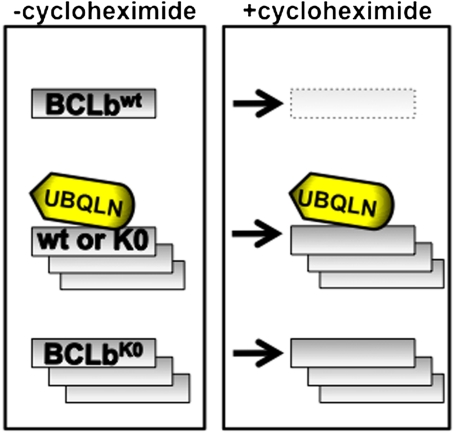
In an effort to elucidate the differences in cooperative potential, we have begun to examine a number of properties of the six anti-apoptotic BCL2 proteins. Most importantly, as reported here, we have found that the three less potent oncogenic BCL2 proteins are less stable proteins, because they disappear rapidly when expressed in cells treated with cycloheximide, an inhibitor of translation. This finding suggested the possibility that a protein’s intrinsic stability may dictate its ability to participate in oncogenic transformation. We further support this observation by demonstrating that a stabilized version of the BCLb protein, from which of all of its lysine residues have been removed (BCLbK0) to prevent addition to ubiquitin and subsequent degradation, is fully stabilized and acts as a more potent oncogenic protein in our model of leukemia (Fig. P1).
Fig. P1.
The regulation of concentrations of BCLb by Ubiquillin and ubiquitination. Treatment of cells expressing BCLbwt or BCLbK0 with cycloheximide leads to varied outcomes of the BCLb proteins depending on whether BCLb contains lysine residues or if Ubiquilin is present. Treatment of cells expressing BCLbwt with cycloheximide leads to rapid degradation of BCLb proteins (top), whereas, there is no affect on BCLbK0 levels following treatment with cycloheximide (bottom). When Ubqln is expressed with either BCLbwt or BCLbK0, BCLb proteins remain stable during prolonged treatment with cycloheximide (middle).
Having shown that BCLb is less oncogenic than other BCL2 proteins and that stabilization of BCLb could increase its oncogenic potential, we set out to identify proteins that interact with BCLb and affect its stability. To this end, we performed immunoprecipitation followed by mass spectrometry to identify BCLb-interacting proteins. One protein that we identified as a BCLb-interacting protein was Ubiquilin1 , a protein with ubiquitin-like and ubiquitin-associated domains. Previous work had identified Ubiquilin1 as a protein capable of interacting with the proteasome and with ubiquitinated proteins, acting as a bridge to bring substrates to the proteasome for proteolytic degradation (5). In addition to this role of Ubiquilin1, it has also been suggested that Ubiquilin1 can play roles in protein trafficking and ER-associated protein degradation. We therefore attempted to determine how the interaction with Ubiquilin1 affects BCLb function.
We observed that Ubiquilin1 interacts specifically with BCLb and not with any of the other five BCL2 proteins. Following interaction with Ubiquilin1, BCLb is altered in multiple ways. Most obviously, the amount of BCLb that is ubiquitinated on multiple lysine residues is increased. Importantly, following interaction with Ubiquilin1, BCLb protein is more resistant to degradation as shown by treatment of cells with cycloheximide (Fig. P1). Coincident with these alterations, BCLb moves from membranes into the cytosol, and the complex containing Ubiquilin1 and ubiquitinated BCLb is exclusively in the cytosolic fraction.
How does the interaction with Ubiquilin1 relate to human cancer? The answer to this question is likely complex and context dependent, but we offer some insight into a potential role of Ubiquilin1 in human lung adenocarcinoma. Examining the expression of Ubiquilin1 from biopsies of primary lung adenocarcinomas and adjacent normal lung tissues from the same patient, we show that Ubiquilin1 mRNA is elevated in cancer samples. Also, if patients are grouped according to the level of Ubiquilin1 mRNA expression in the tumor samples, tumors with higher Ubiquilin1 mRNA levels are associated with shorter survival times than those with relatively lower Ubiquilin1 mRNA levels.
In the future, we hope to examine in more detail the relationship between Ubiquilin1 and BCLb in mouse models of cancer and human cancer. We propose that high expression of Ubiquilin1 in some human cancers may directly regulate the protein levels of BCLb (or other oncogenic proteins). In this situation, inhibition of Ubiquilin1 activity or expression may reduce the levels of oncogenic proteins, resulting in cancer cell death or susceptibility to cytotoxic drugs.
Footnotes
The authors declare no conflict of interest.
This is a Contributed submission.
See full research article on page E119 of www.pnas.org.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281(5381):1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 3.Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103(6):839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 4.Beverly LJ, Varmus HE. MYC-induced myeloid leukemogenesis is accelerated by all six members of the antiapoptotic BCL family. Oncogene. 2009;28(9):1274–1279. doi: 10.1038/onc.2008.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleijnen MF, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6(2):409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
Supplementary Materials
Supporting Information