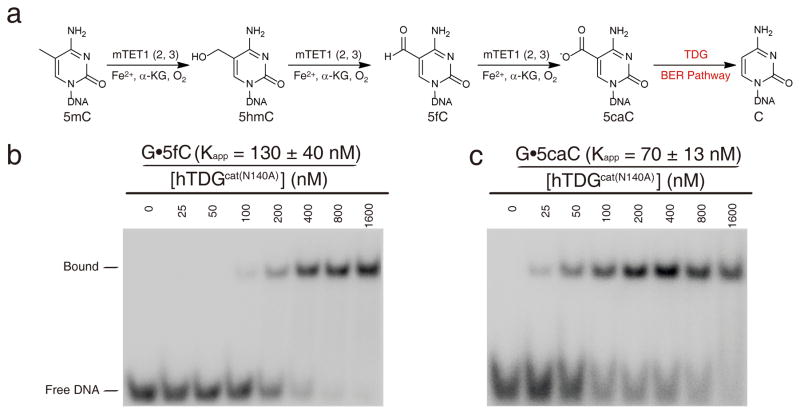
Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA (original) (raw)
. Author manuscript; available in PMC: 2012 Oct 1.
Published in final edited form as: Nat Chem Biol. 2012 Feb 12;8(4):328–330. doi: 10.1038/nchembio.914
Abstract
Human thymine DNA glycosylase (hTDG) efficiently excises 5-carboxylcytosine (5caC), a key oxidation product of 5-methylcytosine in a recently discovered cytosine demethylation pathway. We present here the crystal structures of hTDG catalytic domain in complex with duplex DNA containing either 5caC or a fluorinated analog. These structures, together with biochemical and computational analyses, reveal that 5caC is specifically recognized in the active site of hTDG, supporting the role of TDG in mammalian 5-methylcytosine (5mC) demethylation.
Human thymine DNA glycosylase belongs to the uracil DNA glycosylase superfamily. Enzymes in this family use a base-flipping mechanism to locate damaged bases in double-stranded DNA (dsDNA) and initiate base replacement through the DNA base-excision-repair pathway (BER)1,2. hTDG has been shown to recognize mismatched pyrimidine bases of uracil and thymine in G•U and G•T pairs and perform subsequent cleavage of the glycosylic bond for BER of these DNA base lesions1–4. A crystal structure of the catalytic domain of hTDG (hTDGcat, residues 111–308) bound to dsDNA containing an abasic site (hTDGcat-G•AP, pdb code: 2RBA) has been reported5; however, due to the lack of a base at the lesion site in this structure, base recognition by TDG has not been revealed.
Recently, another major role of TDG has emerged. This protein is involved in epigenetic regulation through an active 5-methylcytosine (5mC) demethylation pathway6,7. Methylation and demethylation at the 5-position of cytosine are critical for transcriptional regulation and genome reprogramming in eukaryotes6,8. Unlike the well-known methylation pathway, the active demethylation pathway is poorly understood, in particular in mammals6. Plants employ 5mC glycosylases that mediate BER as an active demethylation pathway9, one that has not been observed in mammals. However, it was recently shown that 5mC is oxidized to 5-hydroxymethylcytosine (5hmC)10,11, and further to 5-formylcytosine (5fC) and 5caC by the TET family dioxygenases in mammalian cells12–14. The oxidized products of 5caC and 5fC are recognized by TDG and excised through BER to install an unmethylated cytosine (Figure 1a)7. Therefore, the TET-mediated oxidation of 5mC and TDG-mediated BER of oxidized 5mC nucleotides represent a new active demethylation pathway in mammalian cells. This pathway is in agreement with earlier observations revealing that TDG is essential for transcriptional regulation and mouse embryonic development15,16, a property that cannot be explained by the uracil/thymine glycosylase function of the protein.
Figure 1. Electrophoretic mobility shift assay of hTDGcat(N140A) with 23mer dsDNA containing G•T, G•U, G•5fC and G•5caC pairs.
(a) The proposed 5mC demethylation pathway. (b–c) The EMSA assay for hTDGcat(N140A) with dsDNA containing G•5fC and G•5caC pairs. The experiments were performed with 4 nM 32P-labeled DNA (40 pM 32P-labeled DNA for G•AP pairs) and various concentrations of hTDGcat(N140A) as shown.
An alternative pathway has been proposed that involves deamination of 5hmC to 5-hydroxymethyluracil (5hmU) by a family of single-stranded DNA deaminases (Activation-induced cytidine deaminase, AID/Apobac1,3) followed by BER through TDG15,17; however, the presence and involvement of 5hmU in genomic DNA still awaits further establishment. Here we present the crystal structures of hTDGcat in complex with dsDNA containing either 5caC or 1-(2-deoxy-2-fluoro-_β_-D-arabinofuranosyl)-5-carbonylcytosine (_β_-F-5caC). These structures, together with biochemistry and computational analyses, reveal the specific recognition of 5caC by TDG and further confirm that TDG can facilitate 5caC excision in the recently discovered mammalian 5mC demethylation pathway7.
hTDGcat and a corresponding inactive mutant (hTDGcat(N140A)) with the active site residue Asn140 mutated to alanine were cloned, expressed, and purified as previously described3,15,18. We performed activity and electrophoretic mobility shift assays (EMSA) against different substrate candidates using hTDGcat and hTDGcat(N140A), respectively. A series of 23mer dsDNA oligonucleotides containing G•abasic-site (G•AP), G•T, G•U, G•5hmU, G•C, G•5mC, G•5hmC, G•5fC, and G•5caC base pairs were synthesized (Supplementary Methods, Supplementary Results, Supplementary Fig. 1). Glycosylase activity assays showed that hTDGcat cannot excise 5hmC but acted efficiently on both 5fC and 5caC as reported previously (Supplementary Fig. 2)7,19. The single turnover experiment further indicated that 5fC is a better substrate than 5caC for hTDG, which is consistent with a recent report (Supplementary Table 1)19. Surprisingly, EMSA showed that hTDGcat(N140A) preferentially bound to dsDNA containing G•5caC over G•5fC, G•U and G•T with the apparent binding affinity order of: G•AP (_K_app= 440± 50 pM)> G•5hmU (_K_app= 47± 7 nM)> G•5caC (_K_app= 70± 13 nM) > G•5fC(_K_app= 130 ± 40 nM) > G•U (_K_app= 470 ± 40 nM) > G•T(_K_app= 1.3 ± 0.3 μM) (Figure 1b, 1c and Supplementary Fig. 3). In contrast, hTDGcat(N140A) did not bind to G•C-, G•5mC- or G•5hmC-containing DNA (Supplementary Fig. 3). This binding preference is different from the excision activity of hTDG which is in the order of: G•U> G•5hmU > G•5fC > G•5caC19. The binding preference of 5caC suggests that 5caC is specifically recognized by TDG as a cognate substrate.
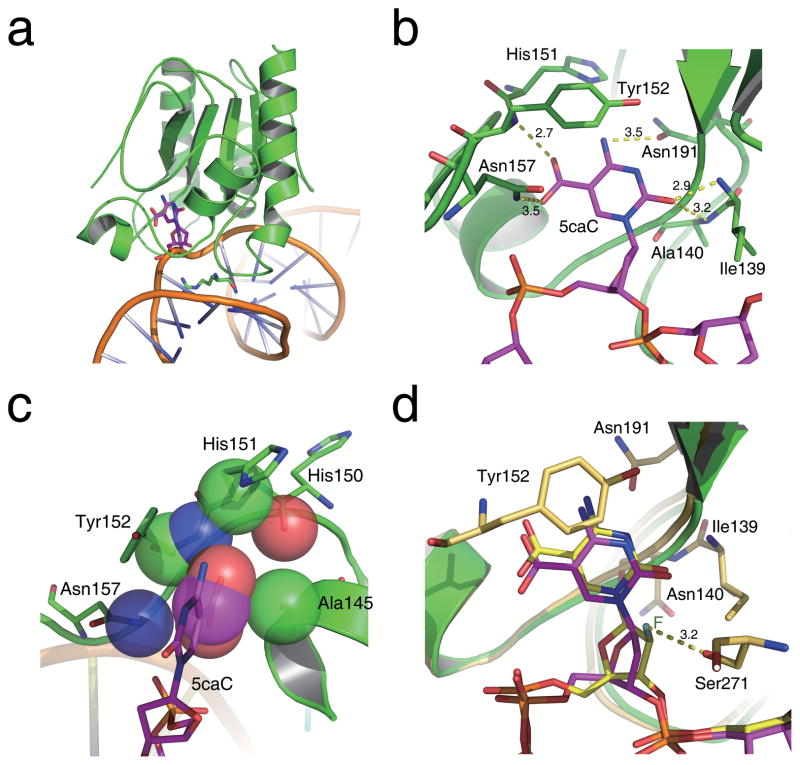
No base-bound TDG structure has been reported4. To reveal how 5caC is recognized by hTDG, we present here the 3.0 Å crystal structure of hTDGcat(N140A) in complex with a 23mer dsDNA containing an A•5caC base pair with an adenine opposite 5caC (hTDGcat(N140A)-A•5caC) (Supplementary Table 2). The structure was solved by molecular replacement using the previous hTDGcat-G•AP structure as the searching model5. The overall hTDGcat(N140A)-A•5caC structure is similar to the model with an overall root-mean-square deviation (r.m.s.d) of 1.04 Å for Cα positions. In the 2RBA structure, two hTDGcat(N104A) molecules bind to DNA with 5caC recognized by one protein while the other hTDGcat binds ten base pairs away nonspecifically (Supplementary Fig. 4a). The hTDGcat interacts with the backbone of the 5caC-containing strand via electrostatic complementarity and bends the dsDNA backbone by ~45° towards the active site. The side chain of the wedge residue Arg275 inserts through the dsDNA minor groove and pushes the 5caC base out of the DNA groove. Concurrently, the 5caC pyrimidine rotates ~40° along the glycosylic bond, while the sugar ring rotates ~45° (compared to 0° in the TDG-abasic site structure5) (Supplementary Fig. 4b), and the whole base penetrates into the active site pocket of hTDG (Figure 2a). The observed conformation is different from that of deoxyuridine previously observed in the UDG-dψU structure20, which shows ~90° rotation of the flipped base.
Figure 2. Schematic diagram of the hTDGcat(N140A)-A•5caC complex structure.
(a) Overall structure of hTDGcat(N140A) bound with 5caC-containing dsDNA. The 5caC and the wedge residue Arg275 are shown as sticks and colored in magenta and green, respectively. (b) A network of hydrogen bonds in the active site of hTDG specifically recognizes 5caC. Residues involved in the interactions are labeled and shown as sticks, and hydrogen bonds are shown as yellow dashes. (c) The interactions involved in the 5-carboxyl-binding pocket. The atoms involved are presented as transparent spheres. (d) Superposition of 5caC and _β_-F-5caC in the active site pocket of the hTDGcat(N140A)-A•5caC and hTDGcat-G• _β_-F-5caC structures. Residues involved in the interactions in the _β_-F-5caC structure are shown as sticks and colored in yellow and orange. The fluorine atom is labeled in dark green. The hydrogen bond interaction between _β_-F-5caC and residue Ser271 is shown in yellow dashes.
Inside the active site pocket where 5caC is bound, the flipped base is locked via polar interactions from surrounding residues21, resulting in a well-observed electron density map of 5caC in the pocket (Figure 2b, Supplementary Fig. 5 and 6). The pyrimidine O2 atom accepts hydrogen bonds from the main chain amide atoms of Ile139 and Ala140, while the pyrimidine N4 atom is located within hydrogen-bonding distance of the side chain of Asn191. The phenol ring of Tyr152 packs with the pyrimidine ring of 5caC through hydrophobic interactions.
In addition to these interactions, the 5-carboxyl moiety of 5caC is specifically recognized in a small pocket formed by the side chains of Ala145 and Asn157 together with the backbone atoms of His150, His151, and Tyr152. The carboxyl group is positioned to form hydrogen bonds with the backbone amide of Tyr152 and the side chain of Asn157, while it is also in van der Waals contact with the side chain of Ala145 (Figure 2c). The two polar interactions may enhance the binding affinity of TDG to 5caC and 5fC over U and T, which is consistent with our binding assay results. Compared to 5fC, these interactions are further enhanced for the negatively charged 5caC. These unique structural features make hTDG the only uracil glycosylase capable of recognizing and excising 5caC and 5fC from dsDNA (Supplementary Fig. 7). Other members of the family do not exhibit such binding (Supplementary Fig. 3) and activities due to the presence of side chain groups (Tyr147 in UDG, Ile449 in MBD4) or main chain atoms (Phe109 in SMUG1, Gly445 in MBD4) that interfere with the potential binding of 5caC or 5fC22–25.
We also obtained and solved a 3.0 Å crystal structure of the wild-type hTDGcat in complex with dsDNA containing a non-hydrolyzable 5caC analog, _β_-F-5caC with a 2′-fluoro substitution on the deoxyribose of 5caC, paired with G, (hTDGcat-G•5caC_β_F) (Supplementary Table 2). The _β_-F-5caC was synthesized as described (Supplementary Methods, Supplementary Scheme 1 and Supplementary Fig. 8). As shown in Figure 2d, the _β_-F-5caC base displays a very similar conformation to 5caC in the active site pocket except that its deoxyribose ring rotates ~15° due to the 2′-fluoro substitution that appears to hydrogen bond to the side chain of Ser271. No direct interaction was observed between the side chain of Asn140 and _β_-F-5caC, indicating that the mutagenesis of Asn140 to alanine does not structurally disturb 5caC binding; Asn140 contributes to cleavage of the glycosylic bond via activating a water molecule that attacks the C1 atom of the flipped base. The opposite G in the _β_-F-5caC structure is engaged in two additional hydrogen bonds with residues on the hTDG insert loop as compared to A in the hTDGcat(N140A)-A•5caC structure, which provides a more stable conformation for the base cleavage and explains the preference of G over A on the opposite strand (Supplementary Fig. 9)2.
To further investigate the binding preference of 5caC by TDG, we modeled different base substrates (C, 5mC, 5hmC, 5fC, U, and 5hmU) into the current structure and performed computational analyses (Supplementary Methods). The calculation results indicate that the positively charged pocket near the C5 substitution (His151 and Tyr152) is well suited to binding a carboxyl group. The empirical binding free energy calculation confirmed that 5caC has a strong binding affinity to hTDG with a low energy score. Cytosine and 5mC yielded the highest energy, reflecting poor binding to hTDG (Supplementary Table 3 and 4). The binding free energy further suggests that His151 and Tyr152 make significant contributions to the binding of 5caC, but the energy contribution decreases significantly when hTDG is “forced” to bind 5mC and 5hmC, revealing that electrostatic interaction between His151 and substrate plays an important role in substrate recognition (Supplementary Table 5, Supplementary Fig. 10). Additionally, the dynamic hydrogen-bonding interactions also show that 5mC and 5hmC lack a hydrogen bond with backbone nitrogen of Asn140. Although they could form hydrogen bonds with the side chain and the backbone nitrogen of Tyr152, these hydrogen bonds are much weaker than that of 5caC due to the lower occupancy rate and longer distance, explaining the high selectivity of hTDG of 5caC over 5hmC and 5mC (Supplementary Table 6).
In contrast to 5mC and 5hmC, the protonated N3 and O4 atoms of U and 5hmU form additional strong hydrogen bonds to the side chains of Asn191 with an occupancy rate of 80.0 and 72.6, respectively (Supplementary Table 6, Supplementary Fig. 11). These interactions may compensate for the lack of negatively charged groups on the 5-position upon binding of thymine, uracil, and 5hmU to hTDG. However, the presence of the 5-carboxyl-binding pocket in hDTG still outweighs some of these factors, and 5caC is preferentially recognized over U and T. hTDG does bind 5hmU tightly and exhibits a high activity against 5hmU in dsDNA. The presence and involvement of 5hmU in 5mC demethylation should be further investigated15,17, as our results do indicate that hTDG is an efficient enzyme capable of recognizing and processing 5hmU, if it is present in the genome.
In summary, we show that hTDG specifically binds 5caC via a well-organized carboxyl-binding pocket over uracil, which is one of the best known physiological substrates for hTDG2. This selective mechanism excludes other common cytosine modifications including 5mC and 5hmC. Our results further confirm hTDG as the first known mammalian protein that selectively binds 5caC and plays a major role in mammalian 5mC demethylation. The current structure also presents a template to develop small molecules that may inhibit the catalytic function of hTDG in the 5mC demethylation process in human cells.
Supplementary Material
1
Acknowledgments
This study was supported by National Institutes of Health (GM071440 to C.H.), the 973 grant (2009CB918502 to H.J.), Chinese Academy of Sciences grant (XDA01040305 to C.L.), and beamlines 23ID-B (General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT)) and 24ID-E (Northeastern Collaborative Access Team (NE-CAT)) at the Advanced Photon Source at Argonne National Laboratory. We thank Dr. Xiaojing Yang from University of Chicago and Dr. Dominika Borek from University of Texas Southwestern Medical Center for structure data procession and discussion.
Footnotes
Accession codes. Protein Data Bank: The atomic coordinates and structure factors for the reported crystal structures are deposited under accession codes 3UO7 and 3UOB.
Author Contributions
L.Z., X.L., and C.H. conceived the original idea. L.Z., X.L., H.L. and C.H. designed the experiments; the 5hmU and _β_-F-5caC phosphoramidites were synthesized by Q.D.; biochemistry assays and structural studies were performed by L.Z., X.L., and H.L.; the computational stimulation was performed by J.L., and C.L.; L.Z., X.L., J.L, C.L., and L.Z. and C.H. wrote the paper; all authors discussed results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Stivers JT, Jiang YL. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 3.Morgan MT, Bennett MT, Drohat AC. J Biol Chem. 2007;282:27578–27586. doi: 10.1074/jbc.M704253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hitomi K, Iwai S, Tainer JA. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Maiti A, Morgan MT, Pozharski E, Drohat AC. Proc Natl Acad Sci USA. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhutani N, Burns DM, Blau HM. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He YF, et al. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klose RJ, Bird AP. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Feng S, Jacobsen SE, Reik W. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahiliani M, et al. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito S, et al. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfaffeneder T, et al. Angew Chem Int Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 13.Ito S, et al. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu SC, Zhang Y. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortellino S, et al. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortazar D, et al. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 17.Guo JU, et al. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett MT, et al. J Am Chem Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiti A, Drohat AC. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parikh SS, et al. Proc Natl Acad Sci USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardeland U, Bentele M, Jiricny J, Schar P. J Biol Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 22.Slupphaug G, et al. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 23.Parikh SS, et al. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang YL, Ichikawa Y, Song F, Stivers JT. Biochemistry. 2003;42:1922–1929. doi: 10.1021/bi027014x. [DOI] [PubMed] [Google Scholar]
- 25.Wibley JE, et al. Mol Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1