An N-terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbate diabetes (original) (raw)
. Author manuscript; available in PMC: 2013 May 25.
SUMMARY
Maintenance of lymphoid homeostasis in a number of immunological and inflammatory contexts is served by a variety of regulatory T (Treg) cell subtypes, and depends on interaction of the transcription factor Foxp3 with specific transcriptional cofactors. We report that a commonly used insertional mutant of FoxP3 (GFP-Foxp3) modified its molecular interactions, blocking Hif1a but increasing Irf4 interactions. The transcriptional profile of these Treg cells was subtly altered, with an over-representation of Irf4-dependent transcripts. In keeping with Irf4-dependent function of Treg cells to preferentially suppress T cell help to B cells and Th2- and Th17-type differentiation, GFP-Foxp3 mice showed a divergent susceptibility to autoimmune disease; protection against antibody-mediated arthritis in the K/BxN model, but greater susceptibility to diabetes on the NOD background. Thus, specific sub-functions of Treg cells and the immune diseases they regulate can be influenced by FoxP3’s molecular interactions, which result in divergent immunoregulation.
The transcription factor FoxP3 plays a central role in specifying the differentiation and function of regulatory T (Treg) cells. FoxP3+ Treg cells help maintain lymphoid homeostasis in a number of immunological contexts: tolerance to self vs autoimmune deviation, responses to pathogens or allergens, interactions with commensal microbes (Campbell and Koch, 2011; Barnes and Powrie, 2009; Feuerer et al., 2009b). The importance of FoxP3+ Treg cells is highlighted by the devastating multi-organ inflammation of FoxP3-deficient scurfy mice or human IPEX patients (Ziegler, 2006). In addition, FoxP3+ Treg cells partake in extra-immune regulatory activities, for instance by dampening inflammation and metabolic activity in visceral adipose tissue (Feuerer et al., 2009a).
Several pathways and molecular mediators of Treg cell function have been described, some involving cell-cell interactions, others soluble cytokines or small-molecule mediators (Vignali et al., 2008; Shevach, 2009). Correspondingly, a number of Treg cell subphenotypes exist, with differential effector functions and tissue localization (Feuerer et al., 2009a; Feuerer et al., 2010; Campbell and Koch, 2011). Although Treg cells generally share a transcriptional program that distinguishes them from conventional CD4+ (Tconv) cells, this program is modified and tuned in relation to Treg cell function. For instance, Treg cells in the adipose tissue express a set of transcripts reflective of homing and adaptation to the adipose environment and to the functions they exert there (Feuerer et al., 2009a and data not shown). These programs appear to be determined in Treg cells by the same transcription factors that are central to the differentiated functions of the Tconv cells they regulate. For instance, Irf4 is required for the differentiation of B cells and of the T helper 2 (Th2)-type cells that help them, and the absence of Irf4 in Treg cells impairs their ability to control Th2 cell responses and antibody production (Zheng et al., 2009)). Similarly, Treg cells expressing T-bet or STAT3 transcription factors optimally suppress inflammatory Th1 and Th17 cell responses, respectively (Koch et al., 2009; Chaudhry et al., 2009).
Underscoring this specialization of Treg cell functionalities, FoxP3 engages in a number of interactions with other transcription factors (Xiao et al., 2010). Through its leucine zipper region, FoxP3 can dimerize with itself or with other forkhead domain proteins (Lopes et al., 2006; Li et al., 2007b), and can homodimerize through the formation of a peculiar domain-swapped dimer of the DNA-binding forkhead domain (Bandukwala et al., 2011). FoxP3 interacts with the transcription factors Runx1, NF-AT, Eos (encoded by Ikzf4), Irf4, RORγt, RORα and phosphorylated Stat3 (Ono et al., 2007; Wu et al., 2006; Pan et al., 2009; Zheng et al., 2009; Zhou et al., 2008; Chaudhry et al., 2009). Several of these interactions have been linked to a particular facet of the Treg cell transcriptional profile; for instance, Irf4 or STAT3 subtly affect, alone or in combination with FoxP3, discrete segments of the Treg cell signature (Zheng et al., 2009; Chaudhry et al., 2009).
This diversity of Treg cell subphenotypes and interacting facets of FoxP3 suggests that shifts in these interactions, occurring physiologically or experimentally, should modulate the range of Treg effector abilities. Here, we report such a situation, where a modification of FoxP3 results in diametrically opposite effects on the severity of different autoimmune diseases.
RESULTS
Divergent autoimmune phenotypes in mice carrying the Foxp3fgfp knockin
Our initial observations were made serendipitously in the course of analyzing the role of FoxP3+ Treg cells in the K/BxN model of inflammatory arthritis. These mice carry the KRN transgene, which encodes a T cell receptor (TCR) reactive against a peptide from the ubiquitous enzyme, Glucose-6-Phosphate Isomerase (GPI), presented by the MHC class II molecule Ag7 (Kouskoff et al., 1996). When the KRN transgene is crossed into an Ag7-positive genetic background, autoreactive T cells promote the massive production of anti-GPI IgG, which rapidly and spontaneously induce arthritis. They can also provoke arthritis after transfer into normal recipients (Korganow et al., 1999). To help identify Treg cells in arthritic mice, we introduced into K/BxN mice the Foxp3gfp (Foxp3tm2Ayr) reporter knockin from Fontenot et al (Fontenot et al., 2005), in which the eGFP coding region is inserted in-frame into the coding region of the Foxp3 locus on Chr. X, yielding a functional chimeric fusion protein with GFP inserted between amino acids 5 and 6 of FoxP3 (Fig. 1A; the locus encoding this fusion protein will be referred to as Foxp3fgfp. It is distinguished from the bicistronic ires-driven reporter Foxp3igfp of Bettelli et al (Bettelli et al., 2006), in which GFP is not fused with FoxP3 but is concomitantly expressed from a bicistronic mRNA, leaving FoxP3 intact, and which was used as a control throughout this study).
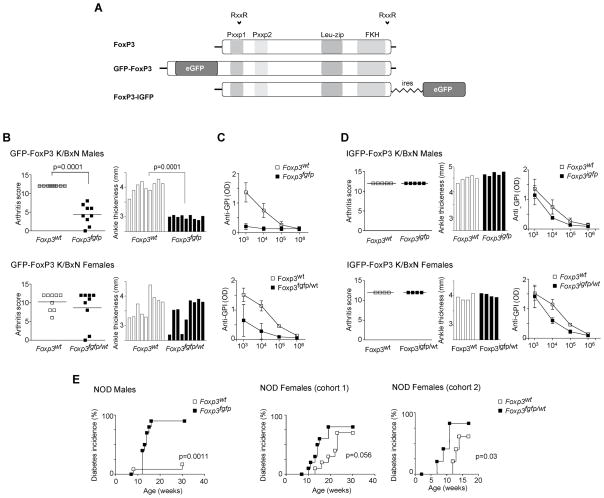
Figure 1. Foxp3fgfp mice exhibit contrasting phenotypes in different autoimmune models.
(A) Schematic representation of FoxP3, GFP-FoxP3, and FoxP3-I-GFP. Protein convertase cleavage sites (RXXR), proline rich regions (Pxxp1 and Pxxp2), leucine zipper (Leu-zip) and forkhead (FKH) domains are represented. (B) Arthritis clinical score and ankle measurements of 8 week-old K/BxN.Foxp3fgfp males, K/BxN.Foxp3fgfp/wt heterozygote females and WT littermate control mice. (C) Anti-GPI IgG reactivity in sera of 8 week-old K/BxN.Foxp3fgfp males, K/BxN.Foxp3fgfp/wt females and WT littermate control mice (mean +/− SD, n=6–8). (D) Arthritis clinical score, ankle thickness, and anti-GPI antibodies in 8 week-old male and female K/BxN.Foxp3igfp and WT controls. (E) Diabetes incidence curves of NOD.Foxp3fgfp males NOD.Foxp3fgfp/wt, females and littermate controls (10 mice/group). Two different cohorts of NOD.Foxp3fgfp/wt heterozygous females were followed, in two different animal facilities.
We noticed that the presence of the Foxp3fgfp reporter modified considerably the course of K/BxN arthritis. Males developed no or very mild arthritis, as judged by both ankle thickening and clinical index (Fig. 1B). Protection in females was less extreme, with a delay in disease onset (Figure S1A) and variable severity. K/BxN.Foxp3fgfp males had low to undetectable amounts of arthritogenic anti-GPI, in contrast to serum of K/BxN littermates (Fig. 1C). K/BxN.Foxp3fgfp heterozygote females also showed a significant reduction in anti-GPI, albeit less dramatic; these titers correlated well with arthritis severity (Fig. S1A). These data imply that protection from arthritis in K/BxN.Foxp3fgfp mice stemmed from a block in autoantibody formation, suggesting an active suppression of the immunologic phase of this model. This idea was further substantiated by the fact that B6._Foxp3fgfp_- animals were sensitive to arthritis induced by transfer of arthritogenic K/BxN serum (Fig. S1B). To address the possibility that protection from arthritis was simply due to GFP expression, we used the FoxP3-ires-GFP reporter line (Bettelli et al., 2006). K/BxN.Foxp3igfp mice developed anti-GPI and arthritis as usual (Fig. 1D), ruling out this interpretation.
Given this protection from arthritis, we next asked whether mice expressing GFP-FoxP3 would also be protected from other autoimmune diseases. Thus, the _Foxp3_fgfp knockin was backcrossed for 12 generations to the NOD/Lt strain, the prototypical model of autoimmune type-1 diabetes (T1D), and evaluated the course of diabetes. Surprisingly, the opposite result was obtained: while male NOD mice are usually resistant to diabetes, NOD.Foxp3fgfp males showed a very high incidence of diabetes (Fig. 1E). Similarly, NOD.Foxp3fgfp heterozygote females exhibited an acceleration of diabetes, in two independent cohorts; some animals became diabetic before 10 weeks of age, which is never seen in our NOD colony.
Foxp3fgfp Treg cells impact disease independent of frequency
Because FoxP3 is exclusively expressed in Treg cells and conditions their differentiation and maintenance, we first asked whether differences in Treg cells between mice expressing GFP-FoxP3 might account for the contrasting phenotypes in the two autoimmune models. Flow cytometric analyses first focused on male mice, in which the Foxp3fgfp phenotypes were most obvious, and on the spleen, where most of the anti-GPI of K/BxN mice are produced. No increase in the fraction of FoxP3+ among CD4+ splenocytes was observed in K/BxN.Foxp3fgfp relative to K/BxN male mice (Fig. 2A), as might have been expected from the suppression of disease. Instead, there was a decrease in the frequency of Treg cells, in mice expressing GFP-FoxP3, most significant in lymph nodes. A similar trend to decreased Treg cell frequency was observed in spleens of NOD.Foxp3fgfp mice, however this observation did not extend to the pancreas (Fig. 2B). Thus, the difference in disease severity imparted by Foxp3fgfp in these two models does not correlate with altered proportions of Treg cells in the key organs.
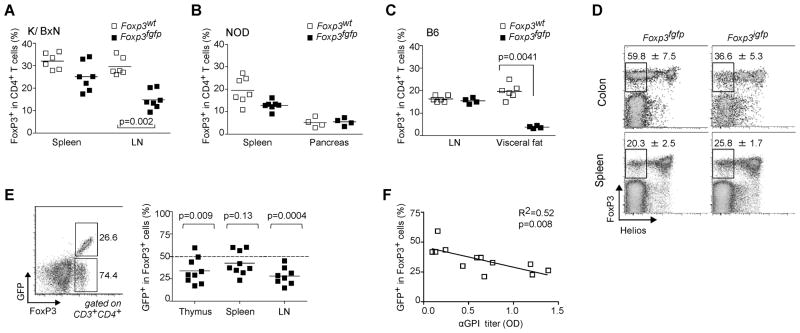
Figure 2. Foxp3fgfp disease phenotype does not correlate with an alteration in Treg frequency.
(A and B) Percentage of FoxP3 positive cells within CD4+ T cells. (A) Spleens and draining lymph nodes (popliteal) of 8 week-old WT and K/BxN.Foxp3fgfp males (each point is an individual mouse). (B) Spleen and pancreas of 9 week-old NOD.Foxp3fgfp males and controls. (C) Proportion of FoxP3+ Treg cells among LN and visceral adipose tissue of 40 week-old B6 males. (D) Representative dot plots showing the percent FoxP3+ Helios− Treg cells in colon lamina propria and spleen of 6 week-old B6.Foxp3fgfp and B6.Foxp3igfp males. Splenocytes from K/BxN.Foxp3fgfp/wt heterozygote females were stained with anti-CD3, -CD4, and –FoxP3 antibodies. (E) Gates and values represent the fraction of GFP positive and negative cells among FoxP3+ population. Summary of data in adjacent plot shows fraction of GFP+ cells among FoxP3+ cells in thymus, spleen and draining lymph nodes of K/BxN.Foxp3fgfp/wt females; the dashed line denotes the 50/50 ratio expected from random X-inactivation (F) Linear regression analysis of serum anti-GPI levels vs the fraction of GFP+ FoxP3+ Treg cells in K/BxN.Foxp3fgfp/wt heterozygote females.
In view of the impact of the chimeric GFP-FoxP3 on distinct Treg cell populations, we asked whether other Treg pools might be similarly affected. We have recently described a particular population of FoxP3+CD4+ T cells that resides in the abdominal adipose tissue, has a distinctive TCR repertoire and transcriptional profile, and impacts metabolic parameters (Feuerer et al., 2009a). In aged males, in which this population is most abundant, mice expressing GFP-FoxP3 showed a marked reduction in numbers of fat Treg cells (Fig. 2C).
Another recently described Treg cell population is found in the colonic mucosa, induced by exposure to components of the gut microbiome, in particular Clostridia species (Atarashi et al., 2010). This population differs from Treg cells in secondary lymphoid organs by not expressing the transcription factor Helios (encoded by Ikzf2), which has been suggested to identify thymus-derived Treg cells (Thornton et al., 2010), although the correlation is likely not absolute (data not shown). We evaluated the representation and Helios expression of FoxP3+ Treg cells in the colonic lamina propria (LP) of B6.Foxp3fgfp males. Significantly more Foxp3fgfp Treg cells lacked Helios than did Treg cells from B6.Foxp3igfp or unmodified B6 mice, but no difference was observed in splenic Treg cells (Fig. 2D). These results suggest that there may be an increased conversion to a Treg cell phenotype in the colon of Foxp3fgfp mice in response to local microbiota.
In heterozygous female mice, we noted a significant imbalance in the fraction of FoxP3+ cells expressing the Foxp3fgfp knockin allele. Random X-inactivation should result in equivalent representation of Foxp3 alleles from both chromosomes, unless one conferred a particular advantage or disadvantage to the cells expressing it. This ratio of GFP+/GFP− among FoxP3+ cells was skewed in splenic Treg cells of most K/BxN.Foxp3fgfp females (Fig. 2E), variably in individual mice, and ranging down to 1:4 (Fig. 2E). This disproportion was evident early in differentiation of the Treg cell lineage in the thymus, likely reflecting a lower efficacy of selection. This thymic imbalance was most evident in K/BxN.Foxp3fgfp females, perhaps because of their self-reactive TCR, as it was more discrete in non-transgenic B6 and NOD females not shown). We used this variability to our advantage by assessing whether there was a relationship between percent Foxp3fgfp Treg cells and anti-GPI titers. Indeed, anti-GPI titers were inversely correlated with the proportion of GFP+ cells among Treg cells in K/BxN females (Fig. 2F).
Thus, the GFP-FoxP3 protein seems to alter in opposite ways the selection or stability of Treg cells that express it. But, whether comparing males to females, or females with different allelic inactivation, there is a strong correlation between the effect on autoimmune disease and the representation of Treg cells expressing the GFP-FoxP3 protein, indicating that Foxp3fgfp Treg cells are directly involved in altering the course of autoimmune diseases described above.
Th17 cells in K/BxN.Foxp3fgfp mice
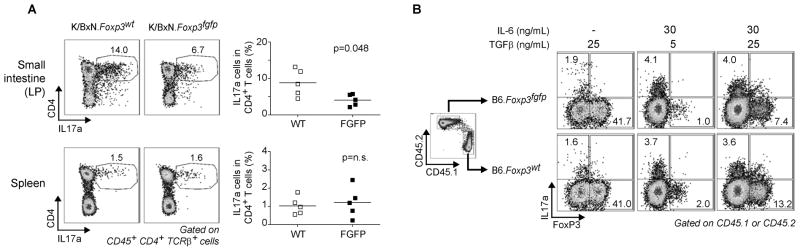
Th17 cells play a central role in K/BxN arthritis, most directly by modulating the amounts of arthritogenic anti-GPI (Jacobs et al., 2009; Wu et al., 2010). Given the low anti-GPI in K/BxN.Foxp3fgfp mice, we assessed the Th17 cell population in these mice. K/BxN.Foxp3fgfp male mice showed a significant decrease in the frequency of Th17 cells in the small intestine lamina propria (SI-LP) relative to control K/BxN.Foxp3igfp mice (Fig. 3A). Preliminary data from NOD.Foxp3fgfp mice follow a similar trend. This difference could be due to an intrinsic defect in Th17 cell differentiation (conceivable in regards to the notion that interactions between FoxP3 and RORγt control the differential towards Th17 or iTreg cells (Zhou et al., 2008)), or to stronger suppression of IL17 production by Treg cells expressing the GFP-FoxP3 protein. We tested the former using an in vitro Th17 cell differentiation system. To control for inter-well variation or indirect effects, congenically marked naïve T cells from B6.CD45.1 and B6.Foxp3fgfp (CD45.2) mice were mixed and cultured together under Th17 cell skewing conditions (Fig. 3B). In cultures lacking IL-6, FoxP3+ cells were similarly induced in Foxp3fgfp or Foxp3wt cells (Fig. 3B, left panels). IL17 producing cells were induced by IL-6 at similar frequencies in Foxp3fgfp or Foxp3wt cells, at either concentration of TGFβ, indicating that there is no intrinsic defect of Th17 differentiation. We also searched for a Th cell bias in gene expression profiles of unstimulated Tconv cells from NOD. or B6.Foxp3fgfp. No bias in typical Th1, Th2 or Th17 cell transcriptional signatures were observed (Fig. S2), indicating that the defective IL17 induction is only revealed in challenged conditions, such as those of the K/BxN LP.
Figure 3. Decrease in Th17 cells in K.BxN.Foxp3fgfp is independent of inherent Th17 differentiation defect.
(A) IL17 producing cells in spleen and small intestine lamina propria from 4–5 week old K/BxN and K/BxN.Foxp3fgfp male mice. Gates and values represent fraction of IL17 positive cells in CD4 population. Data is summarized in adjacent graph. Data is representative of 3 independent experiments. (B) In vitro Th17/Treg differentiation of congenically marked naïve cells from WT (CD45.1) and Foxp3fgfp B6 mice. Data is representative of 2 independent experiments.
Altered characteristics of Foxp3fgfp Treg cells
The impact of GFP-FoxP3 on differentiation of several Treg cell compartments and the correlation between GFP-FoxP3 expression and autoimmune disease suggested that the chimeric protein might modify particular regulatory aspects or phenotypic characteristics in Treg cells, and thereby impact arthritis or diabetes progression. To elucidate these mechanistic underpinnings, we analyzed several aspects of Treg phenotype and function.
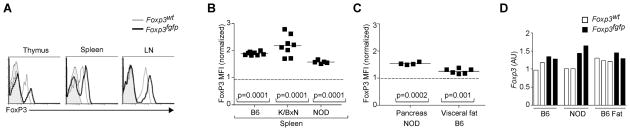
The FoxP3 mean fluorescence intensity (MFI) was increased by 2-fold in Foxp3fgfp Treg cells in K/BxN males (Fig. 4A), evident from early Treg differentiation in the thymus. This increase was also present on the B6 and NOD genetic backgrounds (Fig. 4B), and was equivalent in males and females. The FoxP3 chimeric protein was also over-represented, to a less marked degree, in Treg cells from visceral fat and in the pancreatic infiltrate of NOD mice (Fig. 4C). This over-expression of FoxP3 was not reflected at the mRNA level (Fig. 4D), suggesting that the increase in FoxP3 reflected an effect on the stability of the protein, perhaps through different susceptibility to post-translational modifications. At any rate, it is clear that the addition of GFP does not destabilize FoxP3.
Figure 4. Elevated FoxP3 expression in Foxp3fgfp Treg cells.
(A) Anti-FoxP3 staining profiles of CD3+CD4+ from different organs of K/BxN.Foxp3fgfp male or WT littermates (shaded area is profile from isotype control). Data representative of 3 independent experiments. (B) FoxP3 mean fluorescence intensity (MFI) of splenic Treg cells on 3 different backgrounds. Each dot represents an individual mouse, and values are normalized to the mean FoxP3 MFI of WT mice in each experiment. (C) FoxP3 MFI for LN and visceral fat from 40 week-old B6.Foxp3fgfp mice. (D) Foxp3 mRNA levels in CD4+GFP+ splenic Treg cells from Foxp3fgfp or WT male mice on 2 different backgrounds.
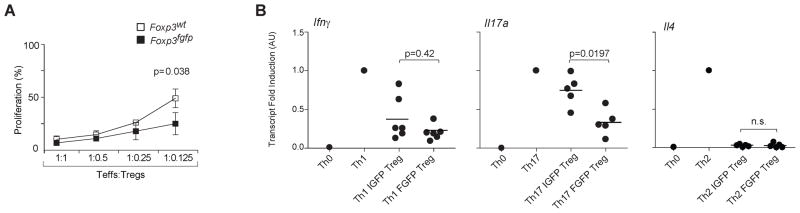
We then tested the functional activity of Foxp3fgfp Treg cells. In the conventional in vitro Treg suppression assay, Foxp3fgfp Treg cells showed a small increase in suppressor activity (Fig. 5A), consistent with the results of Fontenot et al who also observed a small shift (Fontenot et al., 2005). This difference was observed only when responder T cells were activated in the presence of irradiated antigen presenting cells (APCs), since direct activation of responder T cells, using a plate bound anti-CD3 antibodies or anti-CD3 and anti-CD28 conjugated beads, did not reveal this enhanced suppression (data not shown). The limited difference in suppressive activity in vitro was clearly not sufficient to account for the divergent effects on the course of arthritis and diabetes, which could be explained by modifications in the balance of different effector functions of Treg cells. To test this hypothesis, we cultured naïve T cells under Th1, Th2, and Th17 cell-skewing conditions in the absence or presence of GFP-FoxP3 or IGFP-FoxP3 Treg cells. Transcripts of the key indicator cytokines (Ifng, Il17a and Il4) were quantitated by RT-PCR to determine specific Th cell-subset differentiation. Expression of Ifng and Il4 were equally suppressed by both GFP- and IGFP-FoxP3 Treg cells (Fig. 5B), GFP-FoxP3 Treg cells were significantly more efficient at blocking Th17 cell differentiation than their IGFP counterparts. This greater efficiency may explain the decrease in SI-LP Th17 cells in K/BxN.Foxp3fgfp animals.
Figure 5. Enhanced Treg function observed in Foxp3fgfp Treg cells.
(A) In vitro suppressive ability of CD4+CD25+ Treg cells from B6.Foxp3fgfp or WT littermates, titrated at different ratios in co-cultures with CD4+ Tconv responder cells and APCs. Data represent mean proliferation +/− SD of 3 independent experiments. (B) Th17 differentiation suppressed by GFP-FoxP3 (FGFP) and FoxP3-ires-GFP (IGFP) Tregs. Ifng, Il17a, Il4 mRNA expression level assessed by rt-PCR.
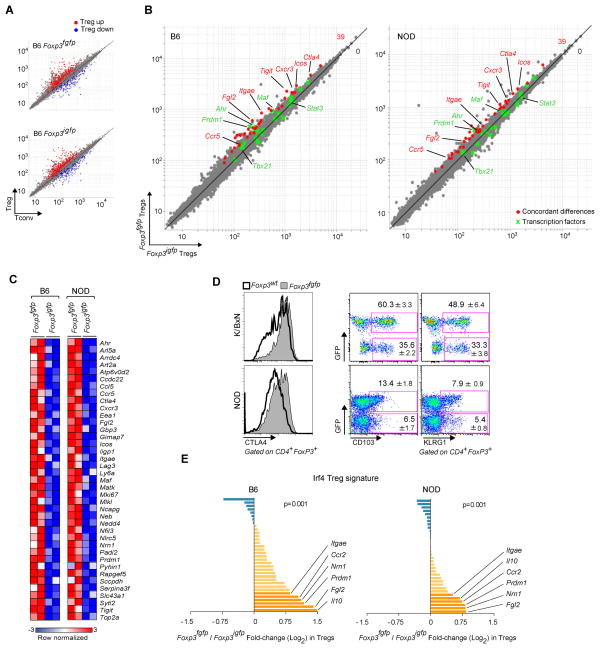
To further shed light on the differences between Foxp3fgfp and WT Treg cells, we compared their transcriptional profiles on gene expression microarrays. To properly compare Treg cell populations, we sorted to high purity CD3+CD4+GFP+ (Treg) and CD3+CD4+GFP− (Tconv) cells from both Foxp3fgfp and Foxp3igfp mice. This comparison was performed on both the B6 and NOD inbred backgrounds, to generate independent datasets and to avoid potential confounders from the strong inflammatory alterations in the K/BxN context. The expression plots of Fig. 6A, which compare Treg and Tconv cells and highlight the canonical Treg cell signature (Hill et al., 2007), give a broad perspective and show that GFP+ cells in B6.Foxp3fgfp mice are indeed Treg cells, with the usual complement of over-and under-expressed transcripts, consistent with the results of Fontenot et al (Fontenot et al., 2005). Next, we sought finer differences between Foxp3fgfp and Foxp3igfp Treg cells, by direct comparison of Treg cell profiles in both strains (Fig. 6B). Treg cells from Foxp3fgfp and Foxp3igfp were very similar, with only 74 to 151 transcripts over-expressed in Foxp3fgfp Treg cells at an arbitrary threshold of 1.5-fold (for comparison, the experimental noise estimated by Monte Carlo randomization and sampling randomization was 60 and 197 hits, respectively, at the same threshold). Many of the transcription factors key in establishing the Treg cell signature were equivalently expressed (green dots in Fig. 6B). In contrast, 39 of these differential transcripts showed a comparable deviation when the Foxp3fgfp vs Foxp3igfp comparison was made on both the B6 and NOD backgrounds, providing cross-confirmation from independent datasets (only 6.54 overlaps on average in randomized datasets, p<10−4). Further substantiating the significance of these discrete differences, _Foxp3fgfp_-differential transcripts were not a random assortment of transcripts but included a number of genes of established relevance to Treg cells (Fig. 6C): effector molecules (Il10, Ctla4, Fgl2), cell-surface receptors (Klrg1, Icos), migration factors (Itgae, Ccr2, Ccr5, Cxcr3), and transcription factors (Prdm1, Maf, Ahr). This signature included several transcripts typically over-expressed in the CD103+Klrg1+ Treg subset (Feuerer et al., 2010). These differences in Klrg1, CD103 and Ctla4 expression were confirmed by flow cytometry in Foxp3fgfp heterozygote K/BxN and NOD females, which allow a direct comparison in the same mouse of Treg cells expressing WT versus chimeric FoxP3 (Fig. 6D).
Figure 6. Biased gene expression in Foxp3fgfp Treg cells.
Gene expression profiles were generated by microarray of Treg and Tconv cells from B6.Foxp3fgfp males, with parallel profiling from B6.Foxp3igfp as a reference. (A) Comparison of expression values in Treg versus Tconv splenocytes from Foxp3fgfp and Foxp3igfp B6 males. The canonical Treg signature is highlighted in red (Treg up-regulated transcripts) and blue (Treg down-regulated transcripts). (B) Comparison of expression values in Foxp3fgfp and control Foxp3igfp Treg splenocytes, on B6 and NOD backgrounds. Genes concordantly over-expressed in Foxp3fgfp relative to Foxp3igfp in both backgrounds are highlighted in red. Transcription factor genes are represented in green. (C) Heatmap representation of transcripts over-represented in Foxp3fgfp on both backgrounds. (D) Flow cytometric confirmation of CTLA-4, CD103, and KLRG1 over-expression in CD4+FoxP3+ Treg cells from spleens of Foxp3fgfp heterozygote females, on the K/BxN and NOD backgrounds. The values shown are the percent CD103 or KLRG1 positive cells within the GFP-positive or -negative fraction of Treg cells (mean +/− SD of 8 mice). (E) Over-representation of the Irf4 Treg signature: The ratio of expression in Foxp3fgfp vs Foxp3igfp of Irf4-responsive transcripts is shown (log2 scale) for both B6 and NOD datasets. The p-value is calculated with a χ2 test for departure from a null hypothesis of random distribution.
We then asked whether this differential representation overlapped with any of the sub-signatures that characterize Treg cell subphenotypes, in particular those linked to Stat3 and Irf4, which are associated with the ability of Treg cells to control Th17 cell- and Th2 cell-related functions. We hypothesized that the over-riding suppression of anti-GPI responses in the arthritis model, or to the higher efficacy at suppressing IL17 responses, might be due to one of these sub-signatures. Using the Irf4- or Stat3-dependent gene-sets previously defined in mutant Treg cells (Zheng et al., 2009; Chaudhry et al., 2009), we observed no bias for Stat3-dependent transcripts (data not shown) but there was a significant over-representation of Irf4-dependent transcripts in Foxp3fgfp Treg cells, on both B6 and NOD backgrounds (Fig. 6E). Thus, Treg cells expressing the chimeric FoxP3 do not show a skewing of the entire Treg cell signature, but only a very focused bias, which overlaps with the imprint of Irf4, previously associated with the control of T helper activity and antibody production.
Altered protein-protein interactions between GFP-FoxP3 and transcriptional co-factors
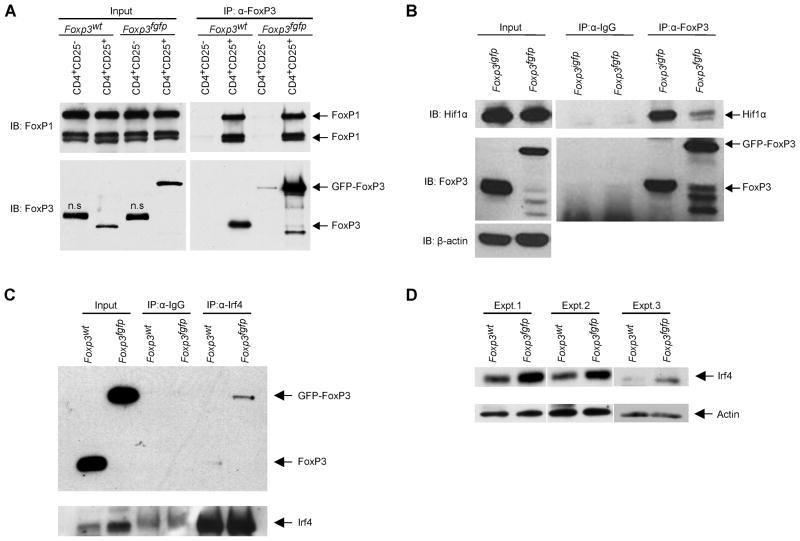
This limited and focused effect of GFP-FoxP3 on the Treg cell signature suggested that the insertion of eGFP into FoxP3 might affect its physical interaction with some of its functional co-factors. We thus performed co-immunoprecipation experiments with Treg cell extracts from Foxp3fgfp, and matched control littermates or Foxp3igfp mice. FoxP3 was immunoprecipitated from nuclear lysates, and associated factors were detected by SDS-PAGE immunoblotting. We first tested the association of FoxP3 with FoxP1, a heterodimer formation known to occur via the leucine-zipper region (Lopes et al., 2006; Li et al., 2007b) and thus presumably away from the GFP insertion site. Co-immunoprecipitation of FoxP1 was equivalent with both FoxP3 and GFP-FoxP3 (Fig. 7A), indicating that the overall structure of GFP-FoxP3 was not grossly perturbed. In contrast, the interaction with the transcription factor Hif1α, recently suggested to antagonize Treg cell differentiation by promoting FoxP3 proteasomal degradation (Shi et al., 2011; Dang et al., 2011), was almost completely abrogated in GFP-FoxP3 relative to WT FoxP3 (Fig. 7B); this loss of interaction may plausibly account for the increased amount of FoxP3 protein observed (Fig. 4A). The interaction of FoxP3 with Irf4 was tested by co-immunoprecipitation with anti-Irf4, revealing a higher amount of FoxP3-Irf4 complexes in Foxp3fgfp Tregs relative to control (Fig. 7C), consistent with the over-representation of Irf4 dependent gene transcripts. In the course of these experiments, we also observed that Irf4 protein expression itself is higher in Foxp3fgfp Tregs (Fig. 7D), which may contribute to the elevated Irf4-GFP-FoxP3 interaction.
Figure 7. GFP-FoxP3 differentially binds transcriptional co-factors.
(A) Association of FoxP3 with FoxP1 determined by co-immunoprecipitation. Anti-FoxP3 was used to immunoprecipitate FoxP3 from nuclear lysates of CD4+CD25− Tconv or CD4+CD25+ Treg cells from B6.Foxp3fgfp or WT mice, and immunoblots were probed for FoxP1 (anti-FoxP3 as control). (B) Nuclear lysates of B6.Foxp3fgfp and B6.Foxp3igfp CD25+ Treg cells were immunoprecipitated with anti-FoxP3 or control IgG, and probed with anti-Hif1a (anti-FoxP3 and anti-β–actin as controls). (C) Anti-Irf4 was used to immunoprecipitate Irf4 from nuclear lysates CD4+CD25+ Treg cells from B6.Foxp3fgfp or WT mice, and immunobloted for FoxP3. (D) Western blot analysis of Irf4 from nuclear lysates of CD4+CD25+ Treg cells from B6.Foxp3fgfp or WT mice, two independent experiments shown.
DISCUSSION
The observations reported here imply that perturbations in Treg cell phenotype and functional characteristics can affect autoimmune diseases in a diametrically opposite manner. These can be traced to a discrete genomic signature, identifying candidates for this biased function, and to perturbations in the interaction between FoxP3 and transcriptional cofactors. These results indicate that specific modifications of FoxP3 and of its interactions may be at play during the unfolding of autoimmune diseases, and could serve as a basis to therapeutically modulate Treg cell function in a qualitative manner.
From a practical standpoint, these results need to be taken into consideration in the interpretation of some experiments performed with the Foxp3fgfp line in recent years. Foxp3fgfp reporter mice have been used by a large number of investigators to track Treg cells. In some cases we would anticipate the discrete phenotypic and transcriptional perturbations reported here to have little or no impact. On the other hand, the insertion does have pleiotropic effects, as it influences Treg lineage commitment, FoxP3 expression, and the interaction with several transcriptional co-factors. The GFP-FoxP3 chimera might be expected to perturb experimental situations dependent on particular facets of Teff responses, and sensitive to the variation in Treg subphenotypes, such as the NOD or K/BxN autoimmune models analyzed here.
From the standpoint of autoimmune diseases, the dichotomous outcome is consistent with the different effector mechanisms involved in NOD diabetes and K/BxN arthritis. Pathogenesis in the former case involves inflammatory infiltration of the target organ directly by autoreactive Th1 cell-like T cells, and it is independent of IL-4 (Wang et al., 1998; Katz et al., 1995; Hung et al., 2005). Although the impact of IL-17 on NOD diabetes has yet to be conclusively established, there are several indications that IL-17 is protective, as demonstrated in several reports showing that mice with high amounts of IL-17 induced by helminth infection, Segmented Filamentous Bacteria (SFB) or NADPH oxidase deficiency are protected from diabetes (Lau et al., 2011; Nikoopour et al., 2010; Tse et al., 2010; Kriegel et al., 2011). In the KxB/N model, in contrast, pathology is due to overwhelming T cell help to B cells, in a process strongly dependent on both IL-4 and IL-17 (Ohmura et al., 2005; Wu et al., 2010). Treg cells in Foxp3fgfp mice are the genomic opposite of Irf4-deficient Treg cells, which are primarily defective in controlling Th2- or Th17-linked B cell responses, resulting in a pathology dominated by uncontrolled production of autoreactive Ig (Zheng et al., 2009): the discrete signature whose expression was lost in the Irf4-deficient cells proved over-expressed in Foxp3fgfp Treg cells. Correspondingly, the in vitro assay showed an enhanced ability of Foxp3fgfp Treg cells to suppress IL-17 production, but not IL-4 (although the assay may have been saturated). This heightened ability of Foxp3fgfp Treg cells to suppress Th17 cell differentiation would translate as more effective suppression of anti-GPI autoantibodies and arthritis in vivo.
One might speculate that imbalances in the ability of Treg cells to suppress different facets of effector cell responses may occur naturally and are involved in the determinism of autoimmune diseases. The consensus is that neither diabetes nor arthritis patients have generic defects in Treg cell numbers or basic functionality. But more focused perturbations in their flavor of suppressor activity may contribute to arthritis or diabetes susceptibility, conferred by genetic variation or environmental exposure.
There is an interesting parallel between the effects reported here and those of gut-resident microbes. We have recently reported that SFB enhance K/BxN arthritis but protect NOD mice from diabetes (Wu et al., 2010; Kriegel et al., 2011), a mirror image of the effect of GFP-FoxP3. In this vein, we have noticed that the protection from arthritis conferred by Foxp3fgfp Treg cells can be more or less marked in different animal facilities (less dramatic in conditions where the disease is inherently more severe). Variations in IL-17 may be the common link, but one might speculate that SFB also acts by influencing Treg subphenotypes.
Underlying these disease changes are the subtle but highly evocative alterations detected in the transcriptome of Foxp3fgfp Treg cells. Of particular interest is the strong overlap with the Irf4-dependent gene-set determined by Zheng et al (Zheng et al., 2009). This Irf4 gene signature includes several of the “usual suspects” amongst effector molecules (Ctla4, Tigit, Icos, Fgl2). It also includes TFs such as Ahr, Blimp-1 (encoded by Prdm1) and Maf that control each other as well as some of the same targets (Apetoh et al., 2010; Marshall et al., 2008). The Irf4 > Blimp1 connection has been previously noted as key elements of a genetic regulatory module in B cell differentiation (Sciammas et al., 2011) and in Treg cells (Cretney et al., 2011). Fittingly, we found a slight positive bias of the Blimp1 signature defined by Cretney et al. But this bias was less pronounced as that of the Irf4 signature, and since Prdm1 transcripts are themselves elevated, the inference is that the GFP-FoxP3 protein impacts on this regulatory module at the level of Irf4, or higher.
Underlying these genomic alterations, likely, are the altered interactions of FoxP3 with its transcriptional partners, as shown here and by Bettini et al 2012. Modifying FoxP3 by insertion of the 238 amino acids of GFP at the N-terminus might be expected, by steric hindrance, to perturb physical interactions occurring through N-terminal motifs of FoxP3 or in its 3-dimensional vicinity, but not others involving distant domains or a different face of the folded protein. The GFP insert has no effect on dimerization with FoxP1, which occurs via leucine zipper domains, some distance away from the N-terminus; correspondingly, interaction with NF-AT via the FKHR domain (Wu et al., 2006) is not affected. But interaction with Hif1α (this paper), Eos, HDAC7 and Tip60 (Bettini et al 2012) are reduced or abolished. Hif1a has recently been shown to directly bind FoxP3, resulting in its proteasomal degradation, and thus affecting Treg cell differentiation (Shi et al., 2011; Dang et al., 2011). This loss of Hf1-FoxP3 interaction, plausibly due to steric hindrance from the bulky domain of GFP, is consistent with the higher levels of FoxP3 in FoxP3fgfp Tregs
How the GFP insert would enhance the interaction with Irf4 is less immediately obvious. Some of this increased representation may simply be due to the increased levels of Irf4 in the cell, although this increase (20–40% by densitometry) may not be sufficient to account for the full increase in FoxP3-Irf4 complex. A simple integrative interpretation is that Hif1α and Irf4 partially compete for binding sites on FoxP3, and the inability of GFP-FoxP3 to interact with Hif1a facilitating Irf4 binding (indeed, IRF4 and FoxP3 synergize to activate Irf4 transcription – data not shown). Tripartite molecular interactions between Hif1α, Irf4 and FoxP3 may also be involved (although interaction between HIF1α and Irf4 has not been reported, HIF1α is stabilized when bound to herpes-virus-derived viral IRF3, which shares significant homology to cellular Irf4 (Shin et al., 2008)). An alternative possibility is that post-translational modifications of FoxP3, mapping to the N-terminus or indirectly affected by the presence of the GFP insertion, would condition FoxP3’s interactions with Irf4. For instance, acetylation of the N-terminal region of FoxP3 results from balanced HAT/HDAC activities (Samanta et al., 2008; Li et al., 2007a). These would very likely be affected by the neighboring insertion, thus indirectly affecting Irf4 binding.
In summary, there is a strong connection between the different facets of these analyses, linking enhanced FoxP3-Irf4 interactions, to higher expression of Irf4-dependent Treg cell genes, to stronger suppression of Th2 and Th17 cell-linked functions and autoimmune disease. It will be important to position these in the broader context of a molecular map of FoxP3’s interactions, but these results already open the way to investigations of FoxP3-related genomics in human patients and avenues for therapeutic modulation.
EXPERIMENTAL PROCEDURES
Mice and autoimmune evaluation
Foxp3igfp mice were provided by Dr. V. Kuchroo. The Foxp3fgfp (aka Foxp3tm2Ayr) trait was crossed to generate K/BxN.Foxp3fgfp, NOD/LtJ.Foxp3fgfp, C57BL/6.Foxp3fgfp, mice (>8 generations on both backgrounds), which were bred and maintained in SPF facilities at Harvard Medical School (IACUC protocol 02954 and 3024). Arthritis scores and ankle thickness measurements, determination of anti-GPI titers were as described (Monach et al., 2008). Mice on the NOD background were monitored biweekly for urine glucose, with confirmation of diabetes by blood glucose determination; mice were considered diabetic with 2 consecutive readings of blood glucose >250 mg/dL.
Treg cell isolation from non-lymphoid tissue
For intestinal lamina propria analysis, mesenteric fat tissue from was removed, the intestine was opened longitudinally, washed in ice-cold PBS and cut into 1 cm pieces, which were incubated in 25 mL DMEM containing 5 mM EDTA, 0.145 mg/mL DTT for 40 min at 37°C at a rotation speed of 200 rpm. After incubation, the epithelial cell layer was removed by vortexing and passing through a 100 μm cell strainer. The remaining intestinal pieces were washed in PBS, cut into 1 mm2 pieces with scissors, and digested in 25 mL of DMEM supplemented with 1 mg/mL Collagenase D (Roche), 0.15 mg/mL DNase I (Sigma), 200 ng/mL liberase TL (Roche), at 37°C for 20 min with rotation (200 rpm), then vortexed for 1 min and passed through a 100 μm cell strainer.
For Treg cells from visceral adipose tissue, blood was flushed from the organs by perfusion of PBS through the portal vein and the heart ventricle, epididymal adipose tissue was removed, cut into small pieces and digested for 20 minutes at 37°C with collagenase type II (Sigma) in DMEM. Cell suspensions were then filtered through a sieve, and the stromovascular fraction was harvested by centrifugation.
In vitro suppression assay
Sorted splenic Teff cells (CD4+GFP−) were cultured, with or without added Treg cells, for 72 hrs in RPMI 1640, 10% FCS, 2 mM L-glutamine, penicillin/streptomycin at 2.5 × 105 cells/well in round bottom 96-well plates, supplemented with 1 × 105 antigen presenting cells (APC) and 1.5 μg/mL anti-CD3. Cell proliferation was measured by incorporation of H3-thymidine (1 μCi added in the last 18 h of culture).
In vitro suppression of Th differentiation
Splenic naïve T cells (CD4+CD25−CD44loCD62Lhi) from B6 mice were sorted and cultured in complete culture medium at 1×105 cells/well in Th1 (anti-CD3/CD28 beads + 0.4 ng/mL of IL12), Th2 (anti-CD3/CD28 beads + 4 ng/mL Il4) or Th17 (anti-CD3/CD28 beads + IL6; 20 ng/mL and TGFβ; 2 ng/mL) skewing culture conditions in the presence or absence of Foxp3fgfp or Foxp3igfp Treg cells at a 1:1 Teff/Treg ratio. After 66 hours in culture, RNA was extracted from flow-sorted Teff cells (CD4+GFP−) for quantitative rtPCR (TaqMan).
In vitro Th17/Treg differentiation assay
Splenic naïve T cells (CD4+CD25−CD62Lhi) from B6.CD45.1 or B6.Foxp3fgfp (CD45.2) mice were purified by flow sorting and co-cultured at a 1:1 ratio under Th17 skewing conditions: 1×106 cells re-suspended in complete culture media and seeded into wells from 48 well plates coated with anti-CD3 (2 μg/mL) and supplemented with soluble anti-CD28 (2 μg/mL), IL-6 (30 ng/mL) and TGFβ (5 or 25 ng/mL). At 96 hours, cells were washed and cultured at 37 °C for 4 hours with 50 ng/ml phorbol 12-myristate 13-acetate (Sigma), 1 mM ionomycin (Sigma), and BD GolgiPlug (1:1000 dilution), then analyzed for IL17a and FoxP3 expression by flow cytometry after intracellular staining.
Microarray analysis
Duplicate samples of Tconv (CD3+CD4+GFP−) and Treg (CD3+CD4+GFP+) splenocytes were double-sorted to achieve > 99.0% purity, from 6 weeks old male Foxp3fgfp and Foxp3igfp mice (B6 and NOD backgrounds). Cells were collected directly into Trizol. RNA was labeled and hybridized to Affymetrix Mouse Genome M1.0 ST microarrays. Raw data were background-corrected and normalized using the RMA algorithm in the GenePattern software package (Heng and Painter, 2008). The cell populations analyzed were generated in duplicate. Data were analyzed with the “Multiplot” or “Heatmap” GenePattern modules. Microarray data are available from the National Center for Biotechnology Information/GEO repository under accession no. GSE37605.
Immunoprecipitation and immunoblotting
Pooled spleen and LN lymphocytes of Foxp3fgfp, Foxp3igfp or Foxp3wt B6 mice were magnetically purified with anti-CD25 antibodies, lysed on ice with hypotonic solution (10 mm HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.05% NP-40 like/IgePal Ca-630) supplemented with EDTA-free complete protease inhibitors (Roche). Nuclear pellets were subsequently treated with nuclear lysis buffer (20 mM HEPES, 300 mM NaCl, 20 mM KCl, EDTA-free complete protease inhibitor cocktail and MNase (Nuclease S7; Roche)). Chromatin digestion was stopped by adding EDTA to 5mM, and post nuclear supernatants were incubated with Protein-G Sepharose beads coupled to anti-FoxP3 antibodies (FJK16, eBioscience; 10 μg/reaction) for 2 hours at 4°C with constant rotation. Bound proteins were eluted by boiling, separated by SDS-PAGE, electro-transferred to PVDF. After blocking (2 hrs in 5% milk/1x PBS 0.02% Tween20), blots were probed overnight with anti-FoxP3 (FJK16, eBioscience), anti-HIF1a (Hif1alpha67, Novus Biologicals), anti-FoxP1 (Cell Signaling). FoxP3/Irf4 co-interactions used the Nuclear complex co-IP kit (Active Motif). Nuclear extract was immunoprecipitated with 2μg goat anti-Irf4 (Santa Cruz), captured on magnetic protein G beads, and Foxp3 was detected after SDS-PAGE and immunoblotting.
Supplementary Material
01
Acknowledgments
We thank Drs Dario Vignali and Matt Bettini for discussing their observations with us prior to publication, Dr Vijay Kuchroo for mice, Kimie Hattori and Adriana Ortiz-Lopez for help with mice, Joyce LaVecchio and Girijesh Buruzula for flow cytometry, Jeff Ericson and Scott Davis for help with microarray analyses and bioinformatics. Catherine Laplace for help with manuscript. This work was supported by grants from the NIH (AI051530, AI065858, DK092541) and the Juvenile Diabetes Research Foundation (JDRF) (4-2007-1057) to DM/CB. JN was supported by an Iacocca Foundation fellowship. JD was supported by an NRSA Fellowship, Postdoctoral Training Program in Cancer Immunology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, Burns EJ, Sherr DH, Weiner HL, Kuchroo VK. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol. 2010;11:854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science. 2010;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandukwala HS, Wu Y, Feuerer M, Chen Y, Barboza B, Ghosh S, Stroud JC, Benoist C, Mathis D, Rao A, et al. Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity. 2011;34:479–491. doi: 10.1016/j.immuni.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31:401–411. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Bettini M. Immunity. 2012 in press. [Google Scholar]
- Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009a;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol. 2009b;10:689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Hung JT, Liao JH, Lin YC, Chang HY, Wu SF, Chang TH, Kung JT, Hsieh SL, McDevitt H, Sytwu HK. Immunopathogenic role of TH1 cells in autoimmune diabetes: evidence from a T1 and T2 doubly transgenic non-obese diabetic mouse model. J Autoimmun. 2005;25:181–192. doi: 10.1016/j.jaut.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Jacobs JP, Wu HJ, Benoist C, Mathis D. IL-17-producing T cells can augment autoantibody-induced arthritis. Proc Natl Acad Sci U S A. 2009;106:21789–21794. doi: 10.1073/pnas.0912152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JD, Benoist C, Mathis D. T helper cell subsets in insulin-dependent diabetes. Science. 1995;268:1185–1188. doi: 10.1126/science.7761837. [DOI] [PubMed] [Google Scholar]
- Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korganow AS, Ji H, Mangialaio S, Duchatelle V, Pelanda R, Martin T, Degott C, Kikutani H, Rajewsky K, Pasquali JL, et al. From systemic T cell self-reactivity to organ-specific autoimmune disease via immunoglobulins. Immunity. 1999;10:451–461. doi: 10.1016/s1074-7613(00)80045-x. [DOI] [PubMed] [Google Scholar]
- Kouskoff V, Korganow AS, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, Lorca G, Li N, Sankar D, Wasserfall C, Neu J, et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol. 2011;186:3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, Basu S, Riley JL, Hancock WW, Shen Y, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A. 2007a;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Samanta A, Song X, Iacono KT, Brennan P, Chatila TA, Roncador G, Banham AH, Riley JL, Wang Q, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol. 2007b;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- Lopes JE, Torgerson TR, Schubert LA, Anover SD, Ocheltree EL, Ochs HD, Ziegler SF. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol. 2006;177:3133–3142. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- Marshall NB, Vorachek WR, Steppan LB, Mourich DV, Kerkvliet NI. Functional characterization and gene expression analysis of CD4+ CD25+ regulatory T cells generated in mice treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Immunol. 2008;181:2382–2391. doi: 10.4049/jimmunol.181.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monach PA, Mathis D, Benoist C. The K/BxN arthritis model. Curr Protoc Immunol. 2008;Chapter 15(Unit 15.22) doi: 10.1002/0471142735.im1522s81. [DOI] [PubMed] [Google Scholar]
- Nikoopour E, Schwartz JA, Huszarik K, Sandrock C, Krougly O, Lee-Chan E, Singh B. Th17 polarized cells from nonobese diabetic mice following mycobacterial adjuvant immunotherapy delay type 1 diabetes. J Immunol. 2010;184:4779–4788. doi: 10.4049/jimmunol.0902822. [DOI] [PubMed] [Google Scholar]
- Ohmura K, Nguyen LT, Locksley RM, Mathis D, Benoist C. Interleukin-4 can be a key positive regulator of inflammatory arthritis. Arthritis Rheum. 2005;52:1866–1875. doi: 10.1002/art.21104. [DOI] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, Jinasena D, Sharma SM, McCadden EM, Getnet D, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta A, Li B, Song X, Bembas K, Zhang G, Katsumata M, Saouaf SJ, Wang Q, Hancock WW, Shen Y, et al. TGF-beta and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc Natl Acad Sci U S A. 2008;105:14023–14027. doi: 10.1073/pnas.0806726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciammas R, Li Y, Warmflash A, Song Y, Dinner AR, Singh H. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol Syst Biol. 2011;7:495. doi: 10.1038/msb.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1{alpha}-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YC, Joo CH, Gack MU, Lee HR, Jung JU. Kaposi’s sarcoma-associated herpesvirus viral IFN regulatory factor 3 stabilizes hypoxia-inducible factor-1 alpha to induce vascular endothelial growth factor expression. Cancer Res. 2008;68:1751–1759. doi: 10.1158/0008-5472.CAN-07-2766. [DOI] [PubMed] [Google Scholar]
- Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse HM, Thayer TC, Steele C, Cuda CM, Morel L, Piganelli JD, Mathews CE. NADPH oxidase deficiency regulates Th lineage commitment and modulates autoimmunity. J Immunol. 2010;185:5247–5258. doi: 10.4049/jimmunol.1001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Gonzalez A, Katz J, Höglund P, Benoist C, Mathis D. Interleukin-4 deficiency does not worsen disease in NOD mice. Diabetes. 1998;47:1207–1211. doi: 10.2337/diab.47.8.1207. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Li B, Zhou Z, Hancock WW, Zhang H, Greene MI. Histone acetyltransferase mediated regulation of FOXP3 acetylation and Treg function. Curr Opin Immunol. 2010;22:583–591. doi: 10.1016/j.coi.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, Deroos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01