The Human Transporter Associated with Antigen Processing: MOLECULAR MODELS TO DESCRIBE PEPTIDE BINDING COMPETENT STATES (original) (raw)
Background: The transporter associated with antigen processing (TAP) is an ABC transporter whose experimental structure is not known.
Results: We have modeled TAP based on the crystal structures of three related ABC transporters.
Conclusion: We identified a possible peptide binding conformation in the transport cycle of TAP.
Significance: These models help interpret experimental data and give information about the transport cycle of ABC transporters in general.
Keywords: ABC Transporter, Molecular Docking, Molecular Dynamics, Molecular Modeling, Multidrug Transporters, Peptide Binding, Transporter Associated with Antigen Processing
Abstract
The human transporter associated with antigen processing (TAP) is a member of the ATP binding cassette (ABC) transporter superfamily. TAP plays an essential role in the antigen presentation pathway by translocating cytosolic peptides derived from proteasomal degradation into the endoplasmic reticulum lumen. Here, the peptides are loaded into major histocompatibility class I molecules to be in turn exposed at the cell surface for recognition by T-cells. TAP is a heterodimer formed by the association of two half-transporters, TAP1 and TAP2, with a typical ABC transporter core that consists of two nucleotide binding domains and two transmembrane domains. Despite the availability of biological data, a full understanding of the mechanism of action of TAP is limited by the absence of experimental structures of the full-length transporter. Here, we present homology models of TAP built on the crystal structures of P-glycoprotein, ABCB10, and Sav1866. The models represent the transporter in inward- and outward-facing conformations that could represent initial and final states of the transport cycle, respectively. We described conserved regions in the endoplasmic reticulum-facing loops with a role in the opening and closing of the cavity. We also identified conserved π-stacking interactions in the cytosolic part of the transmembrane domains that could explain the experimental data available for TAP1-Phe-265. Electrostatic potential calculations gave structural insights into the role of residues involved in peptide binding, such as TAP1-Val-288, TAP2-Cys-213, TAP2-Met-218. Moreover, these calculations identified additional residues potentially involved in peptide binding, in turn verified with replica exchange simulations performed on a peptide bound to the inward-facing models.
Introduction
ATP binding cassette (ABC)3 proteins are one of the largest families of active transporters. They use the hydrolysis of ATP as a source of energy to transfer substrates across membranes. ABC transporters import or export a wide variety of substrates, including sugars, lipids, peptides, metabolites, amino acids, metal ions, and xenobiotics. As a consequence, in humans, mutations in the 48 identified ABC transporter genes are associated with several diseases, including cystic fibrosis, macular degeneration, and lipid and sterol deficiencies (1–3). Despite the different chemistry and size of these substrates, the structural organization and the mechanism of action of ABC transporters are very similar (4).
Each ABC transporter consists of at least four domains: two nucleotide binding domains (NBDs), located on the cytosolic side of the membrane, and two transmembrane domains (TMDs), arranged in membrane-spanning helices to form the translocation pathway. Two ATP binding sites are formed at the interface between the NBDs, characterized by the presence of highly conserved motifs: the Walker A and Walker B motifs, the Signature motif, and the A-, H-, Q-, and D-loop (5). As a consequence of ATP binding, the NBDs form a tightly coupled dimer followed by the hydrolysis of ATP. The energy derived from this process is converted into large conformational changes of the TMDs to transport the substrate from one side of the membrane to the other.
The human transporter associated with antigen processing (TAP) plays a crucial role in the adaptive immune system (6). TAP translocates antigenic peptides derived from proteasomal degradation in the cytosol to the endoplasmic reticulum (ER) lumen, where the peptides are loaded into major histocompatibility complex (MHC) class I molecules to be in turn released and exposed on the cell surface. The inhibition of TAP mediated by viruses that try to evade the immune system as well as deletions or mutations of this transporter seriously impair the antigen presentation mechanism (7). TAP belongs to the ABC-B subfamily. It is a heterodimer formed by two half-transporters, TAP1 (ABC-B2) and TAP2 (ABC-B3). Each monomer consists of a NBD and six transmembrane helices forming the core of the TMD. TAP1 and TAP2 also have an additional N-terminal transmembrane domain (tapasin binding domain), specifically involved in the interactions with tapasin, which is one of the proteins responsible for the peptide-loading process (8, 9). High resolution structures have been published for the human and rat TAP1-NBD both as a monomer (human) and homodimer (rat) (10, 11), whereas the crystal structures of TAP2-NBD and the full-length transporter are not yet available. In the absence of high resolution structures, homology models, supported by experimental data, have been successfully used in the field of ABC transporters to provide feedback and give insights into the structural organization of these transporters (12–28).
In this paper we present molecular models of TAP describing the transporter in three different states. The models have been built based on a multiple sequence alignment taking into account the sequences of several ABC transporters, including mouse P-gp, human ABCB10, and the bacterial transporter Sav1866. The x-ray structures of P-gp (29), ABCB10 (PDB access code 2YL4), and Sav1866 (29, 30) have been solved in different conformations, providing valuable templates to describe different states of the transport cycle of TAP. Like TAP, P-gp is a member of the ABC-B subfamily and consists of one polypeptide chain arranged into two halves connected by a linker. Each half contains a cytosolic NBD and a TMD that is organized into six membrane-spanning helices. The crystal structure of P-gp shows the transporter in an inward-facing conformation, corresponding to an initial state of the transport cycle, with the translocation cavity open toward the cytosol and suitable for drug binding. The crystal structure of human ABCB10 corresponds to an inward-facing conformation of the transporter, but the separation between the two NBDs is smaller than in the P-gp crystal structure. Contacts between the two NBDs are detected between the Walker A motif of one chain and the residues of the D-loop (the serine residue of the SALD motif in particular) of the other chain. The homology models of TAP based on these crystal structures can describe a possible peptide binding competent state of the transporter. Sav1866 is a homodimeric ABC transporter, with a structural organization and membrane topology similar to P-gp and TAP. The crystal structure of Sav1866 corresponds to an outward-facing conformation of the transporter, with the two NBDs tightly coupled, and the cavity of the TMDs close to the cytosol and open toward the opposite side of the membrane. The molecular model of TAP based on this structure can represent a possible final step of its transport cycle, when a peptide is released to the ER lumen. We compared our models with the available experimental data and focused the analysis of the models on the TMDs and the ER-facing and cytosol-facing loops (ELs and CLs, respectively). Based on electrostatic potential calculations, we identified regions of the transporter that could be involved in peptide binding, and we verified our hypotheses with replica exchange simulations performed on a nonamer bound to the inward-facing models. The comparison of the models also provided structural insights into the rearrangements of the TMDs during the translocation cycle of this transporter.
EXPERIMENTAL PROCEDURES
Homology Modeling
To build the homology models of TAP, the sequences of human TAP1 and TAP2 were retrieved from UniProt Knowledgebase (UniProtKB) through the ExPASy molecular biology server (UniProtKB access codes Q03518 and Q03519, respectively) (31, 32). According to the membrane topology of TAP described in Schrodt et al. (9), the core of the TMD in TAP1 and TAP2 is formed by six membrane-spanning helices. For this reason, the first four helices of TAP1 corresponding to the tapasin binding domain and the first three helices of TAP2 were not included in the homology modeling approach. As a result, residues 173–742 for TAP1 and residues 138–686 for TAP2 were only taken into account. The residue numbers correspond to the numbering shown in Schrodt et al. (9).
A protein-protein BLAST (33) search was performed against the Protein Data Bank using the retrieved sequence of human TAP1 and human TAP2 as input. The BLAST search was performed in two steps using as a query sequence first the TMD part of TAP and then the sequence of the NBD. For the TMDs, the BLAST search identified the crystal structure of mouse P-gp (29), human ABCB10, Sav1866 (30), and MsbA (34) as suitable templates for both TAP1 and TAP2. Together with the sequences of mouse P-gp, human ABCB10, Sav1866, and V. cholera, Escherichia coli and Salmonella typhimurium MsbA (UniProtKB access codes P21447, Q9NRK6, Q99T13, Q9KQW9, P60752, and P63359 respectively), the sequence of rat TAP1 and TAP2 as well as the sequence of the heterodimeric ABC transporter TM0287/TM0288 from Thermotoga maritima were also taken into account to generate a multiple alignment with MAFFT (35). For the NBD alignment, from the BLAST search results we considered all the NBD structures with a sequence identity of at least 40% with respect to TAP1 or TAP2 NBD. Thus, in addition to the previously described transporters, we included the sequences of E. coli hemolysin B (36), human ABCB6 (37), and MRP2 from Plasmodium yoelii (38). The alignments are shown in supplemental Figs. S1 and S2. TAP homology models were built by means of MODELLER9v10 using the structures of P-gp, ABCB10, and Sav1866 as templates (39). Among these transporters, P-gp is the only one with two non-symmetric TMDs. For this reason, we built two TAP models based on P-gp, so that in one case TAP1 was modeled on the first halve of P-gp and TAP2 on the second one, and vice versa for the second model. These two P-gp-based models are described in the text as P-gp-based model 1 and model 2, respectively. Per each template, 20 models were generated, and the one with the best score in terms of the discrete optimized protein energy (DOPE) potential implemented in MODELLER was chosen. The assessment of the final structural models was carried out with Molprobity (40, 41), Qmean (42, 43), Procheck (44, 45), and Whatcheck (46) analyses (supplemental Table S1). The TAP models were then inserted into a 1-palmitoyl-2-oleoyl-_sn_-glycero-phosphocoline (POPC) lipid bilayer using the g_membed method (47). Gromacs 4.5.5 (48, 49) was used to optimize the geometry of the final models with the ff54a7 version of the GROMOS 96 force field (50, 51). The geometry optimization of each system was carried out in several steps described in supplemental Table S2. After minimization, the system was equilibrated for 10 ns in the isothermal-isobaric ensemble (NPT) at a temperature of 310 K and pressure of 1 bar, with position restraints (force constant of 10,000 kJmol−1nm−2) on all the non-hydrogen atoms of the protein, followed by a 5-ns run with position restraints (force constant of 100 kJmol−1nm−2) only on the backbone atoms.
We previously published a homology model of TAP based on a sequence alignment that included only the sequences of TAP1, TAP2, and Sav1866, whose crystal structure was used as a template (12). In this study we used a consistent alignment, but we included additional transporters, thus building models of TAP on templates with substantially different conformations.
Electrostatic Potential Calculations
Electrostatic calculations were carried out using APBS software (52). A dielectric constant of 2, 10, and 80 was set for membrane, protein, and water, respectively. All the parameters used for these calculations are listed in supplemental Table S3. To take into account the flexibility of the side chains, the electrostatic potential was calculated for 50 frames with 50-ps spacing during the last 2.5 ns of the equilibration run performed with position restraints only on the backbone atoms. The average of these calculations was then used for analysis.
Replica Exchange Simulations
Replica exchange simulations were performed on the TAP models based on P-gp and ABCB10 structures using Gromacs 4.5.5 (48, 49). The peptide was initially pulled into the cavity by restraining the distance between the α-carbon (Cα) of TAP1-Ser-289 and -Arg-1 of the peptide and between the Cα of TAP2-Ser-254 and -Leu-9 of the peptide (the maximum allowed distance was 6 Å). Replica exchange simulations were performed in implicit solvent with the Amber99sb-ildn force field and GBSA with OBC algorithm (53). 44 temperatures ranging from 298.2 to 433.0 K were initially selected. Short simulations (0.4 ns) were carried out to iteratively change the temperature spacing until the observed acceptance probability between adjacent replicas was between 0.3 and 0.4. The acceptance probability of exchanges between replicas depends on their temperature gap. The acceptance probability of 0.2–0.4 generally provides an optimum balance between number replicas and the temperature range covered by replica simulations (54). The production runs of 15 ns (P-gp based model 1), 20 ns (P-gp based model 2), and 30 ns (ABCB10 based model) were then initiated from those temperatures during which the exchange was attempted every picosecond. The backbone of the transporter was kept frozen during the simulations. As we were interested in sampling only conformations of the peptide in the inward-facing cavity, a flat-bottom quadratic potential was implemented in Gromacs to restrain the peptide center of mass within 1 nm of its initial position.
The algorithm of Daura et al. (55), as implemented in the g_cluster tool of Gromacs, was used to cluster the peptide conformations based on the backbone atoms, using a root mean square deviation (r.m.s.d.) cutoff of 0.2 nm. The r.m.s.d. between the peptide backbone of any two frames was calculated without least square fitting of the structures. The percentage population of each cluster at the lowest temperature (298.2 K) was determined by combining data from all the temperatures using pyMBAR (56).
Figs. 1–3 and 5 and supplemental Figs. S3–S6 were produced with PyMOL, Version 1.4.1 (57), and Fig. 4 and supplemental Fig. S7 were produced with the UCSF Chimera package, Version 1.6.1 (58).
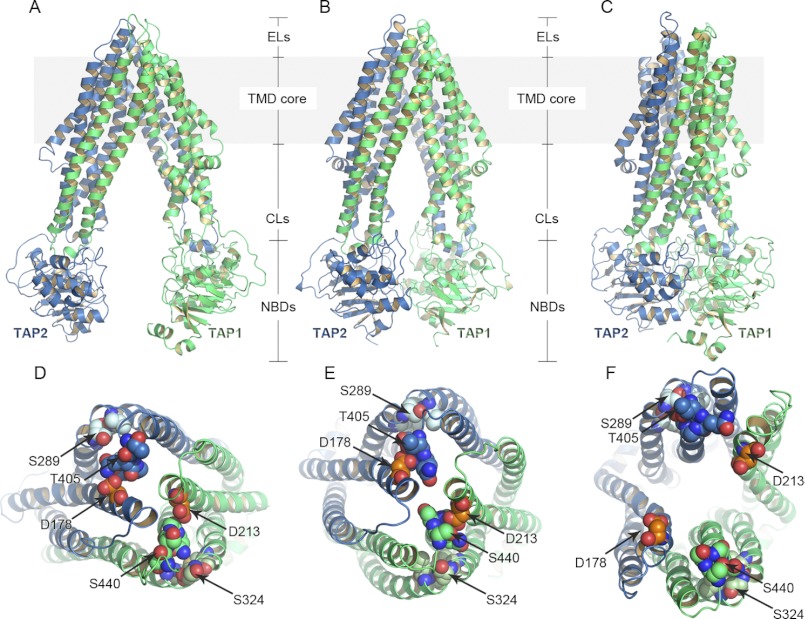
FIGURE 1.
Homology models of TAP-based on P-gp (inward-facing conformation) (A), ABCB10 (inward-facing conformation) (B), and Sav1866 (outward-facing conformation) (C) crystal structures. TAP1 and TAP2 are shown in green and blue ribbons, respectively. The region of the transporter embedded in the membrane (TMD core) is highlighted in gray. Short loops connect the transmembrane helices on the ER lumen (ELs), whereas longer CLs are projected into the cytosol followed by the NBDs. Peptides bind to TAP in the inward-facing cavity, whereas ATP molecules bind to the NBDs, inducing NBD dimerization. The energy derived from ATP hydrolysis is used by TAP to translocate the peptides from the cytosol to the ER. D–F, shown is a top view of the models based on P-gp (D), ABCB10 (E), and Sav1866 (F). The conserved residues of the ELs described under “Conserved Regions in ELs” are shown as spheres. In particular, TAP1-Asp-213 (orange spheres) interacts with TAP1-Ser-440 (blue spheres), and TAP2-Asp-178 (orange spheres) makes contacts with TAP2-Thr-405 (blue spheres) in the inward-facing conformation models (D and E). As a consequence of the cavity opening toward the ER lumen in the outward-facing conformation (F), these interactions are not longer retrieved. The interactions between the conserved residues of the EL2 (TAP1-Met-320, -Ser-324, -Leu-327, -Thr-328, light green spheres; TAP2-Met-285, -Ser-289, -Leu-292, -Thr-293, light blue spheres) are preserved in both the inward-facing and the outward-facing conformations (D–F). For homology modeling, the sequence alignments (supplemental Figs. S1 and S2) were generated with MAFFT (35), and the models were built with MODELLER (39). The TAP models were then inserted into a lipid bilayer as described under “Experimental Procedures.”
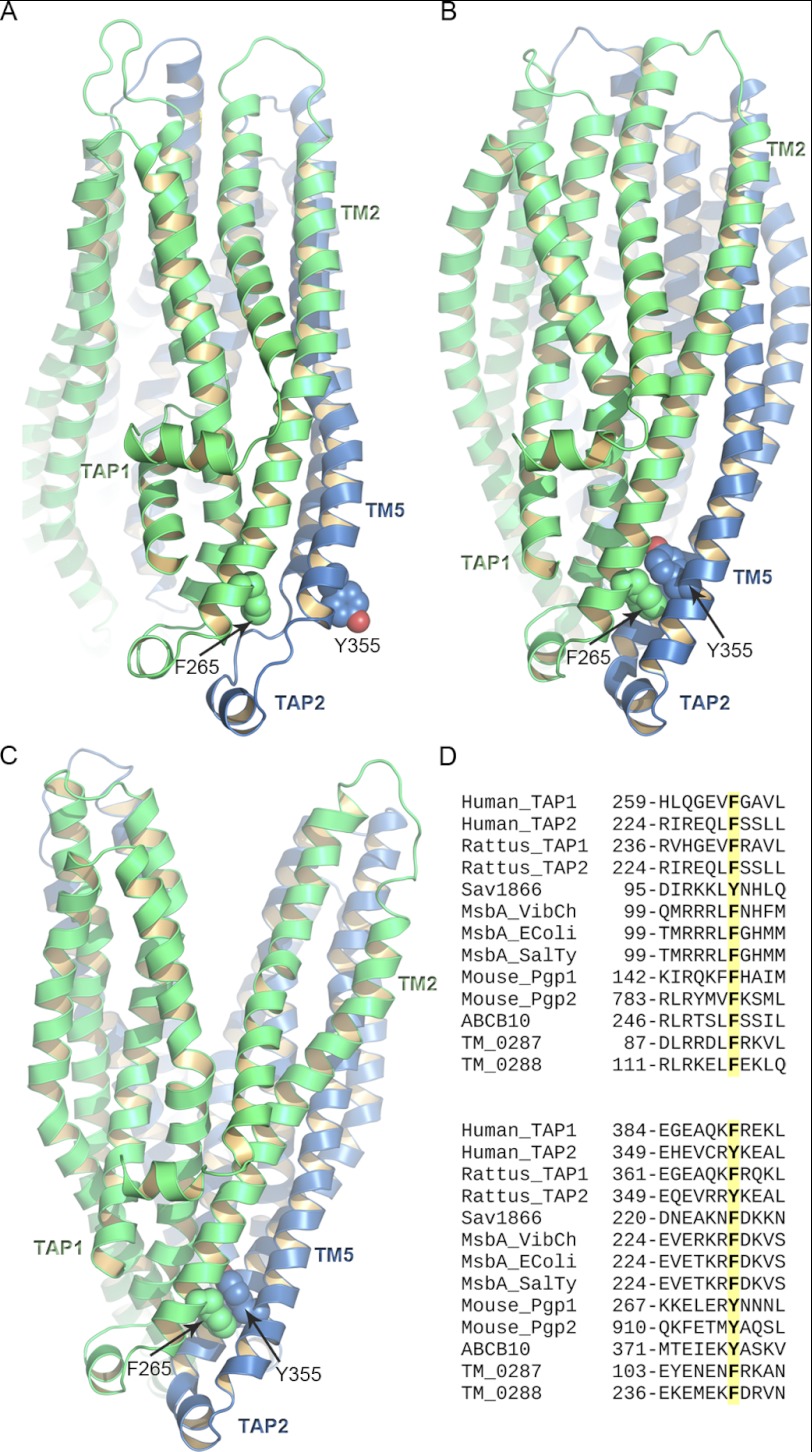
FIGURE 2.
π-Stacking interactions between TM2 and TM5. A–C, TAP1 and TAP2 are shown in green and blue ribbons, respectively. Side view of the TMDs of the TAP models based on P-gp (P-gp-based model 2, A), ABCB10 (B), and Sav1866. TAP1-Phe-265 and TAP2-Tyr-355 involved in the π-stacking interactions in the outward-facing conformation are represented as green and blue spheres, respectively. The NBDs are not shown for clarity. D, the multiple sequence alignment shows that in all the transporters taken into account in this study, the positions corresponding to TAP1-Phe-265 and TAP2-Tyr-355 are conserved. The sequences were retrieved from the UniprotKB data base, and their accession codes are as follow: human TAP1, Q03518; human TAP2, Q03519; Rattus, TAP1, P36370; Rattus, TAP2, P36372; Sav1866, Q99T13 (PDB access code 2HYD); MsbA Vibrio cholerae (Msba_VibCh), Q9KQW9 (PDB access code 3B5X); MsbA E. coli (MsbA_EColi), P60752 (PDB access code 3B5W); MsbA S. typhimurium (MsbA SalTy), P63359 (PDB access code 3B60); mouse P-glycoprotein (Mouse_Pgp1 and Mouse_Pgp2), P21447 (PDB access code 3G5U); ABCB10, Q9NRK6 (PDB access code 2YL4), T. maritima 0287 (TM_0287), Q9WYC3; T. maritima 0288 (TM_0288), Q9WYC4.
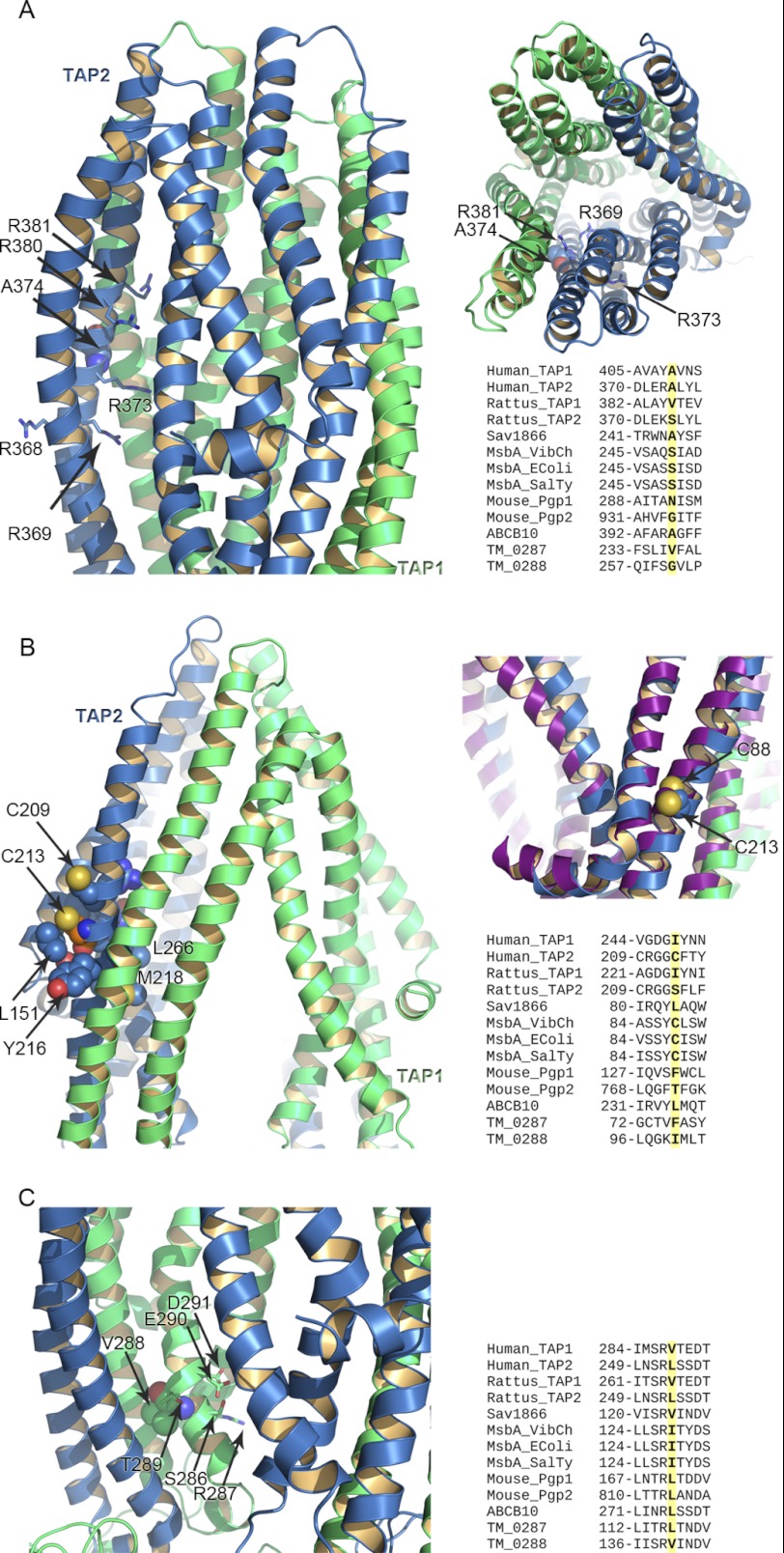
FIGURE 3.
Residues affecting peptide binding and transport. A–C, TAP1 and TAP2 are shown in green and blue ribbons, respectively. A, shown is a side view of the TMDs of the TAP homology model based on ABCB10 (left) and Sav1866 (right) crystal structure to highlight the position occupied by TAP2-Ala-374, represented as spheres. The surrounding TAP2 arginine residues (Arg-368, Arg-369, Arg-373, Arg-380, Arg-381) are in blue sticks. The multiple sequence alignment for the residues surrounding TAP2-Ala-374 is also provided. B, shown is a view of TAP2-Cys-213 in the TAP homology model based on ABCB10. Cys-213 is shown as orange spheres, whereas the neighborhood residues are in blue spheres. The multiple alignment of the region surrounding Cys-213 is also shown. On the right is the superimposition of the homology model of TAP based on Sav1866 and the MsbA crystal structure (PDB access code 3B60) shown in purple ribbons. TAP2-Cys-213 and MsbA-Cys88 are shown as spheres. C, shown is a close-up view of Val-288 of TAP1 in the TAP homology models based on ABCB10. The multiple alignment highlights the residues surrounding Val-288 (Gly-282, Ile-284, Ser-286, Arg-287, and Asp-291) and conserved among other transporters.
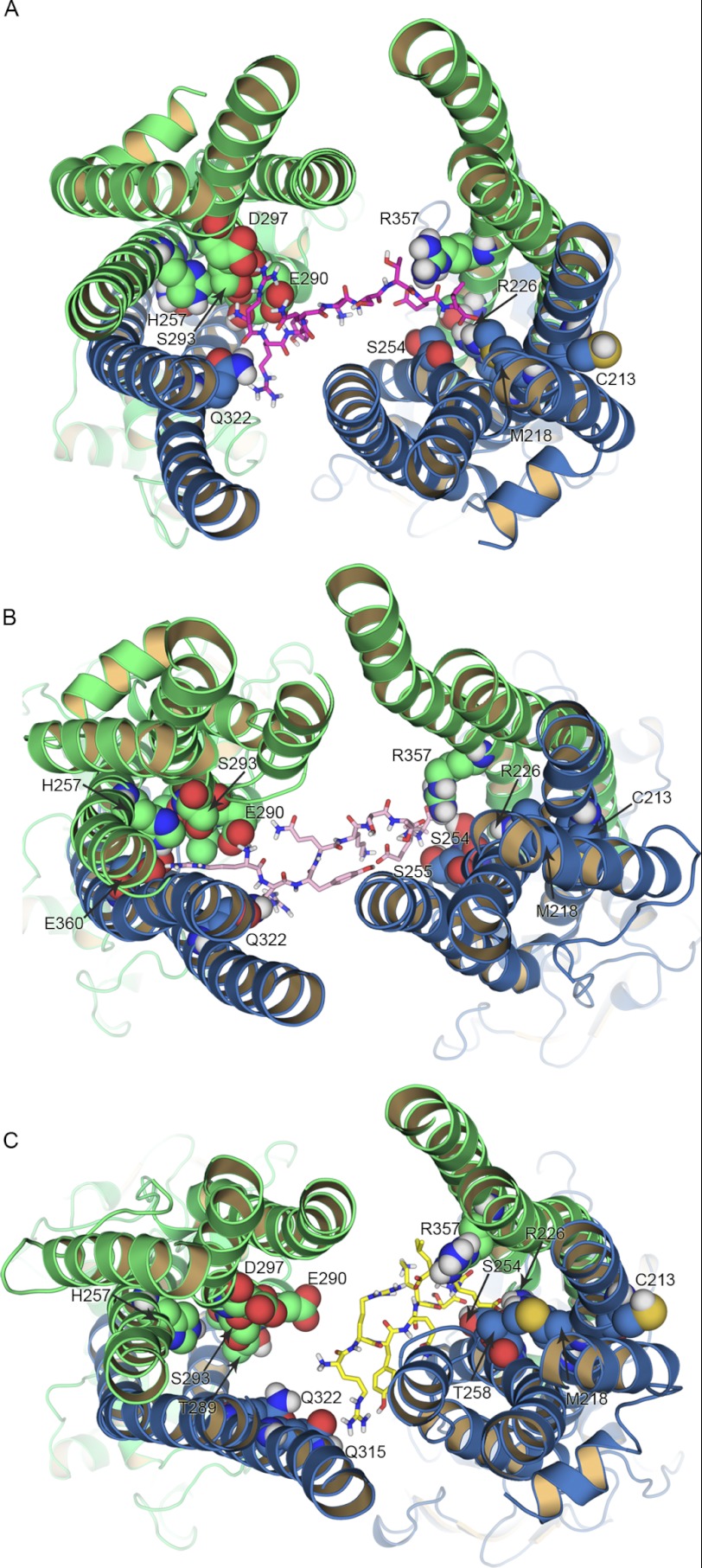
FIGURE 5.
Replica exchange simulations and peptide binding. A–C, TAP1 and TAP2 are shown in green and blue ribbons, respectively. Shown is the top view of the nonamer RRYQKSTEL bound to the inward-facing models of TAP, as it results from the replica exchange simulations and clustering analysis. The peptide conformation corresponding to the cluster with the highest population is shown as sticks for the TAP model based on ABCB10 (A), the P-gp-based model 2 (B), and the P-gp-based model 1 (C). The residues involved in the binding of the N and the C terminus of the peptide are shown as spheres. Only polar hydrogen atoms are shown. For clarity, the loops facing the ER lumen are not shown. Replica exchange simulations were performed in implicit solvent using 44 temperatures ranging from 298.2 to 433.0 K.
FIGURE 4.
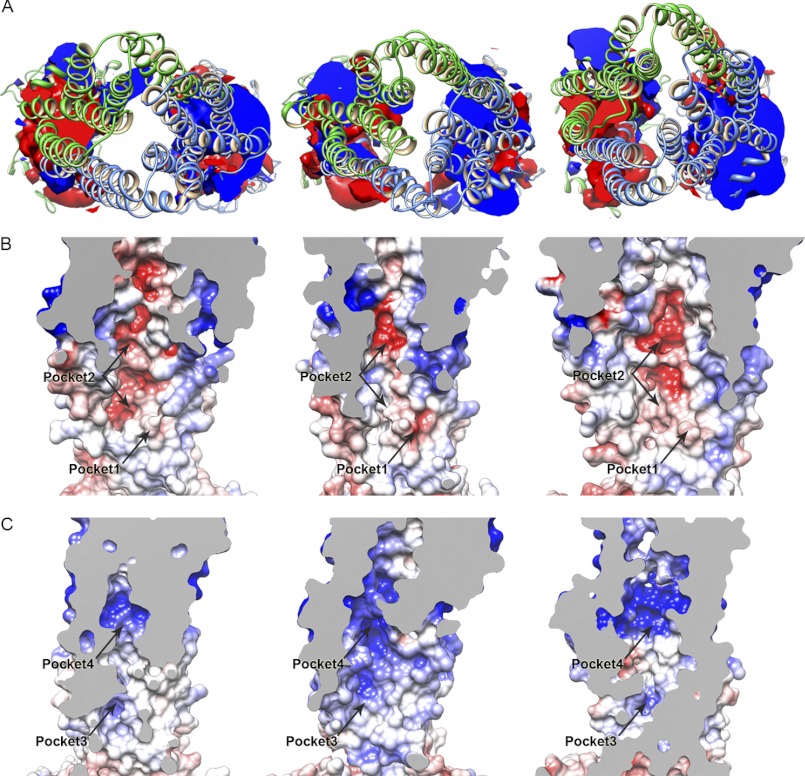
Electrostatic potential of the TAP inward-facing models. A, shown is an isosurface representation of the electrostatic potential of the TAP P-gp-based model 2 (left panel), P-gp-based model 1 (center panel), and ABCB10-based model (right panel). The isosurface has been clipped to highlight the different properties of the lower side of the inward-facing cavity. The isovalue is set to −2 k_B_T/e for the negative potential (red) and +2 k_B_T/e for the positive one (blue). TAP1 and TAP2 are shown in green and blue ribbon, respectively. B and C, per each model (P-gp based model 2 and model 1 on the left and center panel, respectively; ABCB10-based model on the right panel), the electrostatic potential of the inward-facing cavity was mapped on the solvent-accessible surface. Shown are the negatively (B) and positively (C) charged side of the cavity. The isovalue was set to −4 k_B_T/e for the negative potential (red) and +4 k_B_T/e for the positive potential (blue). Pockets 1, 2, 3, and 4 described in the text under “Peptide Binding” are highlighted. Electrostatic potential calculations were performed with APBS software (52) using a dielectric constant of 2, 10, and 80 for membrane, protein, and water, respectively. The solvent-accessible surface was calculated based on the last frame of the 5-ns equilibration run, whereas the electrostatic potential was averaged during the last 2.5 ns, as described under “Experimental Procedures.”
RESULTS AND DISCUSSION
Homology Modeling; Structural Organization of the TAP Models
The transmembrane core of TAP consists of 12 transmembrane helices (8). The same membrane topology characterizes P-gp, ABCB10, and the bacterial transporter Sav1866, chosen as templates to build homology models of TAP. Both TAP1 and TAP2 have an additional N-terminal transmembrane domain required for the binding of tapasin and the assembly of the macromolecular MHC class I peptide loading complex (8, 59–61). TAP constructs lacking these domains are, however, fully functional in terms of peptide binding and transport (59). As a consequence, modeling TAP in the absence of the tapasin binding domains still provides valuable models to analyze and possibly predict the regions of the transporter involved in peptide binding.
The r.m.s.d. calculated on the backbone atoms between the final P-gp-based model 1 and model 2 of TAP and the P-gp structure is 1.6 and 2.3 Å, respectively. The r.m.s.d. between the final ABCB10-based model and the ABCB10 crystal structure is 0.3 Å, whereas the r.m.s.d. between the final Sav1866-based model of TAP and the Sav1866 structure is 0.4 Å. The higher r.m.s.d. that characterizes the P-gp-based models is due to differences in the NBDs. For these models, the r.m.s.d. was calculated separately for TMDs (TAP1 residues 173–483; TAP2 residues 138–448; P-gp residues 33–366 and 694–1009) and NBDs (TAP1 residues 503–743; TAP2 residues 468–686; P-gp residues 388–625 and 1031–1256). For P-gp based model 1 and model 2, the r.m.s.d. for the TMDs is 1.5 and 1.6 Å, whereas the r.m.s.d. for the NBDs is 1.7 and 2.7 Å, respectively. However, the secondary structure of the NBDs of TAP is perfectly preserved in these models, and the higher r.m.s.d. values come from a translation of the NBDs when compared with the crystal structure.
The structural organization of the TAP models after insertion in the membrane and geometry optimization of the side chains is shown in Fig. 1. In the P-gp- and ABCB10-based models, the ER portion of TM1 and TM6 of both TAP1 and TAP2 as well as the ER loop connecting TM1 with TM2 (EL1) and TM5 with TM6 (EL3) close the top of the cavity in the inward-facing conformation. On the other side of the membrane, the cytosolic loop (CL1) between TM2 and TM3 of one of the two half-transporters is in contact with the CL2, between TM4 and TM5, and the NBD of the other half-transporter. In the TAP model based on Sav1866, as a result of the NBD dimerization, new interactions on the intracellular side of TAP1 and TAP2 are established to close the cavity to the cytosol.
NBDs
The NBDs of ABC transporters contain highly conserved key motifs involved in ATP binding and have been extensively characterized from a structural point of view. When the NBDs associate to form a proper dimer, residues of the Walker A, Walker B, A-loop, Q-loop, and Switch motifs from one NBD combine with the signature and the D-loop motifs of the second NBD (62).
TAP and other medically relevant ABC transporters such as the cystic fibrosis transmembrane conductance regulator have two ATP binding sites that are not equivalent, with substitutions in the conserved motifs. As a consequence of this NBD asymmetry, one binding site is highly conserved in a catalytically competent state, whereas the second binding site may have an impaired catalytic activity. In TAP1, the H-loop is replaced with a glutamine, and the highly conserved glutamate of the Walker B motif is replaced by an aspartate. Moreover, the signature motif of TAP2, involved in the hydrolysis occurring at the TAP1-ATP binding site, is characterized by a non-conserved sequence (supplemental Fig. S2). Thus, the ATP binding site in TAP1-NBD is degenerate with respect to the consensus site in TAP2-NBD. Biochemical and crystallographic studies performed on a TAP1-NBD homodimer with mutations introduced to mimic the degenerate site showed that the consensus site is responsible (i) for the ATPase activity needed to promote the transport and (ii) for the NBD dimerization (11). Conversely, the degenerate site has a lower ATPase activity (11).
In the TAP model based on the Sav1866 crystal structure, the NBDs are dimerized (supplemental Fig. S3_A_), and no structural differences were apparent when compared with the TAP1-NBD homodimer with mutations introduced in the conserved motifs (11). This suggests that homodimers like Sav1866 and heterodimers like TAP share the same architecture in their fully dimerized state. The dimer conformation of TAP corresponding to the ABCB10 structure allows the binding of nucleotide analogs, but no NBD closure and no contacts are present between the nucleotide analog and the signature motif. However, it is interesting to note that although in the P-gp-based conformation the NBDs are far apart, in this model the Walker A of one domain is in close proximity of the D-loop serine residue of the second NBD (supplemental Fig. S3_B_). This may imply that NBD disengagement may be reached even without a large separation between the NBDs. Recently, the crystal structure of the heterodimeric ABC transporter TM287/288 has been released (63), providing an additional template to model TAP. In this case, TAP takes an inward-facing conformation, with an ATP analog bound only to the degenerate site, whereas a larger separation between the monomers characterizes the consensus site (supplemental Fig. S3_C_). A superimposition of the TAP2-NBD from the model based on Sav1866 and the model based on TM287/288 shows that the NBD conformational differences between these two states reside mainly in the D-loop and the glutamate residue of the Walker B motif, whereas no differences are observed for the TAP1-NBDs (supplemental Fig. S3, D–E).
Conserved Regions in ELs
Although it is known that the CLs play a significant role in the mechanism of ABC transporters (12, 30, 34), the analysis of the multiple sequence alignment performed in this study shows additional conserved regions in the three loops facing the ER lumen (ELs) in TAP or the extracellular side in other ABC transporters (Fig. 1). The presence of small pools of amino acids that are more conserved than the transmembrane helices might suggest a structural role for these residues. The EL1 can be of different lengths among the transporters taken into account in this study. However, there is an aspartate residue that is highly conserved. This aspartate (TAP1-Asp-213; TAP2-Asp-178) is involved in polar interactions with residues of the EL3 and in particular with a highly conserved serine or threonine residue (TAP1-Ser-440; TAP2-Thr-405). In the inward-facing conformation of TAP, this network of interactions contributes to the closure of the cavity on the ER lumen side of the membrane and reflects the pattern of interactions found in P-gp and ABCB10 (Fig. 1, D and E). The only exception is the second halve of P-gp, where a glycine replaces the conserved aspartate. In the Sav1866-based model of TAP and in the Sav1866 crystal structure, the NBD dimerization and opening of the cavity toward the ER side push the EL1 and EL3 away from each other, breaking all contacts between residues of these loops (Fig. 1F). An additional pool of four residues in the loop between TM3 and TM4 (EL2) is also conserved. In this case the interactions between this pool of residues (TAP1-Met-320, -Ser-324, -Leu-327, -Thr-328; TAP2-Met-285, -Ser-289, -Leu-292, -Thr-293) and the surrounding residues are mainly hydrophobic and are preserved in both the inward-facing and outward-facing conformation (Fig. 1, D–F).
π-Stacking Interactions between TM2 and TM5; Role of TAP1-Phe-265
In P-gp, ABCB10, Sav1866, and TAP, pairs of conserved aromatic residues are located on the intracellular side of TM2 of one halve (or monomer) and TM5 of the other halve (or monomer). In TAP, these pairs of aromatic residues consist of TAP1-Phe-390 (in TM5) and TAP2-Phe-230 (in TM2) and of TAP2-Phe-355 (in TM5) and TAP1-Phe-265 (in TM2). It has been shown that TAP1-Phe-265 mutation affects peptide transport but not peptide binding, and a role in controlling the formation of the cavity has been proposed for this residue (64). Here, changes in orientation of these conserved aromatic residues suggest an explanation of the mechanical role proposed for TAP1-Phe-265 (64); Figs. 2 and supplemental Fig. S4.
In P-gp, TM5 of each halve has a tyrosine residue (Tyr-273 and Tyr-916) facing the external side of the transporter, whereas TM2 helices have phenylalanine residues (Phe-148 and Phe-789) sandwiched between ICL1 of one halve and ICL2 of the other halve. Because of the orientation of the tyrosine residues, π-stacking interactions between the residues of each pair are not allowed in this conformation (supplemental Fig. S5_A_).
In ABCB10, TM5-Tyr-377 of one monomer might interact with TM2-Phe-252 of the other monomer. In this case the two aromatic residues point toward the same cavity, but the bending of TM2 and TM5 still do not allow π-stacking interactions between them (supplemental Fig. S5_B_).
In Sav1866 each pair of aromatic residues is formed by TM2-Phe-226 of one monomer and TM5-Tyr101 of the other monomer. The residues of each pair are involved in π-stacking interactions (supplemental Fig. S5_C_). The TAP models reflect these different states of interaction between the residues of each pair, consistent with the templates; that is, the above-mentioned π-stacking interactions are absent in the P-gp- and ABCB10-based models and are present in the Sav1866-based model (Fig. 2 and supplemental Fig. S4).
Further insights in the conformational changes that involve these residues are given by the available structures of the MsbA transporter (34); supplemental Fig. S5_D_. Its outward-facing conformation shows a π-stacking interaction between Phe-230 of one monomer and Phe-105 of the other monomer, which mimic the interactions shown in Sav1866. In the closed apo structure the distance between the Cα of Phe-230 and Phe-105 is about 12 Å, whereas in the outward-facing conformation it is about 7.6 Å (7.9 Å in Sav1866 crystal structure). This increased distance is due to a partial stretching of the TM5 of one-half and the TM2 of the opposite halve and could represent an additional intermediate state in the rearrangement of the TM helices and the pairs of aromatic residues as well. Once the transporter adopts the outward-facing conformation, it results in a proper orientation for π-stacking interactions.
Combined, the models suggest how TM5 rearranges during the transport cycle of all four related transporters and support the structural role proposed for TAP1-Phe-265. Being part of the aromatic stacking interactions, this residue contributes to the correct packing of the intracellular part of the helices. These results also indicate a similar role for the corresponding residues of P-gp, ABCB10, Sav1866, and MsbA.
Peptide Binding
TAP binds and transports peptides with a preferred length of 8–16 amino acids (65, 66). The peptide binding site was first identified in multiple regions of TAP. In particular, residues 375–420 and 453–487 of TAP1 were shown to be part of the binding site as well as residues 301–389 and 414–433 of TAP2 (6). In both TAP1 and TAP2 these peptide binding site regions include the CH2, described in the previous paragraph.
The mutation A374D in human TAP2 influences the efficiency of binding as well as the peptide transport specificity (67). As described in Armandola et al. (67) and Momburg et al. (68), Ala-374 can directly be part of the peptide binding site or can influence the peptide transport indirectly. Ala-374 belongs to the TM5 and is located in the upper side of the cavity of the transporter (Fig. 3A). In the TAP models, this residue is oriented toward the TM2 of TAP1, with the exception of the P-gp-based model 2, where its side chain is pointing directly to the inward-facing cavity. In the Sav1866-based model, Ala-374 is accessible from the cavity, whereas it appears to be more occluded in both the P-gp-based model 1 and in the ABCB10-based model. We noticed that this residue is surrounded by a pool of positively charged residues (Arg-368, Arg-369, Arg-373, Arg-380, and Arg-381) that belongs to the same transmembrane helix and is part of the peptide binding site of TAP2 described in Nijenhuis and Hämmerling (6). In particular, as it results in the Sav1866-based model, Arg-369, Arg-373, and Arg-381 are oriented toward the cavity lumen. Arg-380 is sandwiched between the TM4, TM5, and TM6 of TAP2, whereas it is exposed to the cavity in the ABCB10-based model and in the P-gp-based model 1. Arg-368 does not appear to be directly part of the peptide binding site because in all the TAP models this residue interacts with the polar groups of the lipids, and it is never oriented toward the cavity lumen. It has been shown that A374D alters the peptide transport specificity (67). Here, based on the analysis of the TAP models, we hypothesize that mutations changing charged side chains in this region could impair the transport of the peptide as a consequence of the altered electrostatic potential.
Cys-213 of TAP2 has been shown to modulate the peptide specificity of the transporter and to contact the peptide in the binding pocket (69). In particular, Baldauf et al. (69) showed that the peptide is oriented with the C terminus facing this residue. Cross-linking was detected between the thiolate of Cys-213 and the peptide RRYQKSTEL with a single cysteine residue at positions 4–9. In the inward-facing models of TAP based on the P-gp and ABCB10 crystal structures, Cys-213, located on TM2, is not directly accessible for a peptide facing the cavity (Fig. 3B); its side chain faces the membrane, and it is surrounded by Leu-151, Leu-152, Ala-154, Ala-155, Cys-209, Tyr-216, and Thr-217. However, the position of Cys-213 is constrained in TM2 because of the presence of some conserved residues (Arg-220, Ile-225, and Arg-226 of TAP2). Moreover, the sequence of MsbA shows a cysteine residue (Cys-88) in this position. In both the inward- and the outward-facing conformations of this transporter (PDB access codes 3B5W and 3B60, respectively (34)), Cys-88 appears to be oriented toward the membrane.
The Cys-213 neighboring residues Thr-217 and Met-218 were shown to affect the peptide binding and transportation (68, 70). In all our TAP models, Met-218 always points into the cavity lumen.
These results suggest that at least for the TAP models based on the inward- and outward-facing states available so far for P-gp, ABCB10, Sav1866, and MsbA, the role of TAP2-Cys-213 in controlling the peptide specificity cannot be fully explained. Two glycine residues are just ahead of Cys-213, and glycine is known to increase the flexibility of α-helices (71, 72). This may indicate that TAP2-Cys-213 might become accessible for a peptide in an intermediate state in between the inward- and outward-facing conformation even though at the end of the cycle (outward-facing conformation) its orientation is restored to the initial one (inward-facing conformation).
An additional peptide contact site is Val-288 of TAP1 (73). Cross-linking experiments showed that this residue is able to sense the central region of a bound peptide. Val-288 is located at the cytosolic side of TM3 in a region that is conserved among the transporters taken into account in this study (Fig. 3C). In P-gp and ABCB10 crystal structures, the polar residues corresponding to Ser-286, Arg-287, and Asp-291 of TAP1 are oriented toward the lumen of the inward-facing cavity. In contrast, hydrophobic residues corresponding to Val-288 and Ile-284 of TAP1 lie in small hydrophobic pockets. Mutagenesis studies showed that these residues in TAP participate in the transport cycle. In particular, Gly-282, Ile-284, and Arg-287 are crucial for coupling the peptide binding and transport, and Val-288 shares the same function (73). In the inward-facing models of TAP, these residues are oriented in agreement with the crystal structures used as templates. Additional details on the role of Val-288 are given in the next paragraph.
Role of Electrostatic Interactions in Ligand Binding
By anchoring the peptides through their N and C termini, TAP is able to translocate peptides of different length. The binding to TAP requires peptides with free N and C terminus (74, 75). Moreover, the first three residues as well as the last one play a critical role in terms of binding affinity (73, 75–79). An example is the nonamer RRYQKSTEL, which binds to TAP with high affinity (KD = 16 nm) (75). Because free N and C termini as well as charged residues are required, electrostatic interactions can represent the driving force for the peptide binding. This is also confirmed by the binding of ICP47 to TAP. ICP47 is a viral protein derived from herpes simplex virus that inhibits TAP (80–82). Its N-terminal domain and in particular residues 1–35 are responsible for TAP inhibition (83). Although residues 4–15 mediate the interaction with the membrane, residues 22–32 bind to TAP (82, 84–86). Charged residues in this region (Asp-24, Lys-31, and Arg-32) are crucial for TAP inhibition and might mimic the interactions of the N and C terminus of the peptides normally translocated by TAP (82).
In protein-protein interactions, electrostatic complementarity can significantly contribute to the binding affinity (87–91). We calculated the electrostatic potential of the TAP models, focusing on the role of charged residues in peptide/substrate binding to TAP.
The electrostatic map of the interior of the inward-facing cavities shows alternating positively and negatively charged sites. The lower side of the cavities (P-gp and ABCB10 based models) is characterized by two regions with opposite electrostatic potential (Fig. 4A). The negative site is mainly formed by residues of TAP1 and surrounds the Val-288 previously described in this paper. To see if this negatively charged site is accessible by a peptide, the electrostatic potential has been mapped on the solvent-accessible surface of the protein. Fig. 4B shows the presence of two negatively charged sites, labeled as pocket 1 and 2 in the three inward-facing models. Pocket 1 is formed by residues of TAP1 (Glu-290, Asp-291, Ser-286, Thr-294) located at the cytosolic side of the TM3. Pocket 2 is a larger groove that extends from the lower side of the cavity to the upper side. Residues common to the three inward-facing models and forming this pocket are mainly from TAP1 (Asp-246, Asn-250, Gln-261, Ser-286, Thr-289, Thr-292, Ser-293, Ser-296, Asp-297, Ser-300, Glu-301).
Using EPR spectroscopy, Herget et al. (92) characterized the conformation of peptides bound to TAP as an extended and kinked conformation, with an average distance between the two termini of a 9-mer of 2.2 nm. These EPR studies suggest that (i) TAP provides two different binding pockets for a peptide, one for the N terminus and one for the C terminus, and that (ii) the distance between these two pockets for the anchor sites is about 2.5 nm. In the TAP models, pocket 1 and 2, previously described, are the main negatively charged sites that could represent the binding site for the N-terminal residues of the peptide. TAP1-Val-288 is located just below these pockets. If pockets 1 and 2 are involved in peptide binding, this could explain why this residue is able to sense a bound peptide.
The opposite site of the cavity shows a different electrostatic map (Fig. 4, A and C). The positive potential extends from the lower to the upper side of the cavity lumen. In particular, we identified a positively charged side (named pocket 3), mainly formed by TAP1-Arg-357 and TAP2-Arg-226. TAP1-Arg-357 is at the edge of the groove, facing the lumen of the inward-facing cavity. In contrast, TAP2-Arg-226 lies deeper inside the groove. TAP2-Met-218 is two helical turns above TAP2-Arg-226. Baldauf et al. (69) proposed that the C terminus of a bound peptide is oriented toward the Cys-213/Met-218 residues of TAP2. If pocket 3 is involved in the coordination of the peptide C terminus, the TAP models might be able to provide a structural context to explain the experimental results (69). Moreover, residues from both TAP1 and TAP2 contribute to the positive potential on the upper side of the cavity (TAP1-Lys-423, TAP2-Arg-210, and TAP2-Arg273), above the TAP2-Cys-213 and TAP2-Met-218 region (pocket 4, Fig. 4C). This further confirms this side of the cavity as more suitable for the binding of the peptide C terminus, as described by Baldauf et al. (69).
Considering the sequence of the nonamer RRYQKSTEL, the results of the electrostatic calculations presented in this study suggest a binding mode in which pocket 1 and 2 might accommodate the free N terminus of the peptide and the side chains of the arginine residues, whereas the free C terminus might make contacts with the positively charged pocket (pocket 3).
The recently published structure of the ABC heterodimeric transporter TM287/288 is characterized by an inward-facing cavity that resembles the ABCB10 one even though TM4 and TM5 show slightly different bent structures. The TAP model derived from this new structure shows properties similar to the model based on ABCB10 (supplemental Fig. S6). The electrostatic potential calculations identified the same pattern of positively and negatively charged sites inside the cavity, thus further supporting our peptide binding hypothesis (supplemental Fig. S7).
We compared the cavity opening of the P-gp- and ABCB10-based TAP models to the available structures of MsbA (34). For this comparison we measured the distance between the Cα of TAP2-Asp-256 (which is adjacent to the positively charged pocket) and TAP1-Asp-291. This distance is 3.0, 2.8, and 2.5 nm for the P-gp-based model 1, model 2, and the ABCB10-based model, respectively. In the MsbA inward-facing conformation (PDB access code 3B5W), with a separation between the two NBDs larger than P-gp, the distance between Asp131-Cα (which corresponds to TAP1-Asp-291 and TAP2-Asp-256) of each chain is 3.9 nm. Such a distance appears to be too large to allow a peptide adopting the extended kinked conformation in TAP. In the MsbA closed apo conformation (PDB access code 3B5X), which is still an inward-facing conformation but with the NBDs in closer contact, this distance is 1.66 nm. In this case the cavity might be too closed to allow peptide binding. Altogether these results and the experimental observation of a kinked bound peptide (92) suggest that the cavity of the transporter in a peptide binding competent state should not be more open than the P-gp crystal structure.
Replica Exchange Simulations
Several programs and servers are available for protein-protein docking (93–97), for docking of small molecules (98–102), or for the refinement of peptide-protein interactions (103). However, these programs are not suitable for the docking of a peptide such as the nonamer RRYQKSTEL because of the large number of rotatable bonds that are involved, the necessity of flexible backbone and side chains of the peptide, and the large dimensions of the inward-facing cavity. For these reasons, we decided to perform replica exchange simulations on the nonamer RRYQKSTEL bound to the inward-facing cavity. Our hypothesis is that pocket 1, 2, and 3 are involved in the coordination of the N and C terminus of the peptide. Based on this assumption, the goal of these simulations was to verify if the cavity opening of the P-gp- and ABCB10-based models is suitable for peptide binding. The replica exchange simulations allowed the peptide to explore the inward-facing cavity lumen, thus identifying potential binding sites for its N and C terminus.
The results of the replica simulations performed on the TAP model based on ABCB10 proposed a model for the binding of the peptide that is in agreement with the experimental data. The average distance between the Cα of Arg1 and Leu-9 of the bound peptide in the cluster with the largest population is 2.2 nm. This distance may be correlated with the 2.5-nm distance proposed by Herget et al. (92) between the two anchor pockets of the transporter. In this model the N terminus of the peptide and the two arginine residues strongly interacts with the residues of the negatively charged pocket (TAP1-Thr-289, -Glu-290, -Ser-293) in close proximity to TAP1-Val-288 (Fig. 5A). Additional interactions are formed with TAP1-Asp-297 and Tap2-Gln-322. Residues of the positively charged groove, and in particular TAP2- Arg-226, -Ser-254, and TAP1-Arg-357, coordinate the C terminus of the peptide in such a way that the side chain of Leu-10 faces the side chain of the previously described TAP2-Met-218.
In the P-gp-based model 2, the N terminus as well as the two arginine residues of the peptide are strongly bound to the negatively charged pocket (Fig. 5B), further confirming that this site might be suitable for coordinating the positive charge of the peptide. Because of the larger opening of the cavity of this model compared with the TAP model based on ABCB10, the C terminus cannot reach TAP2- Arg-226 or -Met-218 but mainly interacts with TAP1-Arg-357 of the positively charged groove, although additional interactions involve TAP2-Ser-254 and -Ser-255. In the case of the P-gp-based model 1, the carboxylic group of Glu-8 of the peptide interacts with TAP2- Arg-226, whereas the C terminus is involved in intramolecular hydrogen bonds, thus preventing the interaction of the positive N terminus with the negatively charged pocket (Fig. 5C).
The different results obtained from the replica simulations on the two P-gp-based models are due to the asymmetry of the TMDs in the P-gp crystal structure, which is reflected in the TAP models. In particular, the accessibility of pocket 1 in the P-gp-based model 1 is limited compared with P-gp based model 2. Moreover, in the P-gp-based model 2 the side chain of TAP2-Glu-360 interacts with Arg-1 of the peptide, whereas in the P-gp-based model 1 this interaction is missing due to a different orientation of TAP2-Glu-360. However, the simulations performed on both these models showed that residues of the positively charged groove constitute the favorite site for the binding of carboxylic groups.
Replica simulations performed on both the ABCB10-based model and the P-gp-based model 2 suggested that the coordination of the N and C terminus of the peptide with the negatively and the positively charged pocket, respectively, is possible, even though only in the ABCB10-based model a close proximity between the C terminus and TAP2-Met-218 was retrieved.
Conclusions
In the absence of high resolution structures, the combination of experimental data and molecular modeling approaches allows the construction of homology models that can be used as a tool to inspect the structural organization and properties of a protein as well as to guide mutagenesis experiments. In this paper we have presented homology models of TAP built based on the P-gp (29), ABCB10, and Sav1866 (30) crystal structures while taking into account the literature data available for all these transporters as well as for MsbA. The TAP models were compared with mutagenesis studies showing specific residues as important for peptide binding and translocation to give structural insights into the role of these residues. Moreover, electrostatic potential calculations and replica exchange simulations combined with experimental data (92) suggested that the inward-facing models of TAP based on P-gp and ABCB10 might represent peptide binding competent states of the transporter.
There is an important difference in the mechanism of action of TAP, P-gp, ABCB10, and MsbA. TAP recruits peptides from the cytosol; MsbA and P-gp acquire their substrates from the membrane, acting as both lipid flippases and drug transporters (104–109); ABCB10 is a peptide transporter in the inner membrane of mitochondria. Nevertheless, the similarity among these transporters and their bacterial homologue Sav1866 allowed us to build molecular models of TAP that can be used to design new experimental studies for a better understanding of the peptide binding and the dynamics of this transporter.
Supplementary Material
Supplemental Data
Acknowledgments
Calculations were carried out in part on WestGrid/Compute Canada facilities. We thank Dr. Robert Tampé and Drew W. F. Bennett for fruitful discussions.
*
This work was supported by the Canadian Institutes for Health Research.
3
The abbreviations used are:
ABC
ATP binding cassette
TAP
transporter associated with antigen processing
ER
endoplasmic reticulum
EL
ER-facing loop
CL
cytosol-facing loop
NBD
nucleotide binding domains
TMD
transmembrane domains
P-gp
P-glycoprotein
r.m.s.d.
root mean square deviation
Cα
α-carbon.
REFERENCES
- 1.Dean M., Rzhetsky A., Allikmets R. (2001) The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 2.Dean M. (2005) The genetics of ATP binding cassette transporters. Methods Enzymol. 400, 409–429 [DOI] [PubMed] [Google Scholar]
- 3.Vasiliou V., Vasiliou K., Nebert D. W. (2009) Human ATP-binding cassette (ABC) transporter family. Hum. Genomics 3, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgins C. F. (1992) ABC transporters. From microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113 [DOI] [PubMed] [Google Scholar]
- 5.Procko E., O'Mara M. L., Bennett W. F., Tieleman D. P., Gaudet R. (2009) The mechanism of ABC transporters. General lessons from structural and functional studies of an antigenic peptide transporter. FASEB J. 23, 1287–1302 [DOI] [PubMed] [Google Scholar]
- 6.Nijenhuis M., Hämmerling G. J. (1996) Multiple regions of the transporter associated with antigen processing (TAP) contribute to its peptide binding site. J. Immunol. 157, 5467–5477 [PubMed] [Google Scholar]
- 7.Lankat-Buttgereit B., Tampé R. (2002) The transporter associated with antigen processing. Function and implications in human diseases. Physiol. Rev. 82, 187–204 [DOI] [PubMed] [Google Scholar]
- 8.Koch J., Guntrum R., Heintke S., Kyritsis C., Tampé R. (2004) Functional dissection of the transmembrane domains of the transporter associated with antigen processing (TAP). J. Biol. Chem. 279, 10142–10147 [DOI] [PubMed] [Google Scholar]
- 9.Schrodt S., Koch J., Tampé R. (2006) Membrane topology of the transporter associated with antigen processing (TAP1) within an assembled functional peptide-loading complex. J. Biol. Chem. 281, 6455–6462 [DOI] [PubMed] [Google Scholar]
- 10.Gaudet R., Wiley D. C. (2001) Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 20, 4964–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Procko E., Ferrin-O'Connell I., Ng S. L., Gaudet R. (2006) Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol. Cell 24, 51–62 [DOI] [PubMed] [Google Scholar]
- 12.Oancea G., O'Mara M. L., Bennett W. F., Tieleman D. P., Abele R., Tampé R. (2009) Structural arrangement of the transmission interface in the antigen ABC transport complex TAP. Proc. Natl. Acad. Sci. U.S.A. 106, 5551–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg M. F., Bikadi Z., Chan J., Liu X., Ni Z., Cai X., Ford R. C., Mao Q. (2010) The human breast cancer resistance protein (BCRP/ABCG2) shows conformational changes with mitoxantrone. Structure 18, 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessadok A., Garcia E., Jacquet H., Martin S., Garrigues A., Loiseau N., André F., Orlowski S., Vivaudou M. (2011) Recognition of sulfonylurea receptor (ABCC8/9) ligands by the multidrug resistance transporter P-glycoprotein (ABCB1). Functional similarities based on common structural features between two multispecific ABC proteins. J. Biol. Chem. 286, 3552–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Mara M. L., Tieleman D. P. (2007) P-glycoprotein models of the apo and ATP-bound states based on homology with Sav1866 and MalK. FEBS Lett. 581, 4217–4222 [DOI] [PubMed] [Google Scholar]
- 16.Globisch C., Pajeva I. K., Wiese M. (2008) Identification of putative binding sites of P-glycoprotein based on its homology model. ChemMedchem. 3, 280–295 [DOI] [PubMed] [Google Scholar]
- 17.Pleban K., Kopp S., Csaszar E., Peer M., Hrebicek T., Rizzi A., Ecker G. F., Chiba P. (2005) P-glycoprotein substrate binding domains are located at the transmembrane domain/transmembrane domain interfaces. A combined photoaffinity labeling-protein homology modeling approach. Mol. Pharmacol. 67, 365–374 [DOI] [PubMed] [Google Scholar]
- 18.Qian F., Wei D., Liu J., Yang S. (2006) Molecular model and ATPase activity of carboxyl-terminal nucleotide binding domain from human P-glycoprotein. Biochemistry 71, S18–S24 [DOI] [PubMed] [Google Scholar]
- 19.Veiga M. I., Ferreira P. E., Jörnhagen L., Malmberg M., Kone A., Schmidt B. A., Petzold M., Björkman A., Nosten F., Gil J. P. (2011) Novel polymorphisms in Plasmodium falciparum ABC transporter genes are associated with major ACT antimalarial drug resistance. PLoS ONE 6, e20212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravna A. W., Sylte I., Sager G. (2007) Molecular model of the outward facing state of the human P-glycoprotein (ABCB1) and comparison to a model of the human MRP5 (ABCC5). Theor. Biol. Med. Model. 4, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravna A. W., Sager G. (2008) Molecular model of the outward facing state of the human multidrug resistance protein 4 (MRP4/ABCC4). Bioorg. Med. Chem. Lett. 18, 3481–3483 [DOI] [PubMed] [Google Scholar]
- 22.Ravna A. W., Sylte I., Sager G. (2008) A molecular model of a putative substrate releasing conformation of multidrug resistance protein 5 (MRP5). Eur. J. Med. Chem. 43, 2557–2567 [DOI] [PubMed] [Google Scholar]
- 23.Ravna A. W., Sylte I., Sager G. (2009) Binding site of ABC transporter homology models confirmed by ABCB1 crystal structure. Theor. Biol. Med. Model. 6, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeGorter M. K., Conseil G., Deeley R. G., Campbell R. L., Cole S. P. (2008) Molecular modeling of the human multidrug resistance protein 1 (MRP1/ABCC1). Biochem. Biophys. Res. Commun. 365, 29–34 [DOI] [PubMed] [Google Scholar]
- 25.Campbell J. D., Sansom M. S., Ashcroft F. M. (2003) Potassium channel regulation. EMBO Rep. 4, 1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serohijos A. W., Hegedus T., Aleksandrov A. A., He L., Cui L., Dokholyan N. V., Riordan J. R. (2008) Phenylalanine 508 mediates a cytoplasmic-membrane domain contact in the CFTR three-dimensional structure crucial to assembly and channel function. Proc. Natl. Acad. Sci. U.S.A. 105, 3256–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander C., Ivetac A., Liu X., Norimatsu Y., Serrano J. R., Landstrom A., Sansom M., Dawson D. C. (2009) Cystic fibrosis transmembrane conductance regulator. Using differential reactivity toward channel-permeant and channel-impermeant thiol-reactive probes to test a molecular model for the pore. Biochemistry 48, 10078–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mornon J. P., Lehn P., Callebaut I. (2009) Molecular models of the open and closed states of the whole human CFTR protein. Cell. Mol. Life Sci. 66, 3469–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aller S. G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P. M., Trinh Y. T., Zhang Q., Urbatsch I. L., Chang G. (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323, 1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dawson R. J., Locher K. P. (2006) Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 [DOI] [PubMed] [Google Scholar]
- 31.UniProt Consortium (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214–D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) ExPASy. The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 34.Ward A., Reyes C. L., Yu J., Roth C. B., Chang G. (2007) Flexibility in the ABC transporter MsbA. Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K., Misawa K., Kuma K., Miyata T. (2002) MAFFT. A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt L., Benabdelhak H., Blight M. A., Holland I. B., Stubbs M. T. (2003) Crystal structure of the nucleotide-binding domain of the ABC transporter hemolysin B. Identification of a variable region within ABC helical domains. J. Mol. Biol. 330, 333–342 [DOI] [PubMed] [Google Scholar]
- 37.Haffke M., Menzel A., Carius Y., Jahn D., Heinz D. W. (2010) Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides. Acta Crystallogr. D Biol. Crystallogr. 66, 979–987 [DOI] [PubMed] [Google Scholar]
- 38.Vedadi M., Lew J., Artz J., Amani M., Zhao Y., Dong A., Wasney G. A., Gao M., Hills T., Brokx S., Qiu W., Sharma S., Diassiti A., Alam Z., Melone M., Mulichak A., Wernimont A., Bray J., Loppnau P., Plotnikova O., Newberry K., Sundararajan E., Houston S., Walker J., Tempel W., Bochkarev A., Kozieradzki I., Edwards A., Arrowsmith C., Roos D., Kain K., Hui R. (2007) Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 151, 100–110 [DOI] [PubMed] [Google Scholar]
- 39.Sali A., Blundell T. L. (1993) Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 40.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benkert P., Tosatto S. C., Schomburg D. (2008) QMEAN. A comprehensive scoring function for model quality assessment. Proteins 71, 261–277 [DOI] [PubMed] [Google Scholar]
- 43.Benkert P., Künzli M., Schwede T. (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res. 37, W510–W514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 45.Laskowski R. A. (2009) PDBsum new things. Nucleic Acids Res. 37, D355–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Errors in protein structures. Nature 381, 272–272 [DOI] [PubMed] [Google Scholar]
- 47.Wolf M. G., Hoefling M., Aponte-Santamaría C., Grubmüller H., Groenhof G. (2010) g_membed. Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 31, 2169–2174 [DOI] [PubMed] [Google Scholar]
- 48.Berendsen H. J. C., van der Spoel D., van Drunen R. (1995) GROMACS. A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 [Google Scholar]
- 49.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) GROMACS. Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 50.Schmid N., Eichenberger A. P., Choutko A., Riniker S., Winger M., Mark A. E., van Gunsteren W. F. (2011) Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 40, 843–856 [DOI] [PubMed] [Google Scholar]
- 51.Chen R., Poger D., Mark A. E. (2011) Effect of high pressure on fully hydrated DPPC and POPC bilayers. J. Phys. Chem. B 115, 1038–1044 [DOI] [PubMed] [Google Scholar]
- 52.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems. Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onufriev A., Bashford D., Case D. A. (2004) Exploring protein native states and large scale conformational changes with a modified generalized born model. Proteins 55, 383–394 [DOI] [PubMed] [Google Scholar]
- 54.Denschlag R., Lingenheil M., Tavan P. (2009) Optimal temperature ladders in replica exchange simulations. Chem. Phys. Lett. 473, 193–195 [Google Scholar]
- 55.Daura X., Gademann K., Jaun B., Seebach D., van Gunsteren W. F., Mark A. E. (1999) Peptide folding. When simulation meets experiment. Angew. Chem. Int. Ed. Engl. 38, 236–240 [Google Scholar]
- 56.Shirts M. R., Chodera J. D. (2008) Statistically optimal analysis of samples from multiple equilibrium states. J. Chem. Phys. 129, 124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 58.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 59.Koch J., Guntrum R., Tampé R. (2005) Exploring the minimal functional unit of the transporter associated with antigen processing. FEBS Lett. 579, 4413–4416 [DOI] [PubMed] [Google Scholar]
- 60.Procko E., Raghuraman G., Wiley D. C., Raghavan M., Gaudet R. (2005) Identification of domain boundaries within the N termini of TAP1 and TAP2 and their importance in tapasin binding and tapasin-mediated increase in peptide loading of MHC class I. Immunol. Cell Biol. 83, 475–482 [DOI] [PubMed] [Google Scholar]
- 61.Leonhardt R. M., Keusekotten K., Bekpen C., Knittler M. R. (2005) Critical role for the tapasin-docking site of TAP2 in the functional integrity of the MHC class I-peptide-loading complex. J. Immunol. 175, 5104–5114 [DOI] [PubMed] [Google Scholar]
- 62.Moody J. E., Thomas P. J. (2005) Nucleotide binding domain interactions during the mechanochemical reaction cycle of ATP binding cassette transporters. J. Bioenerg. Biomembr. 37, 475–479 [DOI] [PubMed] [Google Scholar]
- 63.Hohl M., Briand C., Grütter M. G., Seeger M. A. (2012) Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat. Struct. Mol. Biol. 19, 395–402 [DOI] [PubMed] [Google Scholar]
- 64.Ritz U., Drexler I., Sutter D., Abele R., Huber C., Seliger B. (2003) Impaired transporter associated with antigen processing (TAP) function attributable to a single amino acid alteration in the peptide TAP subunit TAP1. J. Immunol. 170, 941–946 [DOI] [PubMed] [Google Scholar]
- 65.van Endert P. M., Tampé R., Meyer T. H., Tisch R., Bach J. F., McDevitt H. O. (1994) A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity 1, 491–500 [DOI] [PubMed] [Google Scholar]
- 66.Koopmann J. O., Post M., Neefjes J. J., Hämmerling G. J., Momburg F. (1996) Translocation of long peptides by transporters associated with antigen processing (TAP). Eur. J. Immunol. 26, 1720–1728 [DOI] [PubMed] [Google Scholar]
- 67.Armandola E. A., Momburg F., Nijenhuis M., Bulbuc N., Früh K., Hämmerling G. J. (1996) A point mutation in the human transporter associated with antigen processing (TAP2) alters the peptide transport specificity. Eur. J. Immunol. 26, 1748–1755 [DOI] [PubMed] [Google Scholar]
- 68.Momburg F., Armandola E. A., Post M., Hammerling G. J. (1996) Residues in TAP2 peptide transporters controlling substrate specificity. J. Immunol. 156, 1756–1763 [PubMed] [Google Scholar]
- 69.Baldauf C., Schrodt S., Herget M., Koch J., Tampé R. (2010) Single residue within the antigen translocation complex TAP controls the epitope repertoire by stabilizing a receptive conformation. Proc. Natl. Acad. Sci. U.S.A. 107, 9135–9140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deverson E. V., Leong L., Seelig A., Coadwell W. J., Tredgett E. M., Butcher G. W., Howard J. C. (1998) Functional analysis by site-directed mutagenesis of the complex polymorphism in rat transporter associated with antigen processing. J. Immunol. 160, 2767–2779 [PubMed] [Google Scholar]
- 71.Li S. C., Deber C. M. (1992) Glycine and β-branched residues support and modulate peptide helicity in membrane environments. FEBS Lett. 311, 217–220 [DOI] [PubMed] [Google Scholar]
- 72.Jiang Y., Lee A., Chen J., Cadene M., Chait B. T., MacKinnon R. (2002) The open pore conformation of potassium channels. Nature 417, 523–526 [DOI] [PubMed] [Google Scholar]
- 73.Herget M., Oancea G., Schrodt S., Karas M., Tampé R., Abele R. (2007) Mechanism of substrate sensing and signal transmission within an ABC transporter. Use of a Trojan horse strategy. J. Biol. Chem. 282, 3871–3880 [DOI] [PubMed] [Google Scholar]
- 74.Androlewicz M. J., Cresswell P. (1994) Human transporters associated with antigen processing possess a promiscuous peptide binding site. Immunity 1, 7–14 [DOI] [PubMed] [Google Scholar]
- 75.Uebel S., Meyer T. H., Kraas W., Kienle S., Jung G., Wiesmüller K. H., Tampé R. (1995) Requirements for peptide binding to the human transporter associated with antigen processing revealed by peptide scans and complex peptide libraries. J. Biol. Chem. 270, 18512–18516 [DOI] [PubMed] [Google Scholar]
- 76.Momburg F., Roelse J., Howard J. C., Butcher G. W., Hämmerling G. J., Neefjes J. J. (1994) Selectivity of MHC-encoded peptide transporters from human, mouse, and rat. Nature 367, 648–651 [DOI] [PubMed] [Google Scholar]
- 77.van Endert P. M., Riganelli D., Greco G., Fleischhauer K., Sidney J., Sette A., Bach J. F. (1995) The peptide binding motif for the human transporter associated with antigen processing. J. Exp. Med. 182, 1883–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grommé M., van der Valk R., Sliedregt K., Vernie L., Liskamp R., Hämmerling G., Koopmann J. O., Momburg F., Neefjes J. (1997) The rational design of TAP inhibitors using peptide substrate modifications and peptidomimetics. Eur. J. Immunol. 27, 898–904 [DOI] [PubMed] [Google Scholar]
- 79.Uebel S., Kraas W., Kienle S., Wiesmüller K. H., Jung G., Tampé R. (1997) Recognition principle of the TAP transporter disclosed by combinatorial peptide libraries. Proc. Natl. Acad. Sci. U.S.A. 94, 8976–8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Früh K., Ahn K., Djaballah H., Sempé P., van Endert P. M., Tampé R., Peterson P. A., Yang Y. (1995) A viral inhibitor of peptide transporters for antigen presentation. Nature 375, 415–418 [DOI] [PubMed] [Google Scholar]
- 81.Hill A., Jugovic P., York I., Russ G., Bennink J., Yewdell J., Ploegh H., Johnson D. (1995) Herpes simplex virus turns off the TAP to evade host immunity. Nature 375, 411–415 [DOI] [PubMed] [Google Scholar]
- 82.Neumann L., Kraas W., Uebel S., Jung G., Tampé R. (1997) The active domain of the herpes simplex virus protein ICP47. A potent inhibitor of the transporter associated with antigen processing. J. Mol. Biol. 272, 484–492 [DOI] [PubMed] [Google Scholar]
- 83.Galocha B., Hill A., Barnett B. C., Dolan A., Raimondi A., Cook R. F., Brunner J., McGeoch D. J., Ploegh H. L. (1997) The active site of ICP47, a herpes simplex virus-encoded inhibitor of the major histocompatibility complex (MHC)-encoded peptide transporter associated with antigen processing (TAP), maps to the NH2-terminal 35 residues. J. Exp. Med. 185, 1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pfänder R., Neumann L., Zweckstetter M., Seger C., Holak T. A., Tampé R. (1999) Structure of the active domain of the herpes simplex virus protein ICP47 in water/sodium dodecyl sulfate solution determined by nuclear magnetic resonance spectroscopy. Biochemistry 38, 13692–13698 [DOI] [PubMed] [Google Scholar]
- 85.Beinert D., Neumann L., Uebel S., Tampé R. (1997) Structure of the viral TAP inhibitor ICP47 induced by membrane association. Biochemistry 36, 4694–4700 [DOI] [PubMed] [Google Scholar]
- 86.Aisenbrey C., Sizun C., Koch J., Herget M., Abele R., Bechinger B., Tampé R. (2006) Structure and dynamics of membrane-associated ICP47, a viral inhibitor of the MHC I antigen-processing machinery. J. Biol. Chem. 281, 30365–30372 [DOI] [PubMed] [Google Scholar]
- 87.Schreiber G., Fersht A. R. (1993) Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry 32, 5145–5150 [DOI] [PubMed] [Google Scholar]
- 88.Tsai C. J., Lin S. L., Wolfson H. J., Nussinov R. (1996) Protein-protein interfaces. Architectures and interactions in protein-protein interfaces and in protein cores. Their similarities and differences. Crit. Rev. Biochem. Mol. Biol. 31, 127–152 [DOI] [PubMed] [Google Scholar]
- 89.Sheinerman F. B., Norel R., Honig B. (2000) Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 10, 153–159 [DOI] [PubMed] [Google Scholar]
- 90.Norel R., Sheinerman F., Petrey D., Honig B. (2001) Electrostatic contributions to protein-protein interactions. Fast energetic filters for docking and their physical basis. Protein Sci. 10, 2147–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neves-Petersen M. T., Petersen S. B. (2003) Protein electrostatics. A review of the equations and methods used to model electrostatic equations in biomolecules. Applications in biotechnology. Biotechnol. Annu. Rev. 9, 315–395 [DOI] [PubMed] [Google Scholar]
- 92.Herget M., Baldauf C., Schölz C., Parcej D., Wiesmüller K. H., Tampé R., Abele R., Bordignon E. (2011) Conformation of peptides bound to the transporter associated with antigen processing (TAP). Proc. Natl. Acad. Sci. U.S.A. 108, 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dominguez C., Boelens R., Bonvin A. M. (2003) HADDOCK. A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 94.Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 95.Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro. An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50 [DOI] [PubMed] [Google Scholar]
- 96.Chen R., Weng Z. (2002) Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins 47, 281–294 [DOI] [PubMed] [Google Scholar]
- 97.Tovchigrechko A., Vakser I. A. (2006) GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–W314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 99.Trott O., Olson A. J. (2010) AutoDock Vina. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jones G., Willett P., Glen R. C. (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 245, 43–53 [DOI] [PubMed] [Google Scholar]
- 101.Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S. (2004) Glide. A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 102.Halgren T. A., Murphy R. B., Friesner R. A., Beard H. S., Frye L. L., Pollard W. T., Banks J. L. (2004) Glide. A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759 [DOI] [PubMed] [Google Scholar]
- 103.Raveh B., London N., Schueler-Furman O. (2010) Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins 78, 2029–2040 [DOI] [PubMed] [Google Scholar]
- 104.Eckford P. D., Sharom F. J. (2010) The reconstituted Escherichia coli MsbA protein displays lipid flippase activity. Biochem. J. 429, 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smriti, Zou P., McHaourab H. S. (2009) Mapping daunorubicin binding sites in the ATP binding cassette transporter MsbA using site-specific quenching by spin labels. J. Biol. Chem. 284, 13904–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Raviv Y., Pollard H. B., Bruggemann E. P., Pastan I., Gottesman M. M. (1990) Photosensitized labeling of a functional multidrug transporter in living drug-resistant tumor cells. J. Biol. Chem. 265, 3975–3980 [PubMed] [Google Scholar]
- 107.Gatlik-Landwojtowicz E., Aänismaa P., Seelig A. (2006) Quantification and characterization of P-glycoprotein-substrate interactions. Biochemistry 45, 3020–3032 [DOI] [PubMed] [Google Scholar]
- 108.Doerrler W. T., Raetz C. R. (2002) ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 [DOI] [PubMed] [Google Scholar]
- 109.Borst P., Zelcer N., van Helvoort A. (2000) ABC transporters in lipid transport. Biochim. Biophys. Acta 1486, 128–144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data