Extensive alternative polyadenylation during zebrafish development (original) (raw)
Abstract
The post-transcriptional fate of messenger RNAs (mRNAs) is largely dictated by their 3′ untranslated regions (3′ UTRs), which are defined by cleavage and polyadenylation (CPA) of pre-mRNAs. We used poly(A)-position profiling by sequencing (3P-seq) to map poly(A) sites at eight developmental stages and tissues in the zebrafish. Analysis of over 60 million 3P-seq reads substantially increased and improved existing 3′ UTR annotations, resulting in confidently identified 3′ UTRs for >79% of the annotated protein-coding genes in zebrafish. mRNAs from most zebrafish genes undergo alternative CPA, with those from more than a thousand genes using different dominant 3′ UTRs at different stages. These included one of the poly(A) polymerase genes, for which alternative CPA reinforces its repression in the ovary. 3′ UTRs tend to be shortest in the ovaries and longest in the brain. Isoforms with some of the shortest 3′ UTRs are highly expressed in the ovary, yet absent in the maternally contributed RNAs of the embryo, perhaps because their 3′ UTRs are too short to accommodate a uridine-rich motif required for stability of the maternal mRNA. At 2 h post-fertilization, thousands of unique poly(A) sites appear at locations lacking a typical polyadenylation signal, which suggests a wave of widespread cytoplasmic polyadenylation of mRNA degradation intermediates. Our insights into the identities, formation, and evolution of zebrafish 3′ UTRs provide a resource for studying gene regulation during vertebrate development.
Alternative splicing and alternative cleavage and polyadenylation (APA) act in concert to shape the eukaryotic transcriptome (Zhang et al. 2005; Wang et al. 2008; Di Giammartino et al. 2011). APA results from differences in recognition of polyadenylation signals by the pre-mRNA cleavage and polyadenylation (CPA) machinery, which leads to differences in 3′ UTR identity, which can, in turn, influence stability, translation and subcellular localization of mRNAs (de Moor et al. 2005; Hughes 2006; Andreassi and Riccio 2009). Widespread APA and the prevalence of either short or long 3′ UTRs in specific tissues or developmental stages has been reported in human and mouse (Zhang et al. 2005; Evsikov et al. 2006; Liu et al. 2007; Flavell et al. 2008; Sandberg et al. 2008; Wang et al. 2008; Ji et al. 2009; Mayr and Bartel 2009; Salisbury et al. 2009), but the extent and the functional consequences of APA in vertebrate development remain poorly understood. Furthermore, systematic changes in usage of alternative 3′ UTRs are yet to be studied in detail in other vertebrates.
Zebrafish are model vertebrates used for studying RNA biology (Knaut et al. 2002; Giraldez et al. 2006; Choi et al. 2007; Aanes et al. 2011), but their utility for studying various aspects of post-transcriptional regulation, such as targeting by microRNAs (miRNAs), has been hindered by incomplete 3′ UTR annotation. In version 66 of Ensembl zebrafish genes (Feb. 2012), only 64.4% of the transcript models have any 3′ UTR annotation, and only 28.9% of the genes have more than one annotated 3′ end. By comparison, over 96% of the human genes have an annotated 3′ UTR in Ensembl v66, 64.8% of which have multiple annotated 3′ ends. Furthermore, 22.3% of zebrafish transcript models for protein-coding genes end without a stop codon, indicating that both the C terminus of their protein product and the 3′ UTR are not annotated.
Maturation of germ cells and early embryonic development in animals requires extensive post-transcriptional regulation of mRNAs. Preparation of the maternal mRNA cargo that is deposited into the zygote involves deadenylation of many maternal messages, as the cells suspend their metabolic and transcriptional activity after entering a cell-cycle arrest (Tadros and Lipshitz 2009). Shortly after fertilization, maternally deposited mRNAs play key roles in orchestrating the early developmental stages, and the translation, localization, and stability of these mRNAs are regulated, at least in part, through their 3′ UTRs. Eventually, the transcriptome undergoes a maternal-to-zygotic transition (MZT), during which maternal transcripts are degraded, probably in several waves (Giraldez et al. 2006; Schier 2007; Stitzel and Seydoux 2007), alongside the commencement of transcription from the zygotic genome. Zebrafish and Xenopus embryos are excellent models for studying mRNA biology during early embryogenesis, as both pre-MZT and post-MZT embryos can be easily separated and manipulated. A recent study described significant changes in transcript levels occurring between the one-cell and 16-cell stages of early zebrafish development and provided evidence that hundreds of genes undergo cytoplasmic polyadenylation during this period (Aanes et al. 2011), a phenomenon resembling that described previously in Xenopus, mouse, and fly embryos (Vassalli et al. 1989; Simon et al. 1992; Salles et al. 1994; Richter 1999; Oh et al. 2000). Differences in transcript isoforms expressed during those stages, and in particular, differences in 3′ UTR isoforms expressed in oocytes, pre-MZT, and post-MZT embryos remain to be determined.
To address these questions, we used poly(A)-position profiling by sequencing (3P-seq) (Jan et al. 2011) to map poly(A) sites in five different stages of zebrafish development and in three adult tissues. These data allowed us to accurately map 3′ UTRs for over 79% of zebrafish protein-coding genes, define the C-terminal sequence of 640 proteins, and identify widespread alternative polyadenylation affecting transcripts from 55% of zebrafish genes. Comparison of the patterns of 3′ end usage in different stages revealed several significant trends, such as the usage of shorter 3′ UTRs in the ovaries, preferential depletion of short 3′ UTRs in the early embryo, and widespread polyadenylation at noncanonical sites, which appears to occur post-transcriptionally in the early embryo.
Results
3′ UTR reannotation alters most zebrafish gene models
In order to map the poly(A) landscape in the zebrafish genome, we obtained polyadenylated RNA from four embryonic stages (pre-MZT embryo [1.5–2 h post-fertilization (hpf)], post-MZT embryo [4.5–5.5 hpf], 24 hpf, and 72 hpf), mixed gender adults, and three adult tissues (brain, testes, and ovaries) and subjected them to 3P-seq (Jan et al. 2011). Each sample yielded 6,281,147–9,813,481 genome-mapped reads, which mapped to 231,580–954,595 unique genomic regions (Supplemental Table S1). We considered as 3P tags only reads that mapped to no more than four loci in the genome and possessed at least one 3′-terminal adenylate that was not templated in the genome. Positions supported by at least four tags, out of which at least two were either distinct (i.e., had a different number of untemplated adenosines) or came from two different libraries, were carried forward as poly(A) sites.
After combining tags from all eight libraries and consolidating all tags ending within 10 nt of the most frequently implicated site, we obtained 195,199 unique poly(A) sites (Supplemental Table S2). Sequence composition around these sites was highly similar to that reported in mammals, flies, and worms (Fig. 1A; Tian et al. 2005; Retelska et al. 2006; Jan et al. 2011), and 62.3% had the canonical cleavage and polyadenylation signal (PAS) motif AAUAAA or one of ten related variants in a region 10 to 30 bases upstream of the poly(A) site (Fig. 1B). These eleven PAS variants contained nine of the 14 most common variants found in Caenorhabditis elegans (Jan et al. 2011) and all seven of the most common variants reported in human and fly (Retelska et al. 2006). The region between the PAS and the cleavage site had two U-rich segments, and the region downstream from the cleavage site had a GU-rich segment, followed by a U-rich segment (Fig. 1A; Supplemental Fig. S2). The nucleotide composition upstream of the poly(A) sites resembled that of human and worm poly(A) sites. Enrichment of GU-rich/U-rich sequences 5–30 bases downstream from the sites resembled that of human sites and, to a lesser extent, C. elegans sites (Supplemental Fig. S2). Thus, the sequence specificity of the CPA machinery appears to be conserved among fish, mammals, and worms.
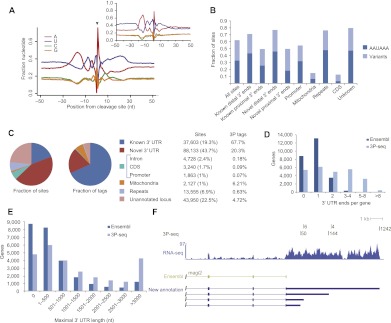
Figure 1.
Reannotation of zebrafish 3′ UTRs. (A) Nucleotide sequence composition around all 197,350 3P-seq–identified poly(A) sites. (Black arrow) Cleavage position. As previously noted (Jan et al. 2011), the sharp adenosine peak at position +1, the depletion of A at position –1, and blurring of sequence composition at other positions was partly due to cases of cleavage after an A, for which the templated A was assigned to the poly(A) tail, resulting in a –1-nt offset from the cleavage-site register. (Inset) Sequence composition around poly(A) sites in C. elegans (Jan et al. 2011), redrawn for comparison. (B) Frequencies of sites containing the canonical PAS motif AAUAAA or one of its ten common variants in the region from –40 to –10 relative to the poly(A) site. Known distal 3′ ends are the distal-most poly(A) sites annotated in Ensembl, and known proximal 3′ ends are all other annotated poly(A) sites. Novel distal 3′ ends are poly(A) sites more distal than the distal-most annotated 3′ end. All other novel 3′ ends were designated as proximal. (C) Classification of poly(A) sites as fractions of sites or as fractions of the 3P tags. The poly(A) site classification scheme is described in Supplemental Figure S1. (D) Genes with alternative 3′ UTR isoforms in Ensembl v66 and following 3P-seq-based annotation. (E) Maximal 3′ UTR lengths in Ensembl v66 and following the 3P-seq-based annotations. For the new models, the longest 3′ UTR was supported by at least 10% of the 3P tags in at least one sample. (F) 3′ UTRs annotated for the magi2 gene. 3P-seq and RNA-seq tracks indicate all tags mapping to this locus. No 3′ UTR was annotated for this gene in Ensembl v66.
When related to the annotated gene models (Fig. 1C; Supplemental Fig. S1; Supplemental Table S2), only 19.3% of the poly(A) sites were within 100 bases of an annotated 3′ end, but these accounted for 67.7% of all the 3P tags, indicating that, when compared to the novel poly(A) sites, the previously annotated poly(A) sites are for the more highly expressed transcripts or are sites that are more efficiently processed. Overall, 88% of the tags could be assigned to a known or novel 3′ UTR of an annotated gene model (Fig. 1C). Another 6.2% of the tags mapped to mitochondrial sequence indicating polyadenylated mitochondrial transcripts and degradation intermediates (Shepard et al. 2011). As expected, the mitochondrial sites rarely followed PAS motifs (Fig. 1B). Poly(A) sites rarely occurred in coding sequence, near transcription start sites, or in repetitive regions; each of these categories accounted for <0.7% of the 3P tags (Fig. 1C). Less than 5% of the tags mapped to previously unannotated regions, some of which were used to identify long intervening noncoding RNAs in zebrafish (Ulitsky et al. 2011).
The 3P tags from all the libraries except for the pre-MZT embryo (which was atypical, as described below) were used for 3′ UTR reannotation (Supplemental Table S3). Although our sequential annotation procedure did not allow assignment of sites to more than one category, sites were often assigned to more than one transcript of the same gene because alternative start sites or alternative splicing generated different transcripts with the same 3′-terminal sequences. Therefore, to prevent double counting of sites corresponding to multiple transcripts from the same gene, our results are typically described with respect to unique gene models (abbreviated as “genes”) rather than to transcripts, even though we recognize that CPA occurs to RNA transcripts, not genes. At least one 3′ UTR was assigned to 79% of the protein-coding genes, and for 63% of the genes, at least one novel 3′ UTR was identified. After combining 3′ ends appearing within 50 nt from each other, 69.7% of the genes with assigned 3′ UTR had at least two alternative 3′ UTR ends, thereby increasing the incidence of APA by threefold over Ensembl annotations. On average, these genes with alternative 3′ UTRs had 2.8 distinct 3′ ends (Fig. 1D).
3P-seq-based annotation substantially extended (>100 nt) the length of the longest 3′ UTR of 11,442 genes when using a cutoff in which the longer UTR must account for at least 10% of the 3P tags in at least one library (Fig. 1E,F; Supplemental Table S4). As a result, the number of genes with 3′ UTRs at least 2 kb long increased from 2104 to 6810. The reannotated 3′ portions of the genes also allowed us to extend the polypeptide sequences of 640 proteins by at least three amino acids, adding an average of 55 amino acids to each (Supplemental Table S5). In 797 cases, two poly(A) sites appeared within 50 nt from each other on opposite strands, and in 336 of these, the distance was 10 nt or less, suggesting that, similar to the situation in C. elegans (Jan et al. 2011), palindromic arrangements of bidirectional elements can lead to close cleavage positions of convergent zebrafish transcripts, leaving little or no intergenic space. Only 192 cases of convergent transcripts ending within 10 bp of each other were annotated in Ensembl v66.
Our expanded and revised set of 3′ UTR annotations provides a rich resource for studying APA, miRNA function, and other types of post-transcriptional regulation in zebrafish. miRNA target predictions based on our revised 3′ UTR collection are presented in TargetScanFish, beginning with TargetScan release 6.2 (http://targetscan.org). Zebrafish 3′ UTRs differed from human 3′ UTRs in several aspects that affect miRNA regulation. First, they were much more likely to overlap repetitive elements (as identified using RepeatMasker), with 44% of the zebrafish 3′ UTR sequence being repetitive compared to just 16% for human 3′ UTRs. The nonrepetitive fraction of zebrafish 3′ UTRs is more A/U-rich than the nonrepetitive human 3′ UTRs (64% vs. 57%). This leads to a higher density of miRNA target sites for miRNAs with A/U-rich seeds (Supplemental Fig. S3), although the overall mean frequency of heptamer miRNA target sites in the nonrepetitive fraction of the 3′ UTR is similar for both species (0.167 sites per kb per miRNA family in zebrafish vs. 0.142 in human).
Prediction of functional miRNA target sites in mammals, flies, and worms has been greatly facilitated by conservation analysis (Brennecke et al. 2005; Lewis et al. 2005; Lall et al. 2006; Ruby et al. 2007; Friedman et al. 2009; Jan et al. 2011). Unfortunately, conservation is currently of limited utility for predicting zebrafish miRNA targets. Due to the large evolutionary distances between zebrafish and other species with sequenced genomes, only 26.1% of zebrafish 3′ UTR bases are alignable to at least two species in the eight-way whole-genome alignment available in the UCSC Genome Browser, which includes four other fish genomes, frog, human, and mouse. The aligned bases are preferentially located near stop codons, suggesting that their alignability is driven primarily by the alignment of coding sequences, which explains why longer 3′ UTRs have significantly less alignable sequence (Pearson correlation r = −0.27 between 3′ UTR length and the fraction of the 3′ UTR that is alignable). Thus, until more relevant genome sequences become available, TargetScanFish will predict zebrafish miRNA targets based on seed-matched sites in 3′ UTRs and will rank them based on features of site context known to correlate with target efficacy in mammals, but it will not consider site conservation because inclusion of conservation as a criterion for miRNA target-site prediction would likely introduce bias against genes with longer 3′ UTRs.
Dynamics of poly(A)-site selection during zebrafish development and organogenesis
The numbers of 3P tags assigned to each transcript was highly correlated with transcript levels, as estimated using RNA-seq data from the same or similar tissue or stage (Spearman correlation coefficients ranging between 0.77 and 0.52) (Supplemental Fig. S4A; Supplemental Table S6), which indicated that the fraction of 3P tags assigned to alternative poly(A) sites for the same gene reflected the frequency of their utilization. Using this metric to estimate the utilization of alternative poly(A) sites, the average 3′ UTR lengths varied as much as 1.8-fold between libraries, ranging from 912 in the pre-MZT embryo to 1624 in the brain (Fig. 2A). Similar trends of extreme 3′ UTR lengths in the brain and gonads and of increasing 3′ UTR length during embryonic development have been observed in mammals using expressed sequence tags (Zhang et al. 2005; Ji et al. 2009).
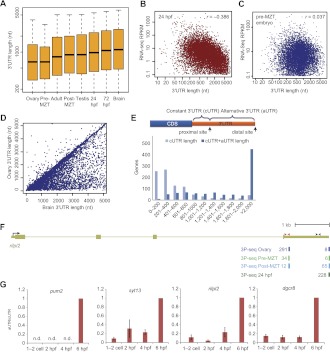
Figure 2.
Changes in 3′ UTR lengths in different developmental stages. (A) Distribution of 3′ UTR lengths in different stages and tissues. In each sample, for each gene with a single annotated or predicted stop codon and 3P-seq data, the mean 3′ UTR length was computed by averaging the lengths of all the 3′ UTRs, weighted by the number of 3P tags supporting each of them. Box plots show the median length, flanked by 25th and 75th percentiles. The whiskers are drawn to the fifth and 95th percentile. (B) Negative correlation between 3′ UTR length and transcript levels 24 hpf. For each gene, the mean 3′ UTR length was computed as in A, and the RPKM was computed using available RNA-seq data from the same developmental stage (SRA accession ERP000016), considering only protein-coding regions. (C) Lack of correlation between 3′ UTR length and transcript levels in the pre-MZT embryo. As in B, except RNA-seq RPKM was computed using available RNA-seq data from the two-cell embryo (SRA accession ERX008924). (D) 3′ UTR lengths of genes expressed in the ovary and in the brain. Lengths were computed as in A. (E) Lengths of 3′ UTRs resulting from proximal and distal poly(A) sites in analysis of genes with substantial differences in isoform fractions (>0.3) when comparing ovary and brain samples. (F) Poly(A) sites of rilpl2. The gene model shown is as annotated in Ensembl v66. 3P-seq tracks show tags from clusters containing at least 10% of the tags in the indicated samples. (Red and black arrows) Position of the qRT-PCR primers for the constant and the alternative regions of the transcript, respectively. (G) qRT-PCR analysis of changes in 3′ UTR usage during early embryogenesis. RT was performed with random primers and expression levels were computed using probes located in the constant and alternative regions of the transcript (cUTR and aUTR, respectively) (Supplemental Fig. S4) and normalized to expression at 6 hpf. (n.d.) aUTR could not be detected at that time point.
In human, transcripts with shorter 3′ UTRs tend to accumulate to higher levels (Chiaromonte et al. 2003). This observation could be explained in part by the presence of destabilizing elements, such as miRNA target sites, in the 3′ UTRs (Sandberg et al. 2008; Mayr and Bartel 2009). We observed a similar trend in some of the zebrafish samples. Strong negative correlations were observed in 24 hpf and 72 hpf embryos (−0.39 and −0.48, respectively) (Fig. 2B) and weaker ones in ovaries and adults (−0.26 and −0.18, respectively). However, there was little correspondence between 3′ UTR length and mRNA accumulation in the brain and the early embryo (−0.08 and 0.037) (Supplemental Fig. S4B; Fig. 2C). In the brain, the lack of correlation could result from relatively high expression of genes with long 3′ UTRs (Supplemental Fig. 4B), whereas in the early embryo, lack of correlation could stem from instability of isoforms with short 3′ UTRs (see below).
Prevalent use of proximal poly(A) sites in the ovaries and pre-MZT embryo
Pairwise comparisons of libraries identified sets of genes with conspicuous changes in preferred alternative 3′ UTR isoforms between samples. For 3111 genes, the difference in the relative usage of at least one poly(A) site differed by at least 30% between two samples, and for 2619 of those, the most common 3′ end differed between two samples (Supplemental Table S7). Consistent with the global trend of shortest 3′ UTRs expressed in the ovary and longest in the brain, the most change in preferred isoforms was observed between these tissues: Compared to brain, 1089 genes preferentially used a poly(A) site that was more proximal to the transcription start site (for simplicity, referred to as proximal sites) in the ovary, whereas only 65 showed the reverse trend and preferentially used the distal sites in the ovary (Fig. 2D). These APA events generated much shorter 3′ UTRs for the affected genes (Fig. 2E). Widespread differences also occurred between the ovary and the testis: Compared to testis, 811 genes preferentially used the proximal site in the ovary, whereas only 104 preferentially used the distal site, indicating that shorter 3′ UTRs were not a shared characteristic of gonads. Finally, comparing pre-MZT and post-MZT, 419 genes preferentially used the proximal site pre-MZT, whereas only 80 had the reverse trend. For some genes, such as rilpl2 (Fig. 2F), the relative change exceeded fivefold, as also confirmed by qRT-PCR (Fig. 2G; Supplemental Fig. S5). In the tested cases, the predominance of transcripts ending at the proximal site appeared to persist until the beginning of the MZT (4 hpf), with a rapid induction of longer isoform during MZT. These results suggest that a less stringent CPA regime, which cleaves the pre-mRNAs at the proximal sites, is prevalent in the ovaries and is replaced during MZT with a more stringent one, which preferentially skips the proximal sites and processes only the distal ones.
Proximal and distal sites were flanked by regions with similar nucleotide preferences, with distal sites having a slightly higher enrichment of A's in the −10 to −30 region and U's downstream (Supplemental Fig. S6A,B). Comparison of the prevalence of 15 CPA-associated motifs in the regions surrounding the proximal and distal sites using the polya_svm algorithm (Cheng et al. 2006) showed that 14 of the motifs were associated with the two site classes with similar frequencies, but that the motif that captured the canonical PAS was associated with proximal sites much less frequently than it was with the distal sites (P = 2.9 × 10−11) (Supplemental Fig. S6C). Direct comparison of defined hexamer motifs 10 to 30 bases upstream of the proximal sites showed a pronounced reduction (>33%) in the use of AAUAAA but not of its common variants (P < 1 × 10−15) (Supplemental Fig. S6D).
If the CPA machinery was more efficient in the ovaries and acted on sites that were not used in other tissues, then intronic poly(A) sites, which would lead to formation of alternative last exons, would also be preferentially utilized in the ovary. Indeed, we found that such sites were preferentially used in both the ovary and the pre-MZT embryo (Supplemental Fig. S5E).
Differential expression and alternative polyadenylation of cleavage and polyadenylation factors in ovaries and pre-MZT embryo
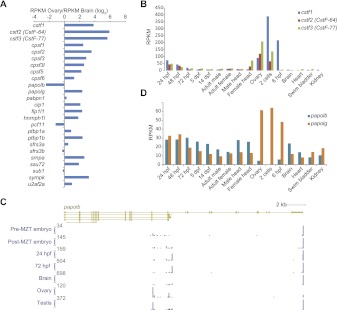
The increased use of weaker poly(A) sites is typically concomitant with increased expression of CPA factors (Liu et al. 2007; Sandberg et al. 2008; Mayr and Bartel 2009). When comparing ovary and brain, most CPA-associated factors (taken from Liu et al. [2007]) were more highly expressed in the ovary (Fig. 3A). The difference was most pronounced for cstf1-3 genes, whose mRNAs were 15- to 60-fold higher in the ovary than in the brain and appeared to be specifically up-regulated in the ovary and the early embryo (Fig. 3B). Adding another dimension to the regulation, some CPA factors underwent alternative polyadenylation. In two cases, s_ub1_ and clp1, an ovary-specific short 3′ UTR isoform accumulated, and in four others, papolb, pcf11, cstf3, and pabpn1, the APA determined the identity of the last exon of the transcript, thereby affecting the coding sequence (Fig. 3C; Supplemental Fig. S7). One of these, papolb, encodes one of the zebrafish poly(A) polymerases. The zebrafish genome has two poly(A) polymerase genes, papolb and papolg, which encode orthologs of mammalian PAP and PAP gamma, respectively. Although the two genes are expressed at similar levels in most tissues and developmental stages, papolg is up-regulated in the ovary and the early embryo, whereas papolb is repressed (Fig. 3D). In mouse, PAP is alternatively spliced and polyadenylated, which generates at least five isoforms, the shorter of which (PAP III, IV, and V) contain only about half of the exons of the full-length mRNA and lack polymerase function (Zhao and Manley 1996). Likewise, in zebrafish, alternative polyadenylation generates three abundant isoforms—a long isoform with 23 exons, and shorter isoforms containing just 11 or 12 exons (Fig. 3C). These isoforms have different expression patterns, with ovary and testis preferentially accumulating the shortest isoform. Thus, in zebrafish ovaries and early embryo, APA-mediated truncation of the papolb coding sequence acts concordantly with altered mRNA levels to shift productive expression nearly exclusively to the papolg homolog.
Figure 3.
Changes in expression and polyadenylation of CPA factors during zebrafish development. (A) Relative expression of CPA-related genes (listed in Liu et al. 2007) in ovary and brain. Mammalian homologs of CstF factors are in parentheses. (B) Expression of mRNA for CstF factors in different stages and tissues. (C) Differential alternative polyadenylation of papolb. The transcript models shown are as annotated in Ensembl v66. The height of each plot indicates the number of 3P tags ending at each position, normalized to the maximum value, which is indicated at the top of each axis. (D) Expression of mRNA for poly(A) polymerases in different stages and tissues.
Preferential loss of ovary transcripts with very short 3′ UTRs
Paucity of transcription in the early embryo allowed for a direct comparison of the post-transcriptional fate of different isoforms transcribed in the ovary, many of which are deposited into the oocytes. We focused on 1351 genes that had a single annotated or predicted stop codon (and thus a unique 3′ UTR 5′ end) and two different poly(A) sites, separated by at least 100 nt, expressed in the ovary (≥20 3P tags each). For most of these genes, the 3′ UTRs resulting from the use of the proximal CPA sites were very short, often <200 nt (Fig. 4A). The proximal sites had a lower propensity for appearing downstream from the AAUAAA hexamer (31.5%), although 43% of them appeared downstream from one of its close variants, indicating that their formation was likely related to the global increase in usage of weaker CPA signals in the ovary. For 576 of the 1351 genes, preference for the longer isoform increased more than twofold in the pre-MZT relative to the ovary (e.g., rnaset2) (Fig. 4B), compared to just 51 genes for which preference for the shorter isoform increased twofold.
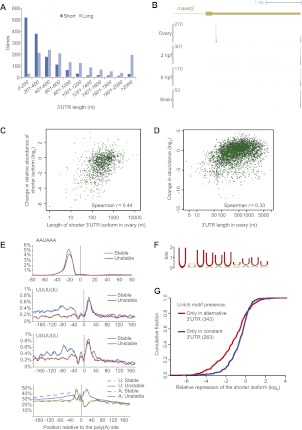
Figure 4.
Differential accumulation of transcripts with short 3′ UTRs in the ovary and pre-MZT embryo. (A) Comparison of 3′ UTR lengths of the short and the long isoforms in genes with exactly two isoforms in the ovary. (B) Poly(A) sites of rnaset2 in the indicated samples. The shown 3′ UTR structure is as annotated in Ensembl v66. The height of each plot indicates the number of 3P tags ending at each position, normalized to the maximum value, which is indicated at the top of each axis. (C) Relationship between length of the shorter isoform and relative abundance of the shorter isoform in the pre-MZT embryo, as inferred from 3P tags for genes with two alternative poly(A) sites in the ovary. (D) Relationship between the length of the 3′ UTR in the ovary and the change in mRNA observed in the pre-MZT embryo relative to that in the ovary, as inferred by 3P tags. Analysis was for genes with a single 3′ UTR supported by at least 20 3P tags in the ovary. (E) Frequency of the indicated motifs or nucleotides flanking poly(A) sites of isoforms that were not reduced in the pre-MZT embryo compared to the ovary (stable) and those that were reduced at least twofold (unstable). For the hexamer motifs, the frequencies shown at each position are averages of a window of 11 consecutive nucleotides centered at that position. (F) The motif identified by Amadeus (Linhart et al. 2008) as significantly enriched in regions upstream of the stable sites. (G) Destabilization in the pre-MZT embryo of shorter isoforms lacking the U-rich motif. Genes with two UTR isoforms in the ovary and a U-rich motif 20–90 nt upstream of only one of the poly(A) sites were stratified based on the location of the motif—upstream of the distal site (red line) or upstream of the proximal site (blue line). A U-rich motif was defined as present if a decamer with up to two mismatches from the UK(U)8 consensus appeared 20–90 nt upstream of the poly(A) site.
A possible explanation for the preferential depletion of the shorter isoforms in the early embryo is that they lack sequence elements required for either long-term stability of the mRNA in the oocyte or protection against degradation of maternal mRNA following fertilization. Indeed, for genes with two 3′ UTR isoforms in the ovary, the drop in relative abundance in the pre-MZT embryo was inversely correlated with the length of the shorter 3′ UTR isoform (Spearman r = −0.44, P < 10−15) (Fig. 4C). Moreover, all 158 distinct mRNAs in the ovary with a single 3′ UTR that was shorter than 50 nt were down-regulated (11-fold on average), compared to 81% of the 381 mRNAs with 3′ UTRs between 50 and 100 nt, and just 41% of the 1187 mRNAs with 3′ UTRs longer than 1000 nt (Fig. 4D). Overall, for single-isoform genes, there was a highly significant inverse correlation between the length of the 3′ UTR in the ovary and the underrepresentation of 3P-tags at the transcript in the pre-MZT embryo (Spearman r = −0.33, P < 10−15). We observed a similar correlation when using RNA-seq data from the ovary and a two-cell embryo and considering only reads that mapped to the coding sequence (r = −0.28). These results suggest that a 3′ UTR of 50–100 bases is required for the stability of a maternal transcript in oocytes or during the first cell divisions after fertilization. For example, 13 zona pellucida genes, which encode components of the fish egg membrane (Wang and Gong 1999), all had 3′ UTRs <50 nt long, were expressed very highly in the ovary (over 750,000 3P-seq tags mapped to two clusters of these genes on chromosomes 17 and 20), and were almost completely absent in the pre-MZT embryo [e.g., the poly(A) site of zp2.6 yielded 114,089 3P tags in the ovary but only 427 in the pre-MZT embryo].
In order to identify regulatory elements that might contribute to mRNA stability, we compared 3′ UTRs of single-isoform genes that were decreased at least twofold (‘unstable') to those that either did not change or increased (‘stable'), as measured by comparing 3P tags in the ovary and pre-MZT embryos (note that by this measure either complete degradation or deadenylation would render transcripts unstable). The two sets had very similar patterns of enrichment for the AAUAAA PAS ∼20 nt upstream of the poly(A) site and similar sequence composition downstream from the poly(A) site (Fig. 4E). The difference came in the region 30–90 nt upstream of the poly(A) site, where a marked enrichment of U's and U-rich hexamers was observed in the stable transcripts compared to the unstable ones (Fig. 4E). This enrichment was observed even when considering only 3′ UTRs at least 200 bases long (to avoid bias due to the proximity to coding sequence). Analysis of sequences from these upstream regions using the de novo motif finder Amadeus (Linhart et al. 2008), which accounts for differences in G/C content, identified a single U-rich motif UKUUUUUUUU (P = 6.3 × 10−19) (Fig. 4F), which, together with its minor variants, was present in 28% of the stable and only 8.5% of the unstable messages. This motif resembled the recently identified motif associated with cytoplasmic polyadenylation in zebrafish (Aanes et al. 2011), and stretches of 12–27 uridines positioned upstream of the PAS have been associated with cytoplasmic polyadenylation after fertilization (but not during oogenesis) in Xenopus (Simon et al. 1992). This poly(U) motif is thought to be distinct from the UUUUUAU CPE element that directs cytoplasmic polyadenylation during meiotic maturation of the oocyte (Richter 1999). A more detailed analysis using a sliding window of 15 bases showed that, in the stable transcripts, the maximal U-enriched window contained nine or 10 uridines (Supplemental Fig. S8A**)** and that the second base in the motif was preferentially G, whereas the other bases were more likely to be A or G in cases in which they were not U (Supplemental Fig. S8B). We did not observe a significant location preference for this motif with respect to the PAS or the poly(A) site, suggesting that its exact position in the region up to 70 bases upstream of the PAS does not have as much influence as its mere presence in the 3′ UTR. Among the 1351 genes with two poly(A) sites in the ovary, the relative change in shorter and longer isoforms when comparing ovary and the pre-MZT embryo was also dependent on the motif, and presence of the U-rich motif exclusively near the distal poly(A) site coincided with less depletion of the longer isoform (P = 1.75 × 10−13) (Fig. 4G). Taken together, our results indicate that the U-rich motif helps stabilize (or prevents deadenylation of) maternally loaded transcripts in the pre-MZT embryo.
A wave of polyadenylation at many noncanonical sites in the early embryo
When each library was separately compared to others, many poly(A) sites appeared exclusive to one sample (the adult sample was excluded from this analysis as it inherently overlapped with other samples) (Fig. 5A). Testis had the largest number of sample-specific poly(A) sites (11,894), many of which could not be explained by the existing gene models. Some testis-specific transcripts are not yet annotated (testis RNA-seq data are not yet available) and presumably account for many of these sites. The testis-specific sites did not differ significantly from other sites in their surrounding base composition (Fig. 5B), or prevalence of an upstream PAS (40.4% followed AAUAAA, and 36.9% followed one of its 10 common variants).
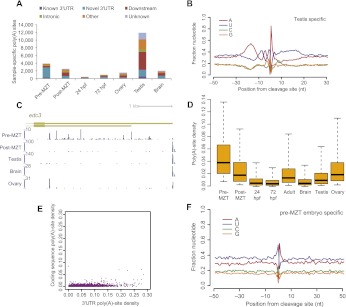
Figure 5.
Many noncanonical poly(A) sites in the pre-MZT embryo. (A) Abundance of sample-specific poly(A) sites. Site classification was as described in Supplemental Figure S1. Downstream sites are those appearing up to 8 kb downstream from the annotated 3′ ends but without the support for connectivity with the stop codon required for assignment as a novel 3′ UTR. (B) Sequence composition near poly(A) sites specific to the testis. (C) Poly(A) sites of edc3. The 3′ UTRs shown are as annotated in Ensembl v66. (D) Density of poly(A) sites occurring within 3′ UTRs from the indicated samples. Poly(A)-site density was defined as the ratio between the number of poly(A) sites and the length of the longest 3′ UTR. (E) Densities of poly(A) sites in the coding sequence and 3′ UTRs plotted for genes expressed in the pre-MZT embryo. (F) Sequence composition near poly(A) sites specific to the pre-MZT embryo.
Another 3913 library-specific events appeared in the pre-MZT embryo, and in contrast to the testis sites, these were predominantly novel sites that could be assigned to existing gene models (Fig. 5A). The poly(A) site density [defined as the number of distinct poly(A) sites as a function of the transcript length] was the highest in this sample (Fig. 5C,D). Although these pre-MZT-specific poly(A) sites were significantly more dense in 3′ UTRs than in the coding sequence (Fig. 5E), they differed dramatically from other sites with respect to surrounding base composition (Fig. 5F) and rarely followed a canonical PAS or its common variants (3.4% and 13.8%, respectively). These observations indicated that many of the pre-MZT-specific poly(A) sites were not generated by the canonical CPA mechanism. Furthermore, these sites were specific to a stage in which transcription is not thought to occur, which suggests that they were formed post-transcriptionally on messages transcribed previously in the oocytes. Thus, we propose that these sites were formed by cytoplasmic polyadenylation of mRNAs undergoing 3′ exonucleolytic degradation. Consistent with the idea that these transcripts underwent degradation in the early embryo, the fraction of poly(A) sites that were pre-MZT-specific was highly correlated with decreased abundance of the transcript in the post-MZT compared to the pre-MZT embryo (Spearman r = 0.28, P < 10−15, expression changes based on RNA-seq data taken from Aanes et al. 2011).
If cytoplasmic polyadenylation generates these isoforms, it would differ from that of previous reports in that previously described cytoplasmic polyadenylation acts by elongating existing short poly(A) tails (Richter 1999). To further characterize the CPA landscape in the pre-MZT embryo, we performed additional sequencing and obtained a total of 39,051,782 3P tags from the pre-MZT sample, which mapped to 2,921,536 unique genomic positions. Focusing on 6838 genes that had at least 50 tags in the pre-MZT embryo and at least 20 tags in the ovary and reducing our stringency to allow a single tag to define a site, we defined pre-MZT-specific poly(A) sites as those that were at least 10 nt away from a position present in any library except that of the ovary and at least 10 nt away from positions covered by at least 10 tags in the ovary. Overall, 641,377 tags mapped to 254,155 pre-MZT-specific poly(A) sites, an average of 2.3 tags per site, compared to the global average of 59.9 in the pre-MZT embryo and 46.6 for all the positions in the ovary. Of these poly(A) sites, 38.9% were defined by a single tag. Thus, the pre-MZT-specific poly(A) sites are typically each used rarely but collectively corresponded to a nontrivial fraction of the polyadenylated messages at the pre-MZT embryo. For 182 genes, most of the 3P tags came from pre-MZT-specific loci (85 loci on average), and for 1942 genes (almost a third of the genes passing our expression cutoffs for analysis), ≥10% of the 3P tags in the pre-MZT embryo were from loci exclusive to that time point. These exclusive positions were 15-fold more likely to appear in the 3′ UTR compared to the coding sequence.
In bacteria, yeast, plants, mammals, and the mitochondria, addition of a short oligo(A) sequence marks an RNA for degradation (Anderson 2005; Slomovic et al. 2006; West et al. 2006; Lange et al. 2009; Shcherbik et al. 2010; Chang and Tong 2011). To test whether the noncanonical poly(A) sites in the pre-MZT embryo have short tails resembling those of marked RNAs or longer poly(A) tails resembling those of typical mRNAs, we developed 3P-PEseq, a variation on 3P-seq that uses paired-end Illumina sequencing to estimate the lengths of poly(A) tails on a global scale (Fig. 6A). 3P-PEseq readouts consist of two reads. The first (read #1) starts at the 3′ end of the RNA (in antisense orientiation), which is typically the 3′ end of the poly(A) tail. The other (read #2) starts within the mRNA and often contains the 3′ portion of the 3′ UTR, followed by untemplated adenylates. 3P-PEseq readouts are informative when read #1 begins with T's, which provides information on the number of terminal adenylates in the amplicon [i.e., an estimate of the length of the poly(A) tail], and read #2 contains a sequence that can be mapped to the genome, followed by untemplated A's, which identifies the mRNA and its poly(A) site. As implemented, 3P-PEseq identified the precise lengths of tails up to 70 bp long and a lower bound on the lengths of longer tails. This dynamic range was more than adequate to distinguish between short tails thought to mark RNAs for degradation and the longer tails typically found on mRNAs.
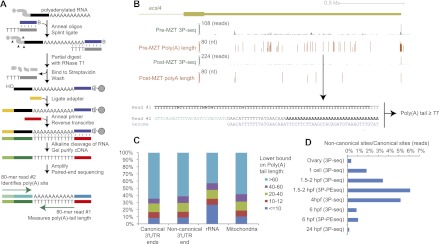
Figure 6.
Genome-wide estimation of poly(A)-tail lengths. (A) Outline of 3P-PEseq. See text for description. (B) Poly(A) sites and poly(A)-tail lengths pre-MZT and post-MZT at the acsl4 3′ UTR. The 3′ UTR shown is as annotated in Ensembl v66. The height of the 3P-Seq plots shows the number of 3P tags at each position, normalized to the maximum value, which is indicated at the top of each axis. The height of the 3P-PEseq plots shows the average poly(A)-tail length measured at each position. The length is zero at positions for which no 3P-PEseq tags were obtained. (Arrow) The poly(A) site corresponding to the paired reads shown below. (Bold) Untemplated nucleotides. In this example, the cleavage position is within three genomically encoded A's, and thus, at least 77 of the 80 T's of read #1 correspond to untemplated A's of the the poly(A) tail. The adapter sequence in read #2 is in green italics. (C) Distribution of pre-MZT poly(A)-tail lengths at poly(A) sites mapping to the indicated loci and RNA classes. Canonical 3′ UTR ends are tallied as 3P-PE tags that map within 20 nt of a 3′ UTR end annotated using samples from other stages. Noncanonical 3′ UTR ends are tallied as 3P-PE tags that map within a 3′ UTR annotated in other stages but not within 20 nt of a poly(A) site defined at any other stage. rRNA and mitochondrial ends are tallied as tags that map within rRNA repeats and the mitochondrial chromosome, respectively. (D) Abundance of noncanonical isoforms. Plotted is the ratio of 3P or 3P-PE tags mapping to noncanonical 3′ UTR ends (defined as in C) relative to those mapping to canonical 3′ UTR ends.
We applied 3P-PEseq to total RNA from pre-MZT (1.5–2 hpf) and post-MZT (6 hpf) embryos and obtained 3,874,353 3P-PEseq readouts from 452,817 sites and 4,803,851 readouts from 365,752 sites, respectively. The fraction of all poly(A) sites that mapped to noncanonical positions within 3′ UTRs pre-MZT was greater than fivefold higher than that fraction in post-MZT embryos, which further indicated the prevalence of noncanonical poly(A) sites pre-MZT (Fig. 6B). Indeed, the 82,280 3P-PEseq pre-MZT sites that mapped within 3′ UTRs but not near poly(A) sites observed in other stages generally appeared in noncanonical sequence contexts with no more than chance association with a PAS (2.9% and 14.6% downstream from the AAUAAA PAS or one of its derivatives, respectively), whereas the 18,598 post-MZT sites were more frequently in canonical contexts (31% downstream from the AAUAAA PAS or one of its derivatives).
The distribution of poly(A) tail lengths at canonical and noncanonical sites appeared to be practically indistinguishable (Fig. 6C), as 61% and 64% of the canonical and noncanonical poly(A) sites, respectively, had poly(A) tails of at least 60 adenylates pre-MZT. This suggests that the mechanism that generates the noncanonical sites ultimately endows them with long poly(A) tails and is not simply the mechanism that marks RNA for degradation.
To further characterize the dynamics of noncanonical sites, we acquired 8,736,478 3P tags from the one-cell embryo and 7,423,152 tags from the pre-MZT embryo at 4 hpf and compared the relative numbers of 3P tags coming from noncanonical sites at different stages (Fig. 6D). The fraction of these sites is highest at 1.5–2 hpf and 4 hpf, which suggests that the bulk of noncanonical poly(A) sites are generated in the pre-MZT embryo rather than inherited as part of the maternal RNA and that transcripts with noncanonical 3′ ends are degraded before or during the MZT.
A correlation between the evolutionary divergence in 3′ UTR length and changes in gene expression
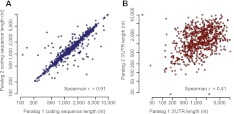
A whole-genome duplication in the teleost lineage, which occurred soon after divergence from tetrapods, generated numerous paralogous gene copies in the zebrafish genome, many of which have diverged in spatial and/or temporal expression during embryogenesis (Talbot et al. 2005; Semon and Wolfe 2007; Kassahn et al. 2009). Although the length of the coding sequence between paralogous pairs was highly consistent (Spearman r = 0.91 across 754 pairs) (Fig. 7A), 3′ UTR length was much less conserved (Spearman r = 0.41) (Fig. 7B). Furthermore, the relative change in 3′ UTR length was inversely correlated with similarity of gene expression profiles across 16 different developmental stages/tissues (Spearman r = −0.12, P = 5 × 10−4), suggesting that sequence divergence in the 3′ UTR, even when evaluated very crudely as change in its length, contributes to divergence of gene expression patterns following gene duplication. Consistent with the typically negative effect of sequence elements in the 3′ UTR on transcript levels, the paralog with the longer 3′ UTR had globally lower expression levels (P = 2 × 10−7, sign test).
Figure 7.
Analysis of paralogous gene pairs generated in the teleost whole-genome duplication. (A) Relationship of coding-sequence lengths for pairs of paralogous genes. Genes of each pair were arbitrarily assigned to each axis. (B) Relationship of 3′ UTR lengths for pairs of paralogous genes. 3′ UTR lengths are the weighted average across all the samples in which the transcript had at least one 3P tag.
Discussion
Comparison of poly(A) sites across diverse developmental stages and tissues allowed us to increase the estimate of the number of zebrafish genes that undergo alternative polyadenylation from 26.1% to over 80%, a major fraction of which appears to be regulated. The most pronounced differences in 3′ UTR length occurred in the ovaries and in the brain, which resembled trends observed in flies and mammals (Zhang et al. 2005; Ji et al. 2009; Hilgers et al. 2011). The efficiency of a poly(A) site is correlated with the strength of the polyadenylation signal (Zhao et al. 1999). A mechanism that takes advantage of this correlation appears to account for the most widespread alternative polyadenylation trends, as differences in sequence composition that are analogous to the ones we identified in zebrafish were previously reported to account for other significant trends of differences in 3′ UTR length (Tian et al. 2005; Zhang et al. 2005; Liu et al. 2007; Sandberg et al. 2008; Ji and Tian 2009; Ji et al. 2009; Mayr and Bartel 2009; Hilgers et al. 2011). In these diverse systems, alternate poly(A) sites that are more proximal to the transcription start site appear to share most of the sequence characteristics of the distal ones but are significantly less likely to appear after the AAUAAA motif. Widespread use of alternative proximal sites in a specific stage or tissue, such as the ovary, thus appears to rely in part on increased activity of the CPA machinery toward weaker poly(A) sites. Specific genes presumably use additional strategies to either enhance or counter the prevailing trend, but these strategies were not apparent in our genome-wide analysis.
Although our analysis provided some hints as to the mechanism responsible for widespread shortening of 3′ UTRs in the ovary, the purpose of this shortening is unclear. Perhaps transcripts with short 3′ UTRs are more suitable for long-term storage of maternally deposited transcripts. Alternatively, the cell might use alternative 3′ UTR isoforms for potent regulation of gene expression during oocyte maturation and pre-MZT development. For example, robust generation of transcripts with very short 3′ UTRs, such as the zona pellucida genes, which then become very unstable once massive degradation of maternal mRNA takes place, could be a useful strategy for providing the first coordinated wave of degradation of maternal gene products.
The thousands of poly(A) sites specific to the pre-MZT embryo represent an unexpected set of transcripts whose 3′ ends appear to be generated post-transcriptionally, as they are rare in the ovary sample, and have no enrichment of the sequence elements associated with other poly(A) sites. These features would be expected if these sites represent either degradation intermediates of a 3′→5′ exonuclease or products of endonucleolytic cleavage, followed by extension of the 3′ end by polyadenylation, most likely by a cytoplasmic polyadenylation activity known to be active at this developmental stage (Aanes et al. 2011). The prevalence of noncanonical sites in 3′ UTRs compared to coding sequence would be explained if loss of the stop codon triggered rapid decay and consequent underrepresentation of mRNA fragments ending in the coding sequence (Frischmeyer et al. 2002), or if actively translating ribosomes protected the coding sequence from degradation. As these readenylated messages might be subject to additional rounds of shortening and readenylation, the levels of maternal transcripts may be governed by a previously unknown competition between mRNA decay and polyadenylation. In this scenario, transcripts with longer 3′ UTRs would persist for longer time periods, which would help explain why we did not observe a negative correlation between 3′ UTR length and expression levels at the pre-MZT embryo. Also helping to explain this lack of correlation is our finding that 3′ UTRs shorter than 100 bases are almost exclusively at reduced levels in the pre-MZT embryo relative to the ovary. Interestingly, a similar trend of preferential degradation of transcripts with short 3′ UTRs has been reported in mouse during GV-oocyte to two-cell stage embryos (Evsikov et al. 2006), suggesting that the underlying mechanisms might extend to other species, including mammals.
Our results also point to the benefits of using a dedicated method for mapping poly(A) sites as part of standard genome annotation. Prior to our work, zebrafish had a rather extensively characterized transcriptome, with more than 1,400,000 ESTs in dbEST (Boguski et al. 1993) (more than for the rat, chicken, and Xenopus genomes), and more than 300 million RNA-seq reads that were already integrated into the Ensembl transcription annotation. Still, eight 3P-seq samples, each with less than 10 million genome-mapping reads were sufficient to identify novel 3′ UTRs for >60% of all annotated protein-coding genes, substantially increasing the 3′ UTR length for over 5000 of them, as well as pointing to novel layers of regulation of 3′ formation at canonical and noncanonical sites. The 3P-seq results were also an important starting point for identifying long noncoding RNAs in zebrafish (Ulitsky et al. 2011). Thus, application of a dedicated method for identifying 3′ ends should become a routine feature of future transcriptome annotation projects.
Methods
3P-seq
Zebrafish were maintained and staged using standard procedures (Kimmel et al. 1995). Ovaries and testes were obtained as described (Gupta and Mullins 2010). 3P-seq was performed as described (Jan et al. 2011). Sequencing was performed using Illumina Genome Analyzer II, except for one of the two additional pre-MZT emrbryo libraries, which was sequenced using Illumina Hi-Seq. Read numbers and genome mapping statistics are presented (Supplemental Table S1). 3P-seq data were processed as described (Jan et al. 2011). Briefly, reads were mapped to the genome using Bowtie (Langmead et al. 2009). Those that mapped to no more than four loci in the genome and possessed at least one 3′-terminal adenylate that was not templated in the genome, were considered 3P tags, unless the 3′ end of the mapped region fell within three bases of an annotated splice site. (Those mapping near splice sites were excluded from further analysis because the reads that supported them could end with adenylates templated in downstream exons). Positions supported by at least four tags, out of which at least two were either distinct (i.e., had a different number of untemplated adenosines) or came from two different libraries, were carried forward as poly(A) sites. To deal with the microheterogeneity of sites, 3P tags were iteratively joined into poly(A) sites that contained all the 3P tags implicating CPA within 10 bases of the most frequently supported site.
Genome annotations and RNA-seq data
Zebrafish genome assembly Zv9 (danRer7) was used throughout this study. Gene models were obtained from Ensembl 66. Predicted transcripts, GenBank cDNAs and ESTs, whole genome alignments, and repetitive elements (excluding simple repeats) were obtained from the UCSC Genome Browser (July 2011). RNA-seq reads were obtained from SRA (accessions ERP000016, ERP000400, and SRP003165) and processed using TopHat (Trapnell et al. 2009) and Cufflinks (Trapnell et al. 2010) with default parameters. Protein homology relationships were taken from Ensembl v66. Genes were considered as paralogs if they were (1) annotated as Clupeocephala paralogs in Ensembl, (2) had exactly one ortholog in mouse, and (3) came from distinct genomic loci.
Poly(A) site classification
Our pipeline for poly(A) site annotation is shown (Supplemental Fig. S1). After clustering to consolidate tags within 10 nt of the most frequently supported site, each poly(A) site was tested for fit to possible categories in the following order:
- Sites mapping to the mitochondria were annotated as mitochondrial poly(A) sites and were not processed further.
- Sites within 100 bases of the 3′ end of a known or predicted gene model were annotated as known poly(A) sites and were not processed further.
- Sites downstream from the 3′ end of a known or a predicted gene, but upstream of the beginning of the coding sequence of the next nonoverlapping gene model on the same strand were considered as potential end points of extended 3′ UTRs. The extension was restricted to 8 kb from the annotated 3′ end. These poly(A) sites were further tested for connectivity with the stop codon (if available) or with the 3′ end of the gene model. Connectivity was tested by screening sets of potential transcript fragments obtained by combining RNA-seq-based reconstructions done using Cufflinks or Exonerate (Slater and Birney 2005; Trapnell et al. 2010) (the latter were obtained from Ensembl v66), ESTs and cDNAs from GenBank. Because the RNA-seq-based reconstructions were based on data that was not strand-specific, the terminal exons of these transcript models could be used to support connectivity on both strands. If connectivity was supported by a fragment spanning the end of the annotated 3′ end and the poly(A) site, the site was annotated as a novel poly(A) site.
- Sites overlapping 3′ UTR exons were annotated as novel poly(A) sites, and sites overlapping coding sequence or 5′ UTRs were annotated as coding sequence and 5′ UTR sites, respectively.
- Sites overlapping introns and appearing downstream from the 3′ end of the coding sequence were tested for connectivity with the 3′ end of the coding sequence. If connectivity was confirmed by a transcript model, the site was annotated as a novel poly(A) site.
- Sites overlapping introns and appearing upstream of the annotated stop codon were tested for connectivity with the end of the preceding exon. If connectivity was supported and the poly(A) site was not annotated as a nonintronic poly(A) site for any other gene model, the site was annotated as the end point of a novel terminal exon. Sites annotated in one of the steps 4–6 were not processed further.
- Sites appearing within 500 bases of the promoter in reverse orientation with respect to the gene model were annotated as promoter-associated poly(A) sites and were not processed further.
- Sites overlapping repetitive elements were annotated as repeat-associated poly(A) sites.
- Sites not fitting any of the categories were classified as unknown poly(A) sites.
Alterative PAS motifs
In order to identify alterative PAS motifs, we focused on regions between 10 and 30 bases upstream of poly(A) sites. We iteratively identified the most common hexamer in this region across all the poly(A) sites, tested its significance, removed from consideration all the sequences containing this hexamer, and repeated the process. The search was terminated when the most common hexamer appeared in <1% of the remaining sequences. Significance was tested by comparing the fraction of sequences containing the hexamer with that found in 100 randomly shuffled sequences with the same dinucleotide frequencies. Eleven hexamers (AAUAAA, AUUAAA, UAUAAA, AGUAAA, UUUAAA, CAUAAA, AAUACA, AAUGAA, AAUAUA, GAUAAA, UGUAAA) each had _P_-values <0.05 and were enriched at least twofold compared to random sequences.
Gene models
For annotation of 3′ UTRs, known protein-coding gene models were obtained from Ensembl v66 and were supplemented with predicted coding genes obtained from alignments of RefSeq gene models from other organisms. We first obtained clusters of known genes by dividing the genes into groups of genes that appeared on the same strand and shared at least one exonic nucleotide. Predicted genes were added to clusters of known genes if they overlapped only one cluster. Predicted gene models that did not overlap any known gene models were then clustered using the same procedure. This procedure resulted in 26,348 clusters.
Alternative polyadenylation between tissues
In order to identify genes with conspicuous differences in usage of alternative poly(A) sites between samples, we first computed for each poly(A) site the number of 3P tags that mapped within 10 bp of it in each sample. These were used to compute fik, the fraction of 3P tags in sample i mapping to poly(A) site k. Genes with poly(A) sites for which |fik_−_fjk| > 0.3 in pairwise comparisons of samples were defined having a difference in usage of poly(A) site k between samples i and j.
qRT-PCR
Total RNA from embryos was isolated using TRI Reagent (Ambion). For each sample, 100 ng of total RNA were used in reverse transcription reactions using oligo-dT primers (IDT) and SuperScript III Reverse Transcriptase (Invitrogen). For each gene, two gene-specific primer sets were designed, a “constitutive” set targeting exons shared between the long and the short isoforms, and the “alternative” set targeting only the 3′ UTR fragment that was alternatively used based on the 3P-seq data. ΔΔCt values were calculated for each gene by normalizing the alternative set against the constitutive one and normalizing this ratio to that observed at 6 hpf.
3P-PEseq
To selectively capture polyadenylated ends for paired-end sequencing, 2.5 μg of total RNA from 2 to 2.2- or 6-hpf zebrafish embryos were splint ligated to a 3′ biotinylated adapter (p-AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGACACATAC-biotin, IDT) in the presence of a bridge oligo (TTCCGATCTTTTTTTTT, IDT) using T4 Rnl2 (NEB) in an overnight reaction at 18°C. Following partial digestion with RNase T1 (Ambion), 115- to 750-nt RNAs were isolated from a denaturing polyacrylamide gel, and ligation products were captured on streptavidin M-280 Dynabeads (Invitrogen). RNAs were 5′ phosphorylated on beads using Polynucleotide Kinase (NEB) and subsequently ligated to an adapter (C3.spacer-GTTCAGAGTTCTAcaguccgacgauc, uppercase, DNA; lowercase, RNA; IDT) using T4 Rnl1 (NEB) in an overnight reaction at room temperature. Complementary DNA was synthesized on beads using SuperScript II (Invitrogen) primed with AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACG, liberated from the beads by base hydrolysis, size-selected (155–790 nt) on a denaturing polyacrylamide gel, and amplified by PCR for 15 cycles. One hundred eighty- to 750-nt products were isolated from a formamide gel and amplified for four additional cycles using primers that contain the Illumina paired-end sequencing primer-binding sites. After a final size selection (220–800 nt) and purification on a formamide gel, 80 × 80 paired-end sequencing was performed on the Illumina Hi-Seq platform.
3P-PEseq data analysis
The 3P-PEseq yielded 219,846,644 and 207,519,359 paired-end 80-nt reads for the pre- and post-MZT samples, respectively. Because of the longer reads compared to 3P-seq and diminishing sequencing quality in longer reads, we used slightly more stringent criteria for defining poly(A) sites. Read #2 began with an adapter of 26 bases. Reads with more than 10 mismatches to the adapter were discarded, and the first 26 bases were removed before further processing. A read pair was considered informative only if read #1 began with T's and read #2 contained 2–39 terminal A's. After leading T's and terminal A's were removed from reads #1 and #2, respectively, they were mapped to the genome using Bowtie, allowing for up to two mismatches and requiring a unique mapping position in the zebrafish genome. When mapping read #2, 7,901,731 and 8,009,618 of the pre-MZT and post-MZT reads, respectively, could be uniquely mapped to the genome. The length of the tail encoded in read #2 was defined as the maximum number of trailing A's, allowing for up to one mismatch. This number was compared to the number of A's in the genome at the corresponding position, accounting for the possibility that an A-rich genomic segment with a single sequencing error might be misclassified as part of a poly(A) tail, and only cases with at least two untemplated A's were carried forward. The poly(A) site was defined as the last non-A base in read #2. The length of the poly(A) tail for that amplicon was estimated using the corresponding read #1 and defined as the maximal i for which >90% of the bases in the first i bases of read #1 were T's. This criterion allowed for some sequencing errors expected when sequencing long homopolymers.
Data access
Sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE37453.
Acknowledgments
We thank Olivia Rissland and Noah Spies for comments on the manuscript, the Whitehead Institute Genome Technology Core for sequencing, Wendy Johnston for technical support, and the Wellcome Trust Sanger Institute for the availability of unpublished RNA-seq data. This work was supported by a grant (GM067031) from the NIH (D.P.B.), an EMBO long-term fellowship (I.U.), a Human Frontiers Science Program long-term fellowship (A.S.), and an NSF predoctoral fellowship (C.H.J.).
Footnotes
[Supplemental material is available for this article.]
References
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, et al. 2011. Zebrafish mRNA sequencing decipher novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res 21: 1328–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT 2005. RNA turnover: Unexpected consequences of being tailed. Curr Biol 15: R635–R638 [DOI] [PubMed] [Google Scholar]
- Andreassi C, Riccio A 2009. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol 19: 465–474 [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM 1993. dbEST–database for “expressed sequence tags.” Nat Genet 4: 332–333 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM 2005. Principles of microRNA-target recognition. PLoS Biol 3: e85 doi: 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JH, Tong L 2011. Mitochondrial poly(A) polymerase and polyadenylation. Biochim Biophys Acta 1819: 992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Miura RM, Tian B 2006. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics 22: 2320–2325 [DOI] [PubMed] [Google Scholar]
- Chiaromonte F, Miller W, Bouhassira EE 2003. Gene length and proximity to neighbors affect genome-wide expression levels. Genome Res 13: 2602–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318: 271–274 [DOI] [PubMed] [Google Scholar]
- de Moor CH, Meijer H, Lissenden S 2005. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol 16: 49–58 [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsikov AV, Graber JH, Brockman JM, Hampl A, Holbrook AE, Singh P, Eppig JJ, Solter D, Knowles BB 2006. Cracking the egg: Molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev 20: 2713–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Gupta T, Mullins MC 2010. Dissection of organs from the adult zebrafish. J Vis Exp 37 doi: 10.3791/1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B 2011. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc Natl Acad Sci 108: 15864–15869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TA 2006. Regulation of gene expression by alternative untranslated regions. Trends Genet 22: 119–122 [DOI] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4: e8419 doi: 10.1371/journal.pone.0008419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA 2009. Evolution of gene function and regulatory control after whole-genome duplication: Comparative analyses in vertebrates. Genome Res 19: 1404–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Knaut H, Steinbeisser H, Schwarz H, Nusslein-Volhard C 2002. An evolutionary conserved region in the vasa 3′UTR targets RNA translation to the germ cells in the zebrafish. Curr Biol 12: 454–466 [DOI] [PubMed] [Google Scholar]
- Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. 2006. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16: 460–471 [DOI] [PubMed] [Google Scholar]
- Lange H, Sement FM, Canaday J, Gagliardi D 2009. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci 14: 497–504 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Linhart C, Halperin Y, Shamir R 2008. Transcription factor and microRNA motif discovery: The Amadeus platform and a compendium of metazoan target sets. Genome Res 18: 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH 2007. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res 35: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB 2000. Timely translation during the mouse oocyte-to-embryo transition. Development 127: 3795–3803 [DOI] [PubMed] [Google Scholar]
- Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F 2006. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics 7: 176 doi: 10.1186/1471-2164-7-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD 1999. Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev 63: 446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J, Hutchison KW, Wigglesworth K, Eppig JJ, Graber JH 2009. Probe-level analysis of expression microarrays characterizes isoform-specific degradation during mouse oocyte maturation. PLoS ONE 4: e7479 doi: 10.1371/journal.pone.0007479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S 1994. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science 266: 1996–1999 [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF 2007. The maternal-zygotic transition: Death and birth of RNAs. Science 316: 406–407 [DOI] [PubMed] [Google Scholar]
- Semon M, Wolfe KH 2007. Rearrangement rate following the whole-genome duplication in teleosts. Mol Biol Evol 24: 860–867 [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG 2010. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep 11: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Tassan JP, Richter JD 1992. Translational control by poly(A) elongation during Xenopus development: Differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes Dev 6: 2580–2591 [DOI] [PubMed] [Google Scholar]
- Slater GS, Birney E 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 doi: 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G 2006. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res 34: 2966–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G 2007. Regulation of the oocyte-to-zygote transition. Science 316: 407–408 [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD 2009. The maternal-to-zygotic transition: A play in two acts. Development 136: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Talbot WS, Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, Kelly PD, Chu F, Postlethwait JH 2005. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res 15: 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli JD, Huarte J, Belin D, Gubler P, Vassalli A, O'Connell ML, Parton LA, Rickles RJ, Strickland S 1989. Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev 3: 2163–2171 [DOI] [PubMed] [Google Scholar]
- Wang H, Gong Z 1999. Characterization of two zebrafish cDNA clones encoding egg envelope proteins ZP2 and ZP3. Biochim Biophys Acta 1446: 156–160 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Norbury CJ, Proudfoot NJ 2006. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell 21: 437–443 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B 2005. Biased alternative polyadenylation in human tissues. Genome Biol 6: R100 doi: 10.1186/gb-2005-6-12-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Manley JL 1996. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol 16: 2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]