Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) (original) (raw)
Abstract
Transcription activator-like effector nucleases (TALENs) are an approach for directed gene disruption and have been proved to be effective in various animal models. Here, we report that TALENs can induce somatic mutations in Xenopus embryos with reliably high efficiency and that such mutations are heritable through germ-line transmission. We modified the Golden Gate method for TALEN assembly to make the product suitable for RNA transcription and microinjection into Xenopus embryos. Eight pairs of TALENs were constructed to target eight _Xenopu_s genes, and all resulted in indel mutations with high efficiencies of up to 95.7% at the targeted loci. Furthermore, mutations induced by TALENs were highly efficiently passed through the germ line to F1 frogs. Together with simple and reliable PCR-based approaches for detecting TALEN-induced mutations, our results indicate that TALENs are an effective tool for targeted gene editing/knockout in Xenopus.
Keywords: genome editing, heritable mutagenesis, mutagenesis detection, reverse genetics, genome engineering
Among current animal models, Xenopus laevis and Xenopus tropicalis are classical animal models widely used in the study of embryonic development. However, because of the lack of methodologies for homologous recombination and embryonic stem cell derivation, it is difficult to perform specific gene targeting in these two models, which has impeded their use in genetic studies. Recently, site-specific gene targeting with transcription activator-like effector nucleases (TALENs) has been successfully applied in several animal models including rat, zebrafish, and Caenorhabditis elegans (1–4). Similar to zinc finger nucleases (ZFNs) (5), TALENs are engineered DNA nucleases that consist of a custom-designed DNA-binding domain and a nonspecific nuclease domain derived from Fok I endonuclease. Binding of adjacent TALENs allows dimerization of the endonuclease domains, leading to double-strand breaks at the predetermined site (6). These double-strand DNA breaks are frequently repaired through nonhomologous end joining (NHEJ) (7, 8), resulting in deletion or insertion (indel) mutations. The DNA binding specificity of TALENs, as distinct from ZFNs, is based on the transcription activator-like effectors (TALEs) from Xanthomonas plant pathogens (9, 10). The TALE proteins consist of an N-terminal translocation domain, a nuclear localization signal, and various numbers of tandem 34-aa repeats that determine the DNA binding specificity. Each repeat in the tandem array is identical except for two variable amino acid residues at positions 12 and 13 called repeat variable di-residues (RVDs), through which each repeat independently determines the targeted base (11, 12). It is known that the RVDs NI, NG, HD, and NN preferentially recognize adenine (A), thymine (T), cytosine (C), and guanine (G)/adenine (A), respectively (13). With a given repeat combination, the TALE recognizes a specific target sequence predicted by this code. A pair of TALENs can then cleave double-strand DNA between the two targeting sites upon dimerization of the Fok I nuclease domain. Here, we evaluated whether this technology can be applied for gene targeting in Xenopus embryos. Because ZFNs have been successfully used to disrupt the noggin in X. tropicalis embryos (14), we used ZFNs as positive controls to evaluate the efficiency of TALENs in this system. We found that TALENs were highly efficient in targeted genes disruption, resulting in mostly short indel mutations in Xenopus embryos. Importantly, such TALEN-induced mutations were passed efficiently to the next generation through the germ line. Our study indicates that TALENs are robust tools for targeted gene disruption in Xenopus embryos.
Results
TALENs Effectively Induce Targeted Gene Disruption in Xenopus Embryos.
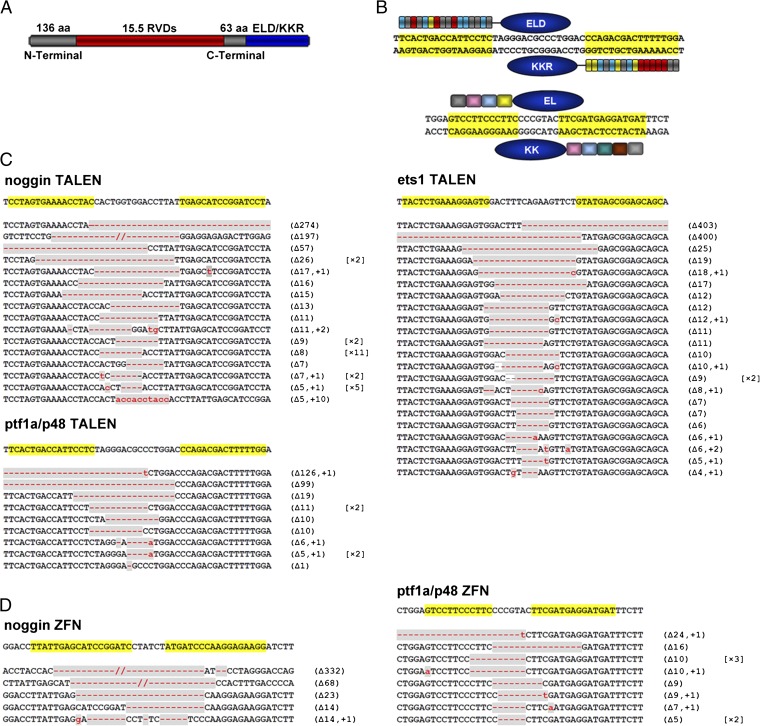
We generated TALEN constructs using the Golden Gate assembly method with the following RVD cipher codes: NI for A, NG for T, HD for C, and NN for G (13). The C-terminal TALE repeat was shortened to 63 aa (15). The TALE repeats targeting upstream and downstream effector binding elements (EBEs) were then subcloned into pCS2+ vector harboring the modified Fok I nuclease domains ELD and KKR (16), respectively (Fig. 1_A_ and Fig. S1). ELD and KKR have less off-target effects and higher efficiency because of compulsory heterodimer formation (16, 17). Thus, through dimerization of ELD and KKR, a pair of TALENs that binds to upstream and downstream EBEs of a given site is expected to cleave the DNA in the spacer region between the two EBEs (Fig. 1_B_).
Fig. 1.
Schematic drawing of TALENs and ZFNs and sequences of somatic mutations induced in X. tropicalis G0 embryos. (A) Schematic drawing of TALEN with 15.5 RVDs located between 136-aa N-terminal and 63-aa C-terminal regions. The ELD and KKR Fok I nuclease domains are linked to the C terminus of the TALE monomer. RVDs are shown in red and Fok I domains in blue. (B) Schematic drawing of TALENs (Upper) and ZFNs (Lower) that target ptf1a/p48. Recognition sequences are highlighted in yellow. (C) DNA sequences targeted by noggin-, ptf1a/p48-, or ets1-TALENs, and somatic mutations induced in Xenopus embryos. Sequenced mutations are listed. The largest forward deletion (400 bp) and the largest backward deletion (403 bp) were found in ets1 of X. tropicalis. (D) noggin- and ptf1a/p48- ZFNs targeting sites and somatic mutations induced by these pairs of ZFNs. In C and D, mutated regions are marked in gray, with red dashes indicating deletions (Δ) and lowercase letters in red indicating insertions (+). The numbers in parentheses show the number of deleted or inserted base pairs, whereas numbers in square brackets show the frequencies of the mutation in the sequenced samples.
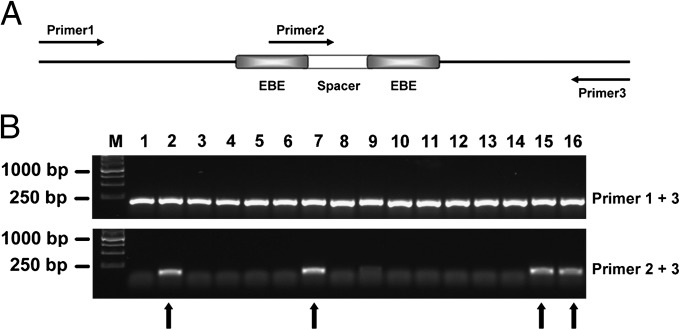
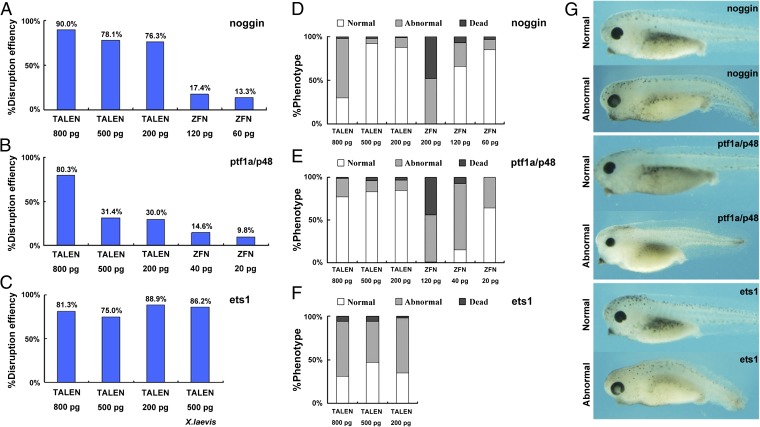
First, we targeted three genes of X. tropicalis, noggin, ptf1a/p48, and ets1 (Fig. 1). noggin (14) and ptf1a/p48 were also targeted by the corresponding ZFNs for a side-by-side comparison with TALENs. The TALEN EBE sequences of noggin and ptf1a/p48 were located in the adjacent regions to the corresponding ZFN targeting sequences (Fig. S2). The target site in the third exon of X. tropicalis ets1 is identical to the corresponding site in X. laevis (Fig. S2), and therefore we evaluated the targeting efficiency of ets1-TALENs in both species. The mix containing a pair of TALEN or ZFN mRNAs was injected into one-cell stage embryos of X. tropicalis or X. laevis. Forty-eight hours after injection, we randomly pooled five embryos injected with each pair of TALENs/ZFNs, extracted genomic DNA, amplified the targeted region, subcloned the fragments, and examined multiple colonies by PCR. As illustrated in Fig. 2_A_, primers 1 and 3 bridge both EBE regions, whereas primers 2 and 3 link the spacer region and the downstream EBE. PCR products of primers 1 and 3 were transferred into the vector pMD18-T (Takara) by TA cloning, and single colonies were examined by PCR using both 1/3 and 2/3 primer pairs. If primer pair 1/3 generates a PCR fragment, whereas primer pair 2/3 fails to do so, it suggests that the targeted gene is disrupted by the TALENs (Fig. 2_B_). Putative disrupted colonies were checked by DNA sequencing, and the targeting efficiency was determined as the ratio of mutant to total colonies. All tested TALENs and ZFNs were effective at disrupting the targeted genes in Xenopus embryos (Fig. 1 C and D). We found that the targeting efficiency of all pairs of TALENs was profound with the highest TALEN-induced mutation ratio of 90.0% (18/20) for noggin when 800 pg of mRNA (400 pg of left and 400 pg of right monomer mRNA) was injected (Figs. 1_C_ and 3_A_). Similarly, the targeting efficiency for ptf1a/p48 was about 80.3% (37/46) (Figs. 1C and 3_B_). By contrast, at the same injection dose, the corresponding ZFNs caused higher levels of dead and deformed embryos, whereas at a lower dose the targeting efficiency was lower (Figs. 1_D_ and 3 A and B). The targeting efficiency of ets1_-_TALENs in X. laevis injected with 500 pg of mRNA was similar to that in X. tropicalis (Figs. 1_C_ and 3_C_). Notably, a 403-bp fragment was deleted in ets1 of X. tropicalis, which was the largest deletion induced by TALENs in this study (Fig. 1_C_).
Fig. 2.
Mutagenesis detection by PCR. (A) Schematic drawing of primer pairs used for detecting mutagenesis by PCR (Materials and Methods). (B) A representative DNA gel displaying results of colony PCR with the two primer pairs to detect mutagenesis in X. tropicalis embryos injected with ets1-TALENs. In the upper panel, the ∼200-bp fragments amplified with primer pair 1, 3, indicates the presence of the targeted sequence in the plasmids. In the lower panel, the absence of PCR products for primer pair 2, 3, indicates the presence of targeted mutations, whereas a ∼140-bp fragment is amplified from wild-type sequences (indicated by arrows).
Fig. 3.
Frequencies of mutations and abnormal embryos induced by the indicated TALENs or ZFNs in Xenopus. Mutation frequencies were assayed as shown in Fig. 2. Only the mutation ratios induced by lower doses of ZFNs are shown because higher doses (200, 500, and 800 pg) resulted in dead or malformed embryos. (A–C) Frequencies of targeted mutagenesis induced by TALENs or ZFNs for the genes and at the doses indicated in the panels. All data refer to X. tropicalis except in C where X. laevis results are also shown. (D–F) Percentage of normal, abnormal, and dead X. tropicalis embryos injected with the indicated doses of TALEN or ZFN mRNAs. The injected embryos were inspected at 48 h postfertilization (about stage 41). (G) Overall morphology of X. tropicalis embryos injected with TALEN mRNAs directed against the indicated gene at stage 41. Curled axis, repression of head structures including eyes, and loss of pigments were observed. Such abnormal tadpoles usually could not complete metamorphosis.
We next designed TALENs to specifically disrupt the hhex, vpp1, foxd3, sox9, and grp78/bip genes (Figs. S3 and S4). These TALENs were also highly efficient in the generation of somatic mutations at the targeted loci with 82.6% (19/23) for hhex, 87.0% (20/23) for vpp1, 95.7% (22/23) for foxd3, 85.0% (17/20) for sox9, and 61.9% (13/21) for grp78/bip, respectively (Fig. S3). Collectively, these data indicate that TALENs are powerful and effective tools for conducting mutagenesis in both X. tropicalis and X. laevis. The high frequencies observed suggested that both alleles were disrupted in many cells of the injected embryos.
High-dose injection of TALEN mRNA may cause nonspecific defects in Xenopus embryos. As shown in Fig. 3 D–F, high-dose injections of TALENs led to abnormal morphology in a portion of the embryos, which could be reduced by decreasing the injection dose. Abnormalities apparently due to the toxicity of TALENs include curled axis, repression of head structures including eyes, and loss of pigment (Fig. 3_G_). Such abnormal tadpoles usually could not complete metamorphosis.
Specificity is essential for any gene-editing approach. To examine whether the TALENs have off-target effects, we used e-PCR (www.ncbi.nlm.nih.gov/sutils/e-pcr) to scan the X. tropicalis genomic sequence to identify potential off-target sites potentially targeted by noggin, ets1, and ptf1a/p48 TALENs (Fig. S5). The criteria for determining off-target sites were that up to six nonidentical bases occur in the two EBEs, 2-bp gaps in the two EBEs, and the spacer between the two putative EBE regions is <100 bp because it was suggested that longer spacers interfere with Fok I dimerization (15). PCR was used to amplify the identified potential off-target regions using genomic DNA from TALEN-injected embryos as template; no mutations were found at these sites by DNA sequencing. These data suggest that TALENs have high specificity for their target sequences.
Phenotypes of Somatic Mutations Induced by TALENs in X. tropicalis.
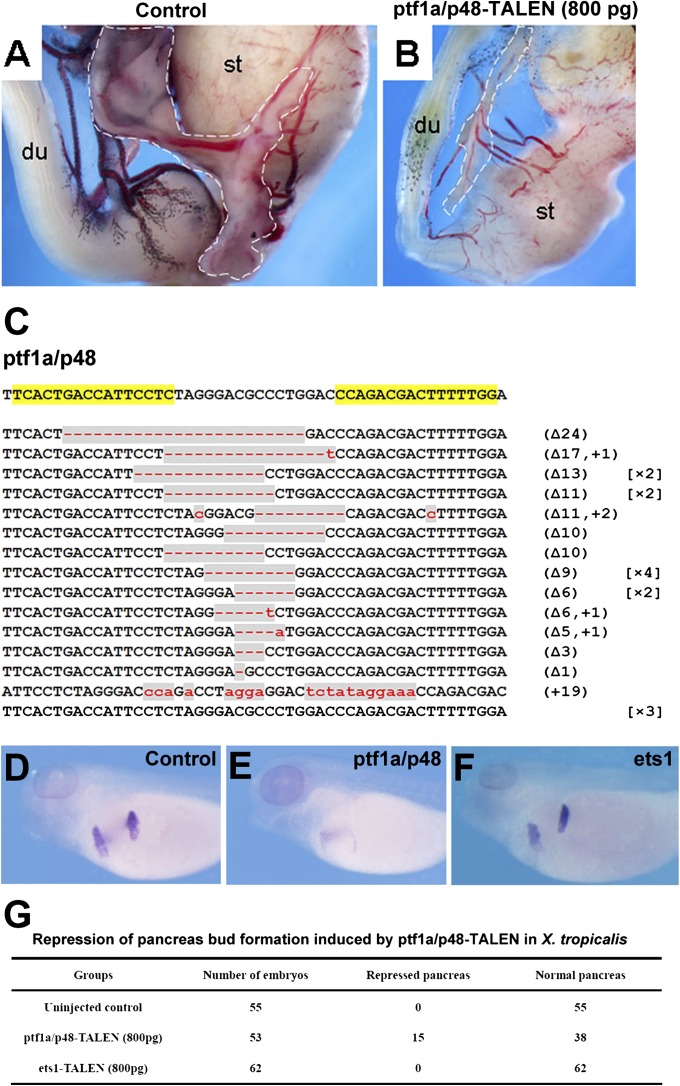
We followed growth and development of X. tropicalis embryos injected with TALENs to monitor functional consequences of the gene disruption. The ptf1a/p48-TALEN targeted froglets (800-pg injection dose) showed smaller body size compared with wild-type froglets. Twelve froglets were dissected for anatomical analysis, and all of them showed agenesis of the pancreas (Fig. 4 A and B). The observed phenotypes of pancreas are reminiscent of those seen in _ptf1a/p48_−/− mice (18). PCR analysis and further DNA sequencing of genomic DNA extracted from hindlimb tissues of a G0 X. tropicalis froglet showing the described phenotypes confirmed the TALEN-induced mutations at ptf1a/p48 loci (Fig. 4_C_). It also indicated a high ratio of ptf1a/p48 disruption (87.0%; 20/23) in somatic cells, suggesting that both alleles of ptf1a/p48 were disrupted in this frog. Furthermore, multiple alleles were observed, indicating that this G0 founder animal was highly mosaic for the disrupted locus. The high ratio of gene disruption induced by TALENs could provide an immediate phenotype assessment for gene-specific knockout that is usually time consuming. In line with our observation of pancreas agenesis in adult ptf1a/p48-TALEN-injected G0 frogs, the expression of pdip, an early marker gene specific to dorsal and ventral pancreatic buds (19), was reduced in a portion of ptf1a/p48-TALEN-injected embryos compared with those in control uninjected or ets1-TALEN-injected embryos (Fig. 4 D–G), further suggesting the specificity of pancreatic phenotype induced by direct injection of ptf1a/p48 TALENs.
Fig. 4.
Phenotypes of ptf1a/p48-TALEN targeted X. tropicalis. (A and B) Anatomical analysis revealed visceral abnormalities such as much reduced pancreas in ptf1a/p48-TALEN-injected G0 froglets. The pancreas is outlined by dashed lines. (C) DNA sequencing of genomic DNA extracted from hindlimb tissue dissected from a froglet showing a phenotype similar to that in B confirmed the gene disruption at the ptf1a/p48 locus (20/23) (du, duodenum; st, stomach). (D–F) Whole-mount in situ hybridization of pancreas marker pdip in X. tropicalis tadpoles injected with the indicated TALEN mRNAs. ptf1a/p48-TALENs, but not ets1-TALENs, induced repression of pancreas bud formation. (D) Uninjected control embryos, (E) ptf1a/p48-TALEN-injected embryos (800 pg), and (F) ets1-TALEN-injected embryos (800 pg). (G) Summary of the phenotypes shown in D–F. The TALEN mRNAs were injected into the animal pole region at the one-cell stage; embryos were analyzed at stage 40.
Mutations Induced by TALENs Are Heritable in X. tropicalis.
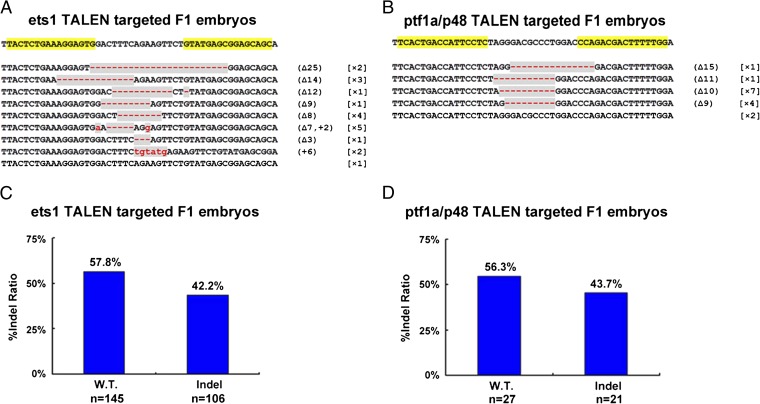
Successful germ-line transmission is essential for establishment of knockout lines. The ets1- and ptf1a/p48-TALEN-targeted G0 frogs were mated with wild-type frogs to examine germ-line transmission. Twenty embryos for ets1 from three independent crosses and 15 embryos for ptf1a/p48 from two independent crosses were collected, and genomic DNA was extracted from each individual embryo to assess mutagenesis at the TALEN-targeted site by the PCR and subcloning-sequencing assays described above. Nineteen of 20 or 13 of 15 F1 embryos carried TALEN-induced mutations in the ets1 or ptf1a/p48 gene (Fig. 5 A and B and Fig. S6). This high proportion indicates that a majority of gametes in the G0 frogs was mutant. As can be predicted for viable F1 offspring, the gene-disrupted embryos were heterozygous as revealed by PCR (ets1-TALEN targeted F1 embryos; Fig. S6), or by DNA sequencing (ptf1a/p48_-_TALEN-targeted F1 embryos). These results indicate that TALEN-induced gene disruption in Xenopus is heritable.
Fig. 5.
Sequence and frequency of mutant alleles inherited in F1 embryos. (A and B) DNA sequencing confirmed germ-line transmission of ets1 and ptf1a/p48 mutants induced by the TALENs. Disrupted sequences are shaded in gray. Red dashes indicate deletions (Δ). The numbers of deleted base pairs are shown in parentheses, and the number of embryos showing the particular mutation is given in square brackets. (C and D) The percentage of wild-type and the TALEN-disrupted alleles in F1 embryos derived from ets1- or ptf1a/p48-TALEN-targeted frogs, determined either by colony PCR (ets1-TALEN-targeted F1 embryos) or DNA sequencing (ptf1a/p48-TALEN-targeted F1 embryos). Twenty F1 embryos for ets1 from three crosses and 15 F1 embryos for ptf1a/p48 from two crosses of G0 by wild type were examined. The n in C and D indicates the number of bacterial clones that were examined after TA cloning.
Discussion
The X. laevis and X. tropicalis are important model organisms for studies in cell and developmental biology. However, the lack of methodologies for targeted mutagenesis has impeded their application to studies on the genetic control of development. In this study, we have established modified procedures for generating targeted mutations in Xenopus embryos using TALENs and found that such mutations are heritable.
Based on the Golden Gate method (13), we assembled TALEN arrays to target selected gene sequences, and used heterodimeric ELD/KKR Fok I variants for gene editing. ELD/KKR are derived from the Fok I nuclease domain and are reported to have higher specificity (16) and less toxicity (17) than Fok I homodimers. Although it was reported previously that these modified Fok I domains exhibited decreased cleavage efficiency in human cells, recent studies indicated that ELD/KKR showed higher activities than wild-type Fok I in zebrafish embryos, and that TALENs containing EL/KK or ELK/KKR induced a similar rate of deformed and dead embryos in zebrafish (17). The ZFN pairs we tested contained EL and KK, respectively. High-dose injection of these ZFNs mRNA led to high proportions of abnormal and dead embryos, whereas low doses of ZFN mRNAs had reduced efficiency of targeted gene disruption. In contrast, Xenopus embryos seemed tolerant to quite high doses of TALEN mRNAs (Fig. 3).
A number of studies suggested that T at site 0 is significant for target site recognition and binding by TALENs (11, 12, 20), and that a 14- to 16-bp spacer is optimal for DNA cleavage (15). The RVDs of HD, NI, and NG have a high preference for C, A, and T, respectively, but NN can recognize both G and A (11, 21). We therefore chose EBEs with fewer G residues to avoid the potential off-target effects caused by this TALE ambiguity. We routinely chose TALEN targeting sequences by the following criteria: (i) nucleotide T is at position 0 and the EBE sequence follows this T; (ii) the length of both EBE sequences is 16 bp, and the spacer sequence is around 16 bp; (iii) minimize G residues in EBE sequences; (iv) select EBEs in an exon. Following these rules, we designed eight pairs of TALENs to target eight genes, and all showed high efficiencies of up to 95.7% in generating somatic mutations at the targeted sites in Xenopus embryos. Thus, our results strongly suggest that TALENs can target most of loci in the Xenopus genome.
X. tropicalis is a diploid, containing two alleles of each gene. Using our PCR detection method, we should be able to amplify the TALEN-targeted regions from both alleles of the examined tropicalis embryos. After TA cloning of the PCR products, we found 80–90% inserts bearing mutations (Fig. 3 A–C) when 800 pg of TALEN mRNAs were injected, suggesting two alleles in most of cells were disrupted. Such high mutagenesis rates make some applications of TALENs possible. Thus, TALENs might be used for gene knockdown as achieved by morpholino. We could also regionally disrupt one gene by microinjecting TALEN mRNAs into a particular blastomere of developing Xenopus embryos.
Moreover, the germ-line transmission rate of TALEN-induced mutations was efficient at the two examined loci, ets1 and ptf1a/p48, indicating that TALEN-induced mutations are heritable in Xenopus. Taken together, these results indicate that these procedures allow simple, robust, and efficient generation of targeted mutations in Xenopus.
Although the PCR-based method for detecting mutagenesis is reliable, it may underestimate the mutation rate. For example, if the indel mutations were outside the primer 3 binding site, the PCR would miss such mutations. An additional primer 4 that overlaps the joint region of the downstream EBE site will help overcome this shortcoming. Two sets of PCRs with primers 2 and 3, and primers 1 and 4 might be able to detect all indel mutations. Because the mutation rates induced by our TALEN system are generally high, direct sequencing may also be feasible after TA cloning.
A number of studies indicate that TALENs are highly specific for targeted gene editing (3, 21). In this study, using e-PCR to perform BLAST searches, we identified potential off-target sites, but no mutation was found at these sites by DNA sequencing. However, a more detailed investigation on the possible off-target effects of TALEN will need to be addressed in the future.
Although the TALEN-targeted G0 embryos were mosaic (Fig. 1), the high frequency of somatic mutation induced by our TALEN system resulted in obvious phenotypes in G0 founders. We observed agenesis of the pancreas in ptf1a/p48-TALEN targeted G0 frogs, which was similar to that of ptf1a/p48 knockout mice. It would be interesting to compare phenotypes of G0 embryos with those induced by morpholinos. However, the high mutation load may interfere with derivation of a mutant line, which could be avoided by injection of TALENs at lower doses. In addition, studies using TALENs to mediate homologous recombination in Xenopus genome are also worth exploring.
We expected the high mosaicism in G0 adult frogs, which was confirmed by Fig. 4. TALENs can disrupt both alleles of a gene and generate two different mutant alleles after NHEJ in somatic cells. Because indel mutations usually are found in the spacer region, the TALEN pair could bind to their EBE sites again even after NHEJ-mediated DNA repair, inducing additional mutations as cells proliferate during development. Therefore, it is possible to observe a large diversity of mutants at the targeted loci of G0 frogs. At present, it is not clear whether the mutation frequencies in the soma and germ line are similar. However, it is clear that the G0 germ line of TALEN-targeted frogs was mosaic in our experiments, as seen from the fact that the progeny of a single G0 frog yielded several different mutations at the same locus (Fig. 5 A and B). To balance mutagenic efficiency with survival of offspring, we believe that TALEN injection doses may need to be varied and possibly reduced from those we used when aiming to generate mutant strains.
Because of fecundity, large size of embryos, suitability for embryo manipulation, and fast embryonic development, X. tropicalis is a good research model for vertebrate development and for the study of disease. Our results demonstrate that TALENs are robust tools for genetic engineering in X. tropicalis. Application of this technology will help pave the way for more comprehensive genetic studies using this excellent research organism.
Materials and Methods
Construction of TALENS.
TALEs targeting endogenous genes are constructed through Golden Gate TALEN Assembly (13). ELD/KKR (22) derived from Fok I nuclease domains were used for constructing TALENs. The plasmids for TALEN assembly were obtained from Addgene. The DNA fragment encoding N-terminal to C-terminal in pTAL1 vector was transferred into pCS2+KKR and pCS2+ELD, and these two vectors became feasible to TALEN assembly (Fig. S1). The plasmids containing ptf1a/p48-ZFN coding sequences were purchased from Sigma, and the DNA encoding noggin-ZFNs were synthesized according to Yong et al. (14) and then subcloned into pCS2+KK and pCS2+EL.
Manipulation of Xenopus Embryos.
X. tropicalis and X. laevis were treated with human chorionic gonadotropin as described (23, 24). The TALEN and ZFN plasmids were linearized by NotI, and mRNAs were synthesized by the mMessage mMachine kit (Ambion). TALENs and ZFNs mRNAs were microinjected into Xenopus embryos at the one-cell stage.
Detection of Somatic Mutations in TALEN-Targeted Xenopus Embryos.
Forty-eight hours after microinjection, TALEN or ZFN-targeted embryos were pooled for genomic DNA extraction (five embryos for each pool). PCR was performed using primers 1 and 3 (Fig. 2 and Table S1). Amplicons harboring targeted gene fragments were subcloned into pMD-18T by TA cloning (Takara). Colony PCR was performed to examine the occurrence of mutations at the targeted sites and to determine mutagenesis rate, using primer pair 1, 3, and primer pair 2, 3, respectively (Fig. 2 and Table S1). If mutations are generated in the spacer region, no amplicon can be detected using the primer pair 2, 3. PCR-positive plasmids were verified by DNA sequencing.
Detection of Heritable Mutations.
TALEN-injected X. tropicalis embryos were raised to sexual maturity. Male TALEN-targeted G0 frogs were crossed with wild-type females, and F1 embryos were collected at 48 h postfertilization. Genomic DNA was extracted from each embryo individually to assess mutagenesis at the TALEN-targeted site by PCR-based assays described above for detecting somatic mutations. The individual positive clones were confirmed by DNA sequencing. For F1 embryos derived from ptf1a/P48-TALEN-injected G0 frogs, ptf1a/P48 mutations were detected by direct DNA sequencing after the TA cloning of the DNA fragments.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dr. Thomas Cermak, Dr. Daniel Voytas, and Addgene for making the Golden Gate TALEN constructs accessible to the research community. This work was supported by grants from the Research Grants Council of Hong Kong (CUHK480709) (to H.Z.); and in part by funds from the National Basic Research Program of China (2009CB941202) and funds from the Key Project of Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-R-083) (to Y. Chen), the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health (I.B.D.), and the National Natural Science Foundation of China (31172394) (to C.H.K.C.). Y. Lei is supported by the Graduate Studentships from The Chinese University of Hong Kong.
Footnotes
The authors declare no conflict of interest.
3Plasmid requests should be addressed to C.H.K.C. at chkcheng@cuhk.edu.hk.
References
- 1.Huang P, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 2.Tesson L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 3.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sander JD, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mani M, Smith J, Kandavelou K, Berg JM, Chandrasegaran S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun. 2005;334:1191–1197. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 11.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 12.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 13.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young JJ, et al. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2011;108:7052–7057. doi: 10.1073/pnas.1102030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 16.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 17.Cade L, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40(16):8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krapp A, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afelik S, Chen Y, Pieler T. Pancreatic protein disulfide isomerase (XPDIp) is an early marker for the exocrine lineage of the developing pancreas in Xenopus laevis embryos. Gene Expr Patterns. 2004;4:71–76. doi: 10.1016/s1567-133x(03)00150-9. [DOI] [PubMed] [Google Scholar]
- 20.Mahfouz MM, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholze H, Boch J. TAL effector-DNA specificity. Virulence. 2010;1:428–432. doi: 10.4161/viru.1.5.12863. [DOI] [PubMed] [Google Scholar]
- 22.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Han D, Dawid IB, Pieler T, Chen Y. Homeoprotein hhex-induced conversion of intestinal to ventral pancreatic precursors results in the formation of giant pancreata in Xenopus embryos. Proc Natl Acad Sci USA. 2012;109:8594–8599. doi: 10.1073/pnas.1206547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, Tanegashima K, Ro H, Dawid IB. Lrig3 regulates neural crest formation in Xenopus by modulating Fgf and Wnt signaling pathways. Development. 2008;135:1283–1293. doi: 10.1242/dev.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information