Conserved Bacterial RNase YbeY Plays Key Roles in 70S Ribosome Quality Control and 16S rRNA Maturation (original) (raw)
. Author manuscript; available in PMC: 2014 Feb 7.
Abstract
Quality control of ribosomes is critical for cellular function since protein mistranslation leads to severe physiological consequences. We report the first evidence of a ribosome quality control system in bacteria that operates at the level of 70S to remove defective ribosomes. YbeY, a previously unidentified endoribonuclease, and the exonuclease RNase R act together by a process mediated specifically by the 30S ribosomal subunit, to degrade defective 70S ribosomes but not properly matured 70S ribosomes or individual subunits. Furthermore, there is essentially no fully matured 16S rRNA in a Δ_ybeY_ mutant at 45°C, making YbeY the first endoribonuclease to be implicated in the critically important processing of the 16S rRNA 3' terminus. These key roles in ribosome quality control and maturation indicate why YbeY is a member of the minimal bacterial gene set and suggest that it could be a potential target for antibacterial drugs.
Keywords: ribosome quality control, rRNA maturation, RNase
INTRODUCTION
Given the intricacies of rRNA and ribosome biogenesis, quality control mechanisms are critically required to eliminate defective ribosomes and thus ensure proper protein translation. Studies to date have led to the conclusion that ribosome quality control in bacteria acts mainly at the level of the unassembled 30S and 50S subunits (Basturea et al., 2011; Deutscher, 2009). Although a late-stage ribosome quality control system [non-functional rRNA decay (NRD)] that can act at the level of fully assembled ribosomes has been characterized in eukaryotes (Cole et al., 2009; LaRiviere et al., 2006), a similar system by which bacteria can specifically eliminate defective 70S ribosomes has not been reported (LaRiviere et al., 2006). In this paper, we show that a highly conserved, previously unidentified RNase, YbeY, plays critical roles in a hitherto undescribed mechanism of late-stage 70S ribosome quality control in bacteria.
YbeY (UPF0054 protein family) is found in nearly every sequenced bacterium (Davies and Walker, 2008; Sonnhammer et al., 1997). Also, ybeY is one of the 206 genes postulated to comprise the minimal bacterial genome set (Gil et al., 2004). ybeY is essential in some bacteria (Akerley et al., 2002; Kobayashi et al., 2003), whereas in others (e.g. Escherichia coli and Sinorhizobium meliloti), ybeY is not essential but its loss sensitizes cells to a wide variety of physiologically diverse stresses, including heat (Davies et al., 2010; Davies and Walker, 2008; Rasouly et al., 2009). Structural studies of E. coli YbeY and its orthologs have led to the suggestion that YbeY is a metal-dependent hydrolase. Despite these structural insights and extensive screening, the biochemical activity of YbeY and its orthologs has remained elusive (Oganesyan et al., 2003; Penhoat et al., 2005; Zhan et al., 2005).
Our recent studies of the physiological roles of YbeY led us to consider the possibility that it might be an RNase, rather than a protease as has been previously suggested (Oganesyan et al., 2003; Penhoat et al., 2005; Zhan et al., 2005). We recently found that YbeY is involved in the processing of all three rRNAs (Davies et al., 2010). Furthermore, ybeY shows strong genetic interactions with rnc, rnr and pnp, whose products RNase III, RNase R and PNPase play important roles in both rRNA maturation and RNA degradation (Bollenbach et al., 2005; Davies et al., 2010; Deutscher, 2009; Purusharth et al., 2007; Walter et al., 2002). The almost complete lack of properly matured 16S rRNA 3' termini in the Δ_ybeY_ Δ_rnr_ and Δ_ybeY_ Δ_pnp_ mutants (Davies et al., 2010) was of particular interest because no RNase has yet been implicated in this critically important processing step (Deutscher, 2003). Additional observations that tie YbeY to RNA metabolism include our finding that the S. meliloti YbeY ortholog SMc01113 affects the regulation of some sRNAs and their mRNA targets, and the structural similarities that bacterial YbeYs share with the MID domain of the eukaryotic Argonaute protein (Pandey et al., 2011).
Here, we show that YbeY is a single strand specific endoribonuclease that plays key roles in two crucial physiological functions, a hitherto unrecognized late-stage 70S ribosome quality control system that is particularly important under stress, and in processing of the 16S rRNA 3' terminus. These critical roles of YbeY account for its presence in most bacterial genomes and its inclusion in the minimal bacterial gene set.
RESULTS
YbeY is a metal-dependent ribonuclease
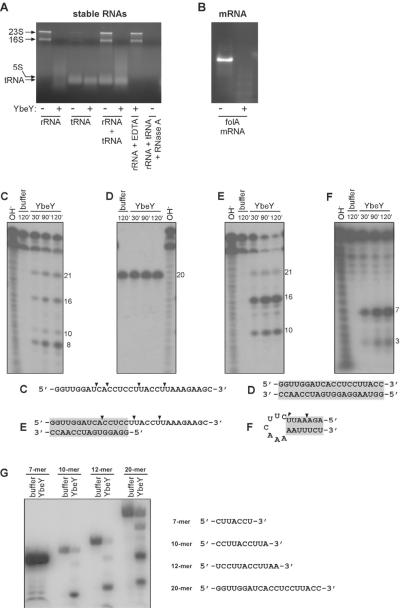
YbeY belongs to the UPF0054 family characterized by a three histidine H3XH5XH motif that coordinates a metal ion thought to be zinc. We report that purified YbeY is an RNase that degrades total rRNA and mRNA effectively (Figure 1A and 1B). YbeY is substantially less effective at degrading total tRNA in vitro (Figure 1A). For example, when a mixture of rRNA and tRNA was used as a substrate, rRNA was degraded but tRNA was not (Figure 1A). However, partial degradation of tRNA was observed when YbeY was approximately in 2-fold excess compared to the substrate (Figure S1A). RNase A, used at the same concentration as YbeY, efficiently degraded both rRNA and tRNA (Figure 1A). As expected for an RNase, YbeY was unable to degrade either double or single stranded DNA (Figure S1B and S1C). Consistent with its predicted metal-dependent hydrolase activity, the RNase activity of YbeY was inhibited by 50 mM EDTA (Figure 1A).
Figure 1. YbeY is a metal-dependent single strand specific endoribonuclease.
Purified YbeY was used at a concentration of 5 μM in all assays unless mentioned otherwise. (A) YbeY is able to degrade total rRNA (3.8 μM) isolated from E. coli. tRNA (4.5 μM) is a relatively poor substrate for YbeY. When rRNA (3.8 μM) and tRNA (4.5 μM) were mixed, YbeY preferentially degraded the rRNA. YbeY is inhibited by 50 mM EDTA. RNase A (5 μM) was used as control to show degradation of the mixture of rRNA (3.8 μM) and tRNA (4.5 μM). The positions of the 23S, 16S, 5S rRNA and tRNA are indicated. (B) YbeY cleaves folA mRNA (4.0 μM) generated by in vitro transcription. Digestion products were analyzed by Synergel/agarose gel electrophoresis. (C)–(G) In vitro cleavage assay to identify the substrate requirement of YbeY using short synthetic oligoribonucleotides. Assays were performed using YbeY (5.0 μM) and 5'-32P-labeled oligoribonucleotides (5.0 μM); (C) ssRNA, (D) dsRNA, (E) dsRNA containing a single stranded extension at the 3', (F) hairpin substrate with perfectly base paired blunt ends, (G) ssRNAs 7, 10, 12, 20 and 30 nt long. Digestion products were analyzed by polyacrylamide gel electrophoresis. OH−, alkali ladder. ▼ indicates sites of cleavage by YbeY on the substrate (see also Figure S1 and S2).
YbeY is a single strand specific endoribonuclease
To further characterize the RNase activity of YbeY, we used a synthetic 30 nucleotide (nt) RNA substrate that mimics the 3' terminus of 16S rRNA in its unprocessed form. It contains 18 nt of the mature 3' terminus of 16S rRNA and 12 nt of the 3' terminal precursor sequence. YbeY can bind to this 30 nt single stranded RNA (ssRNA) in a concentration-dependent manner in a gel shift assay (Figure S1D), albeit weakly. We attribute this weak binding due the assay being carried out at 4°C and not 37°C at which YbeY efficiently degrades RNA. To elucidate the substrate requirement of YbeY, equimolar amounts of protein and synthetic oligonucleotide substrates was used. YbeY cleaved the 30 nt ssRNA substrate, producing a distinct pattern that indicates a preference for cleavage after U (Figure 1C). However, YbeY was unable to degrade double stranded RNA (dsRNA) (Figure 1D). We also examined YbeY's ability to degrade a partially dsRNA substrate containing a ssRNA extension at the 3' end. YbeY cleaved the ssRNA 3' extension with a major cut after a U +1 from the beginning of the duplex. It also cleaved weakly inside the double stranded portion of the RNA substrate, even though it could not cleave dsRNA without a ssRNA 3' extension (Figure 1E). We attribute this weak cleavage to transient melting of the dsRNA allowing bound YbeY access to this portion of the RNA substrate. YbeY cleaved a hairpin substrate with perfectly paired blunt ends after a U at the junction between the stem and the loop (Figure 1F) and also weakly in the stem region, again possibly due to transient melting of the dsRNA stem. The ability of YbeY to cleave hairpin structures explains its capacity to degrade free total rRNA, in which hairpins are the most abundant secondary structural element. Furthermore, YbeY cannot degrade a synthetic RNA substrate in which all 2'-hydroxyl groups were substituted with 2'-O-methyl groups (Figure S2A).
To investigate the nature of the termini generated by YbeY's endoribonuclease activity, we treated an RNA substrate blocked at both ends with YbeY. We found that we could label the cleavage products with T4 polynucleotide kinase (PNK) and γ-32P-ATP, but not with T4 RNA ligase and 32P-pCp (Figure S2B), an observation indicating that YbeY cleavage generates products containing 5' hydroxyl and 3' phosphate termini. Furthermore, the treatment of YbeY-digested 5'-32P- labeled RNA with T4 PNK in the absence of ATP to promote removal of the 3' phosphate (Cameron and Uhlenbeck, 1977), resulted in a shift in mobility of the RNA products (Figure S2B), supporting the conclusion that YbeY cleaves RNA to generate a 3' phosphate.
YbeY cleaved ssRNA substrates 10 nt or longer but did not cleave a 7 nt substrate despite the presence of a site that is cleaved in the context of larger oligoribonucleotides (Figure 1G). The length requirement of YbeY is different from that of a typical oligoribonuclease, which prefers 2–5 nt long substrates (Ghosh and Deutscher, 1999), and from a degradative RNase like RNase A that degrades oligoribonucleotides of varying lengths to mononucleotides (Arraiano et al., 2010; Condon and Putzer, 2002). Consistent with being a heat shock protein (Rasouly et al., 2009), YbeY has good RNase activity at 37°C and 45°C, although it decreases at 65°C (Figure S2C).
Conserved histidine H114 and arginine R59 are required for the RNase activity of YbeY
In previous work we had shown residues H114 and R59 to be important for the in vivo function of YbeY, specifically for survival under heat shock (Davies et al., 2010). H114 is part of the conserved histidine triad involved in metal binding, while R59 is part of a highly conserved cluster of amino acids on the other side of the cleft that have been suggested to be important for YbeY activity (Pandey et al., 2011; Zhan et al., 2005) (Figure 2A). Using site directed mutagenesis, we generated single mutants H114A, R59A and R59E as well as the multiple mutants H114A H124A and R59E K61E (Figure S3A).
Figure 2. RNase activity of YbeY requires the histidine H114 residues and conserved amino acid residue R59.
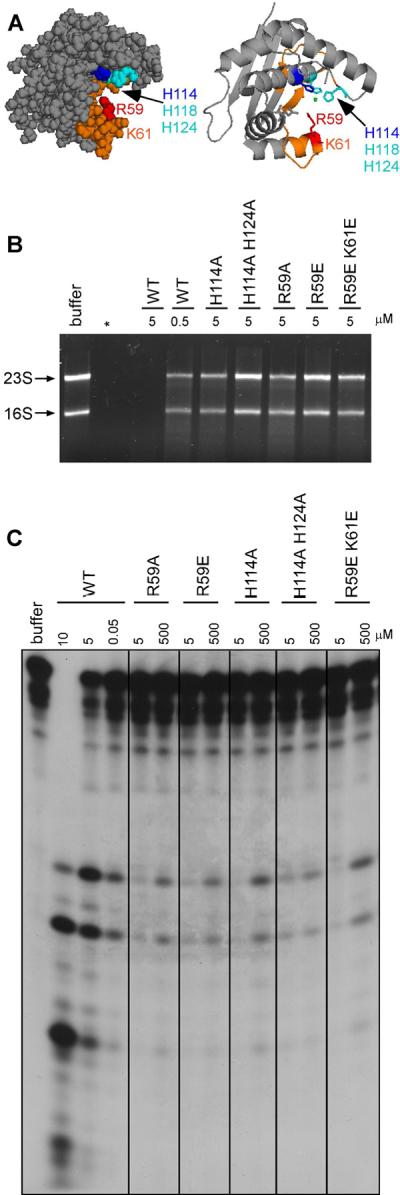
(A) Model of YbeY generated in PyMOL (http://www.pymol.org) using Swiss Prot Entry P77385 (Zhan et al., 2005), showing the positions of conserved residues R59, K61, H114, H118 and H124 in the proposed RNA channel (highlighted in orange). (B) – (C) RNase activities of YbeY wild type and mutant proteins (H114A; H114A H124A; R59A; R59E; R59E K61E) on (B) total rRNA (3.8 μM) isolated from E. coli and (C) a short 30 nt ssRNA substrate (5.0 μM) as shown in Figure 2A. Digestion products were analyzed by Synergel/agarose or polyacrylamide gel electrophoresis, respectively. RNase concentrations between 0.05 and 500 μM were used as indicated (see also Figure S3).
The mutant enzymes were purified to homogeneity and tested at a final concentration of 5 μM for RNase activity on total rRNA (Figure 2B) and the short 30 nt ssRNA substrate (Figure 2C). All mutant enzymes have extremely low RNase activity. To obtain a degree of RNA cleavage seen with 0.05 μM wild type YbeY, we required 10,000 fold more of any of the mutant enzymes (Figure 2C). Prolonged incubation of the substrate with 5 μM concentration of mutant enzymes also did not yield a degree of cleavage comparable to that with 100-fold less of the wild type YbeY (Figure S3B). Finally, the mutant proteins are inactive on dsRNA, dsRNA with a ssRNA 3′ extension, and hairpin substrates (Figure S3C).
The results clearly show that H114 and R59 are required for the RNase activity of YbeY. Since the single mutants H114A and R59E showed such a drastic decrease in RNase activity on their own, we were unable to detect any further decrease from additionally mutating H124 or K61. The triple histidine mutant (H114A H118A H124A) could not be purified because it was very poorly expressed.
YbeY together with RNase R is involved in the removal of defective non-translating and translating 70S ribosomes in vitro: a new mechanism of ribosome quality control in E. coli
Since the Δ_ybeY_ mutant accumulates 70S ribosomes containing substantial amounts of misprocessed RNA (Davies et al., 2010), the impaired translational efficiency and fidelity of these defective 70S ribosomes could account in part for the highly pleiotropic phenotype of the Δ_ybeY_ mutant. Furthermore, RNase R, with which YbeY shows a strong genetic interaction (Davies et al., 2010), has been implicated in rRNA degradation/RNA quality control as well as in rRNA maturation (Bollenbach et al., 2005; Deutscher, 2009; Purusharth et al., 2007). Together with our discovery that YbeY is an RNase, these in vivo results led us to consider the possibility that YbeY, either by itself or in concert with RNase R, participates in a ribosome quality control system.
To test this hypothesis, 30S ribosomal subunits and 70S ribosomes were purified from wild type E. coli and the Δ_ybeY_ mutant. The 30S subunits and 70S ribosomes from the wild type strain contain mainly mature 16S rRNA (Figure 3A). The 30S subunits isolated from the Δ_ybeY_ mutant contained considerably less mature 16S rRNA and, in addition, several faster migrating rRNA species (Figure 3B). Furthermore, the 70S ribosomes purified from the Δ_ybeY_ mutant showed the presence of the 17S rRNA precursor, low amounts of mature 16S rRNA, and a faster migrating rRNA species distinct from 16S rRNA (Davies et al., 2010) (Figure 3B).
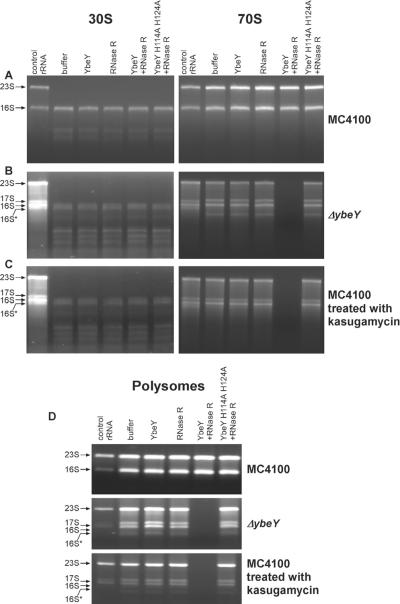
Figure 3. YbeY and RNase R together eliminate defective non-translating and translating ribosomes.
Polysomes, 70S ribosomes and 30S and 50S ribosomal subunits were isolated from wild type MC4100 and Δ_ybeY_, and wild type MC4100 treated with 200 μg/ml kasugamycin. Ribosomal subunits (4.3 μM), 70S ribosomes (5.5 μM) and polysomes (3.9 μM) were incubated with equimolar amounts (5 μM each) of (i) YbeY, (ii) RNase R, (iii) YbeY and RNase R, and (iv) YbeY H114A H124A and RNase R as indicated. rRNA was thereafter extracted from these samples and analyzed on a Synergel/agarose gel. (A) rRNA from 30S ribosomal subunits and 70S ribosomes from MC4100 after incubation with YbeY and/or RNase R. Total rRNA isolated from wild type cells was used as a marker. (B) rRNA from 30S ribosomal subunits and 70S ribosomes from the Δ_ybeY_ mutant strain after incubation with YbeY and/or RNase R. Total rRNA isolated from the Δ_ybeY_ mutant strain was used as a marker. (C) rRNA from 30S ribosomal subunits and 70S ribosomes from kasugamycin-treated MC4100 after incubation with YbeY and/or RNase R. Total rRNA isolated from kasugamycin-treated wild type cells was used as a marker. (D) rRNA from polysomes isolated from MC4100, the Δ_ybeY_ mutant strain and MC4100 treated with 200 μg/ml kasugamycin after incubation with YbeY and/or RNase R. Total RNA from wild type cells, the Δ_ybeY_ mutant strain and wild type treated with 200 μg/ml kasugamycin, respectively, was used as marker (see also Figure S4).
30S subunits and 70S ribosomes isolated from wild type and Δ_ybeY_ cells were incubated with (i) YbeY, (ii) RNase R, (iii) equimolar amounts of YbeY and RNase R, and (iv) equimolar amounts of YbeY H114A H124A and RNase R. RNA was isolated from these samples and the rRNA profile was analyzed. rRNA from 30S subunits and 70S ribosomes isolated from the wild type strain remained unaltered under all conditions (Figure 3A). Similarly, rRNA from 30S subunits isolated from the Δ_ybeY_ mutant remained unaffected under all conditions (Figure 3B).
A strikingly different result was obtained with the defective 70S ribosomes isolated from the Δ_ybeY_ mutant. Neither YbeY nor RNase R on its own affected the rRNA profile of these 70S ribosomes. However, when YbeY and RNase R were added together, the rRNA in these 70S ribosomes was completely degraded (Figure 3B). No degradation was seen when YbeY H114A H124A was substituted for YbeY, indicating that the catalytic activity of YbeY is required. Decreasing YbeY while keeping RNase R constant reduced the degradation of defective 70S ribosomes (Figure S4A). When normal (MC4100 wild type) and defective (Δ_ybeY_ mutant) 70S ribosomes were mixed together and co-incubated with YbeY and RNase R, only a portion of the rRNA, most likely derived from the defective ribosomes, was degraded (Figure S4B).
To ascertain whether this phenomenon is specific to ribosomes isolated from Δ_ybeY_ or is instead reflective of a general rRNA/ribosome quality control mechanism, we repeated the same set of experiments with ribosomes isolated from wild type E. coli cells treated with the aminoglycoside antibiotic kasugamycin, a drug to which the Δ_ybeY_ mutant is very sensitive (Figure S4C). Kasugamycin inhibits canonical translation by preventing the binding of initiator tRNA to the 30S ribosomal subunit and induces the formation of 61S ribosomes that lack several proteins of the small ribosomal subunit (Kaberdina et al., 2009). 30S subunits and 70S ribosomes isolated from wild type E. coli treated with 200 μg/ml of kasugamycin showed the presence of 17S and 16S rRNA. Similar to the Δ_ybeY_ mutant, several truncated species of rRNA were also present in the 30S subunits (Figure 3C). The 30S subunits and 70S ribosomes from the kasugamycin-treated cells were incubated with YbeY and RNase R and analyzed as described above. rRNA from 30S subunits remained unaffected under all conditions tested. The rRNA profile of 70S ribosomes remained unaltered in the presence of YbeY or RNase R, individually. However, when these two enzymes were added together, rRNA from 70S ribosomes from kasugamycin-treated cells was completely degraded (Figure 3C). The mutant YbeY enzyme lacking RNase activity had no effect on its own or when added together with RNase R, thus demonstrating that the RNase activity of YbeY is required in this case as well.
To determine whether YbeY and RNase R together can distinguish between misassembled non-translating 70S ribosomes and defective 70S ribosomes that are part of the translating pool, we carried out the above assay on polysomes isolated from MC4100, MC4100 Δ_ybeY_ and MC4100 treated with kasugamycin. The amount of ribosomes and polysomes isolated from kasugamycin-treated cells were approximately two fold lower than from untreated wild type cells. Similar to the results obtained with 70S ribosomes, YbeY and RNase R together efficiently degraded rRNA in polysomes from the Δ_ybeY_ mutant and MC4100 treated with kasugamycin, but did not degrade the rRNA in polysomes isolated from wild type cells (Figure 3D). These data indicate that YbeY and RNase R together not only recognize and eliminate non-translating 70S ribosomes; they can also eliminate defective 70S ribosomes that are part of translating polysomes.
In summary, our in vitro results suggest a general physiological role for YbeY together with RNase R in the specific removal of defective 70S ribosomes from the cellular pool, thereby allowing translation to proceed effectively under standard growth conditions and under stress. The striking sensitivity of ybeY mutants to a wide variety of harmful agents (Davies et al., 2010; Davies and Walker, 2008), including kasugamycin, is consistent with this YbeY-dependent system of 70S ribosome quality control being especially important under stressful conditions.
In vitro degradation of defective ribosomes by YbeY and RNase R requires the ribosomes to be in the assembled 70S state and is mediated by the defective 30S ribosomal subunit
We tested whether 50S and 30S subunits obtained by dissociating the defective 70S ribosomes from the Δ_ybeY_ mutant are substrates for YbeY and RNase R. As above, YbeY and RNase R degraded the rRNA of 70S ribosomes isolated from the Δ_ybeY_ mutant efficiently. However, the RNA profiles of 30S and 50S subunits derived from the very same pool of 70S ribosomes were not affected by the combination of these two RNases (Figure 4A). These observations provide strong additional support for our suggestion that YbeY and RNase R function together in a hitherto unrecognized quality control mechanism that acts at the level of assembled 70S ribosomes.
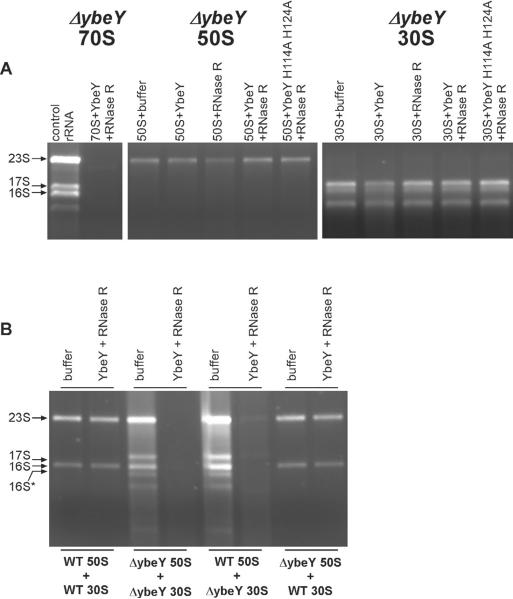
Figure 4. Ribosome quality control requires 70S ribosome complex formation and is mediated by defective 30S ribosomal subunit.
70S ribosomes isolated from the Δ_ybeY_ mutant were dissociated by fractionation on a sucrose gradient run at low magnesium concentration. (A) 50S and 30S ribosomal subunits (4.3 μM each) were subjected separately to digestion with YbeY and RNase R and the rRNA was analyzed on a Synergel/agarose gel. rRNA isolated from the Δ_ybeY_ mutant 70S ribosome was used as a marker (see also Figure S4). (B) 50S and 30S ribosomal subunits isolated from MC4100 and the Δ_ybeY_ mutant were mixed together in different combinations as indicated at equimolar concentrations (3.8 μM each) and subjected to digestion with YbeY and RNase R. rRNA was analyzed as described above.
The Δ_ybeY_ mutant is reported to have both defective 30S and 50S subunits (Davies et al., 2010; Rasouly et al., 2010). To determine whether this unique 70S ribosome quality control is mediated specifically by either the defective 30S or 50S subunit, we purified subunits from wild type and the Δ_ybeY_ mutant. The subunits were mixed at equimolar concentrations in different combinations and incubated with YbeY and RNase R (Figure 4B). As expected, 70S ribosomes resulting from the mixing of wild type 30S and 50S subunits were not degraded, whereas 70S ribosomes resulting from the mixing of 30S and 50S subunits from the Δ_ybeY_ mutant were effectively degraded. When the 30S subunit from wild type was mixed with 50S subunit from the Δ_ybeY_ mutant no degradation of the resulting 70S ribosome was observed, indicating that defective 50S from the mutant does not initiate 70S ribosome degradation. In contrast, when the 30S subunit from the Δ_ybeY_ mutant was mixed with the 50S subunit from the wild type, the resulting 70S ribosome was completely degraded. Taken together these results provide strong evidence that this YbeY/RNase R-dependent 70S ribosome quality control recognizes the presence of a defective 30S subunit in a 70S ribosome and not the 50S ribosomal subunit. Interestingly, the presence of a defective 30S subunit in a 70S ribosome leads to degradation of its partner 50S subunit, even if that 50S subunit is not defective.
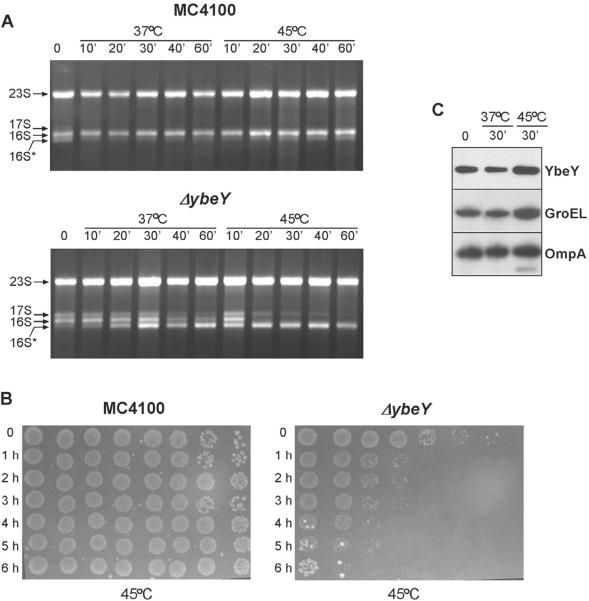
A heat-stressed Δ_ybeY_ mutant is deficient in processing of the 16S rRNA 3′ terminus and accumulates 17S rRNA and 16S* rRNA
As mentioned above, prior to our discovery that YbeY is an RNase we had obtained data indicating that YbeY is involved in the processing of all three rRNAs (Davies et al., 2010). Since YbeY is a heat shock protein and the Δ_ybeY_ mutant is temperature-sensitive (Davies et al., 2010; Rasouly et al., 2009), we sought to gain additional insights into the roles of YbeY in rRNA maturation and ribosome quality control by investigating the effects of subjecting a Δ_ybeY_ mutant to heat stress.
There was virtually no change in the rRNA profile of the wild type strain shifted from 37°C (Davies et al., 2010) to 45°C. In contrast, the rRNA of the Δ_ybeY_ strain changed remarkably from that seen at 37°C (Davies et al., 2010), as the amount of 16S rRNA decreased drastically and was barely visible after 60 min of exposure to heat, while the levels of 17S and 16S* rRNA remained relatively stable throughout (Figure 5A). Mapping of the 16S rRNA 5' and 3' termini confirmed the drastic reduction of mature 3' terminus to nearly undetectable levels in the Δ_ybeY_ strain (Figure 5C). Both termini of the 17S rRNA precursor and a small amount of the mature 5' terminus, most likely derived from 16S* rRNA, were still present at 45°C (Figure S5A). An rRNA profile similar to that of wild type was restored to the Δ_ybeY_ strain at 45°C by ectopically expressing YbeY (Figure 5B and 5C). Northern hybridization using a probe complementary to 33 nucleotides of the mature 3' terminus of 16S rRNA did not show hybridization to the 16S* rRNA (Figure S5B), consistent with the truncation of the 3' end of 16S rRNA by several nucleotides. This would make 16S* rRNA essentially incompetent for translation of canonical mRNAs (Shine and Dalgarno, 1975; Vesper et al., 2011).
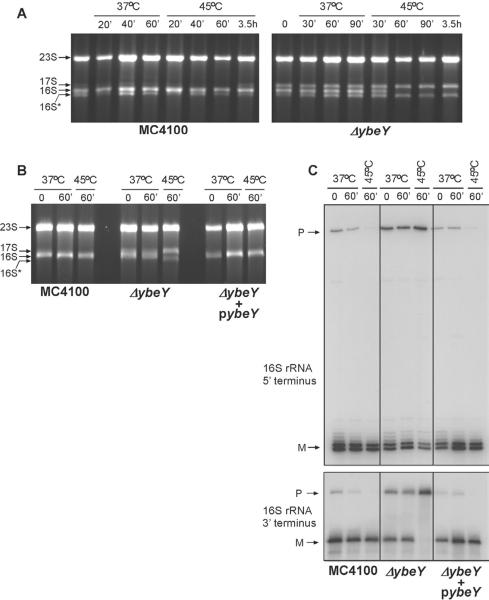
Figure 5. Thermosensitivity of the Δ_ybeY_ mutant can be attributed to the absence of mature 16S rRNA and strong defects in processing of the 16S rRNA 3' terminus.
Cells were grown to an OD600 of 0.3 at 37°C and shifted to 45°C. Aliquots of cells were taken at time points indicated. (A) Total RNA extracted from MC4100 and the Δ_ybeY_ mutant grown at 37°C and 45°C at different time points after temperature shift was analyzed on a Synergel/agarose gel. Positions of the 23S, 17S, 16S and 16S* rRNA are indicated. (B) – (C) Ectopic expression of YbeY rescues the 16S rRNA phenotype in the Δ_ybeY_ mutant at 45°C. (B) Total RNA extracted from MC4100, MC4100 Δ_ybeY_ and MC4100 Δ_ybeY_ pYbeY grown at 37°C and 45°C was resolved on a Synergel/agarose gel. (C) The 5' and 3' termini of the 16S rRNA were mapped by primer extension and specific RNase H cleavage followed by Northern hybridization, respectively, as described (Davies et al., 2010). “P” and “ M” indicate the positions of bands derived from the precursor and mature forms of the rRNA (see also Figure S5).
When new RNA synthesis was blocked with rifampicin just prior to the temperature shift, a constant level of mature 16S rRNA was present in the wild type strain, both at 37°C and 45°C (Figure 6A). For the Δ_ybeY_ mutant, at 37°C, early time points showed the characteristic rRNA pattern previously reported (Davies et al., 2010) with similar amounts of 17S, 16S and 16S* rRNA. However, after 40 min, the amounts of 16S and 17S rRNA species decreased and 16S* rRNA was the major species present (Figure 6A). Strikingly, when the Δ_ybeY_ mutant was shifted to 45°C, there was a complete loss of 16S rRNA and a steady decrease in 17S rRNA within 30 min along with constant levels of 16S* rRNA throughout. Mapping of the 5' and 3' termini of 16S rRNA isolated from rifampicin-treated cells corroborated these observations (Figure S6). This complete loss of 16S rRNA at 45°C explains why approximately 99% of Δ_ybeY_ cells die within an hour of being shifted from 37°C to 45°C (Figure 6B). As expected (Rasouly et al., 2009), YbeY expression in wild type cells increased upon a shift to 45°C to a degree similar to that of the heat shock protein GroEL (Figure 6C).
Figure 6. Severe 16S rRNA maturation defects of the Δ_ybeY_ mutant at 45°C are correlated with a substantial decrease in viability.
(A) In the absence of new rRNA synthesis, almost no mature 16S rRNA is observed 20 min after the shift to 45°C in the Δ_ybeY_ mutant. MC4100 and the Δ_ybeY_ mutant were grown at 37°C to O.D600 of 0.3. Prior to temperature shift, rifampicin was added to the cultures at a concentration of 400 μg/ml to block transcription of new rRNA and cells collected at different time points after temperature shift from treated cells grown at 37°C and 45°C. Total RNA was extracted and analyzed on a Synergel/agarose gel. (B) – (C) Decrease in viability of the Δ_ybeY_ mutant within 1h exposure to 45°C and a two fold increase in YbeY levels under heat shock. (B) A detailed time course was performed on wild type E. coli MC4100 and the Δ_ybeY_ mutant at 45°C. Cells were grown to an OD600 of 0.3 at 37°C and shifted to 45°C. Aliquots of cells were taken at time points indicated, 10-fold serially diluted and 10 μl of each dilution were spotted on LB agar plates and incubated at 37°C. The Δ_ybeY_ mutant is severely compromised for growth at 45°C and there is a substantial decrease in viable cells within 1 hour of exposure to heat shock. (C) Levels of YbeY at 37°C and 45°C in MC4100 cells carrying a FLAG tagged genomic copy were determined by Western blot analysis using an anti-FLAG antibody. GroEL was used as control to ascertain the heat shock response and OmpA was used as a loading control (see also Figure S6).
These results suggest that when Δ_ybeY_ cells are subjected to heat stress, the absence of YbeY's endoribonuclease activity results in a failure to carry out correct processing of the 3' terminus of the 17S rRNA precursor to generate the mature 16S rRNA; instead 17S rRNA is converted to 16S* rRNA (Figures 5 and 6). Furthermore, the observation that the 16S* rRNA species accumulates in the Δ_ybeY_ mutant under these conditions is consistent with the aforementioned role of YbeY in ribosome quality control.
DISCUSSION
YbeY is a single strand specific endoribonuclease with unique substrate specificity
We have discovered that the highly conserved E. coli protein YbeY, which is present in most sequenced bacterial genomes and is part of the minimal bacterial gene set, is a single strand specific metallo-endoribonuclease. YbeY has the unusual ability to degrade free rRNAs, a property of E. coli RNases that it shares only with RNase R and RNase A (Arraiano et al., 2010; Deutscher, 2009). Furthermore, YbeY is the first endoribonuclease to be described that has the remarkable ability to specifically cleave rRNA in defective 70S ribosomes but not in dissociated 30S and 50S subunits derived from these defective 70S ribosomes. This unique substrate specificity of YbeY distinguishes it from currently known RNases. Furthermore, YbeY is also the first endoribonuclease to be implicated in the critically important maturation of the 3' terminus of 16S rRNA.
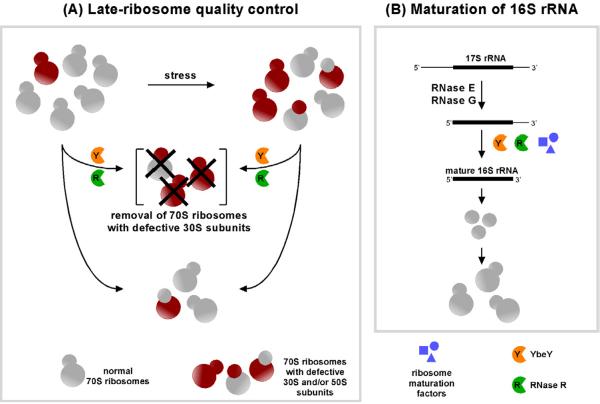
A model for YbeY-dependent 70S ribosome quality control
We propose a model for ribosome quality control that can function at the level of both non-translating and translating 70S ribosomes in E. coli (Figure 7). YbeY acts as a sensor of defective 70S ribosomes mediated by defective 30S subunits and initiates degradation of 70S ribosomes by making one or more endonucleolytic cuts in the rRNA. In principle, YbeY could recognize rRNA perturbations in defective 70S ribosomes through an exposed single stranded portion of rRNA due to misprocessing or misfolding of rRNA, damage to rRNA by environmental factors, absence of certain base modifications, or absence or unbalanced amounts of certain ribosomal proteins (Liang et al., 2009; Roy-Chaudhuri et al., 2010; Siibak et al., 2011). After the primary cut(s) by YbeY, RNase R could unwind the rRNA with its helicase activity (Awano et al., 2010) and continue the degradation of rRNA exonucleolytically, which may be further assisted by YbeY. Recent evidence in vivo (Basturea et al., 2011; Davies et al., 2010) is consistent with PNPase also contributing to this quality control-associated degradation once the single stranded RNA ends have become accessible. Additionally, the ability of YbeY and RNase R to degrade polysomes from the Δ_ybeY_ mutant and kasugamycin-treated cells indicates that defective 70S ribosomes in these cells can participate in translation and that this late-stage quality control is an important check point to avoid protein mistranslation. Our results suggest that 70S ribosome quality control is extremely important for cell survival under a wide variety of stresses, which could in part explain the high conservation of YbeY and RNase R throughout evolution.
Figure 7. Proposed model for ribosome quality control and 16S rRNA maturation by YbeY and RNase R. (A) Role of YbeY and RNase R in 70S ribosome quality control.
YbeY acts as a sensor for defective 30S ribosomal subunits and, along with RNase R, exerts a key role in a unique quality control mechanism involving 70S ribosomes, but not individual 30S and 50S ribosomal subunits. After YbeY initiates the degradation of defective 70S ribosomes by making endonucleolytic nick/s in exposed single stranded portion/s of the rRNA, RNase R unwinds the RNA with its helicase activity (Awano et al., 2010) and continues the degradation of rRNA exonucleolytically, assisted further by YbeY.
(B) Role of YbeY and RNase R in 16S rRNA maturation and/or rRNA stability: YbeY cleaves the 17S rRNA precursor endonucleolytically generating a 3' phosphate terminus at or near the final maturation site. Processing may be completed by the activity of additional RNases, such as RNase R, and/or a 3' phosphatase and may be further modulated by other ribosome maturation factors including Era, KsgA, RbfA or RsgA (Campbell and Brown, 2008; Inoue et al., 2006; Tu et al., 2009; Xu et al., 2008). Maturation of the 16S rRNA 3' terminus can stabilize and protect the rRNA making it inaccessible to further degradation by other housekeeping RNases.
Nonfunctional rRNA decay (NRD) is an important late-stage ribosome quality control system in Saccharomyces cerevisiae, that detects and degrades rRNA with deleterious point mutations in the peptidyl transferase or the decoding centers present in fully assembled ribosomes or ribosomal subunits (Cole et al., 2009; LaRiviere et al., 2006). Although the YbeY-dependent late-stage 70S quality control system shares the characteristic with NRD of acting at the level of fully assembled ribosomes to detect and degrade rRNA present in defective ribosomes, its specificity appears to differ from that of eukaryotic NRD (Muth et al., 2000; Powers and Noller, 1990, 1993; Thompson et al., 2001). Moreover, unlike NRD that is triggered in the yeast proteasome, the bacterial YbeY-mediated 70S ribosome quality control is initiated by a nuclease.
The defective ribosomes utilized in this study were obtained from the Δ_ybeY_ mutant and from wild type cells treated with kasugamycin, a drug to which the Δ_ybeY_ mutant is extremely sensitive. In both cases, polysomes, 70S ribosomes, and the ribosomal subunits contained increased amounts of misprocessed rRNAs, most notably misprocessed 16S rRNA. Additionally, some ribosomes from kasugamycin-treated cells have been reported to lack several ribosomal proteins yielding 61S ribosomal particles (Davies et al., 2010; Kaberdina et al., 2009)] that were likely included in our 70S ribosome preparation. Our electrophoretic analyses suggest the defective 70S ribosomes obtained from both the Δ_ybeY_ mutant and kasugamycin-treated cells are mixtures of abnormal ribosomes containing 17S rRNA precursor or misprocessed 16S rRNAs along with normal ribosomes containing mature 16S rRNA. Nevertheless, all of the rRNAs present in 70S ribosomes was degraded by YbeY and RNase R in vitro. Thus, the subset of apparently normal ribosomes carrying mature 16S rRNA obtained from the Δ_ybeY_ mutant and from kasugamycin-treated cells, must in fact contain abnormalities that can be detected by the YbeY/RNase R 70S ribosome quality control system. Furthermore, the substantially lower amount of ribosomes obtained from kasugamycin-treated cells suggests the YbeY/ RNase R quality control system has been saturated so that the defective 70S ribosomes and polysomes we isolated are those that have escaped this critical checkpoint.
Importantly, our results indicate this YbeY-dependent system of 70S ribosome quality control is mediated exclusively by a defective 30S ribosomal subunit and not by a 50S subunit. In fact, when normal 50S subunits are mixed with defective 30S subunits, YbeY and RNase R effectively degrade the resulting 70S, even though the 50S subunits on their own cannot be degraded. Whether this degradation of the 70S complex formed by normal 50S and defective 30S subunits is due to improper assembly of the subunits because of inherent defects in the 30S subunits isolated from the Δ_ybeY_ mutant and from kasugamycin-treated cells or due to destabilization of the 70S ribosomes subsequent to the degradation of the defective 30S subunits is under investigation.
The substrate and enzyme were used at approximately equimolar concentrations in our in vitro assays. While this could be considered a high concentration for an RNase, given that YbeY is involved in highly specific, controlled processing and degradative events in living cells, it is not surprising that its specific activity is relatively low compared to some degradative RNases. Also, it is extremely likely that YbeY action in vivo is subject to multiple layers of regulation like that of RNase R, which also degrades free RNA substrates relatively non-specifically (Awano et al., 2010; Chen and Deutscher, 2010; Liang and Deutscher, 2010, 2012). Such in vivo regulation could limit the extensive degradation of the defective 70S ribosomes that is observed in vitro, thereby allowing a sub-population of these ribosomes to participate in translation.
A model for participation of YbeY in 16S rRNA maturation
In E. coli, the 16S, 23S, and 5S rRNAs are cotranscribed as a single RNA molecule that is then converted to three precursors, each of which undergo further 3' and 5' processing to yield the final mature rRNAs. Although the extensive rRNA maturation defects observed in the Δ_ybeY_ mutant in vivo (Davies et al., 2010) do not distinguish between a direct or indirect involvement of YbeY, our identification of YbeY as an endoribonuclease suggests that it could be playing a direct role in 16S rRNA processing that becomes essential at 45°C. Correct processing of the 3' terminus of 16S rRNA is critical (Inoue et al., 2003; Sato et al., 2005; Wireman and Sypherd, 1974), whereas mutations affecting the maturation of the other rRNA termini do not have strong phenotypic consequences (Li and Deutscher, 1995; Li et al., 1999, 1999a, 1999b).
Our results suggest that the endoribonuclease YbeY (Figure 7) acts at or near the 16S rRNA 3' terminus maturation site, generating a 3' phosphate. Additional enzymes, possibly acting in a redundant fashion, would be required for the final maturation step. Since very little mature 16S rRNA 3' terminus can be detected in either a Δ_ybeY_ Δ_rnr_ mutant or a Δ_ybeY_ Δ_pnp_ mutant grown at 37°C (Davies et al., 2010), RNase R and PNPase are possible candidates to carry out this additional 3' end processing. RNase R can cleave RNA substrates that have a 3' phosphate (Cheng and Deutscher, 2002) and has been implicated in the processing of the 3' terminus of 16S rRNA in other organisms (Bollenbach et al., 2005; Purusharth et al., 2007), while PNPase plays a role in the 3' processing of the 23S rRNA in Arabidopsis thaliana chloroplasts (Walter et al., 2002). Another candidate is YbeZ, a phoH paralog, usually found in an operon with ybeY, that could potentially function as a phosphohydrolase (Kazakov et al., 2003) to remove the 3' phosphate generated by YbeY.
If the RNase activity of YbeY does play a direct role in 16S rRNA 3' maturation, then some other factor(s) would be necessary to restrict YbeY's relatively non-specific endoribonuclease activity to the appropriate region of the 16S rRNA precursor. In addition to ribosomal proteins that are present in the newly assembled ribosomal particles still containing immature rRNA (Mangiarotti et al., 1974), Era, an essential highly conserved cellular GTPase implicated in ribosome maturation (Verstraeten et al., 2011), is an attractive candidate. Era interacts with the 3' terminus of the 16S rRNA in a manner that leaves three nucleotides of the mature 3' terminus of 16S rRNA protruding out of the Era/16S rRNA complex (Tu et al., 2009). Interestingly, era is found in the same operon as ybeY in many bacteria, while in Clostridiales BVAB3 str. UP119-5 ybeY and era are fused into a single gene.
Finally, our observations suggest that the sensitivity of a Δ_ybeY_ mutant to a variety of stresses [this work; (Davies et al., 2010; Davies and Walker, 2008; Rasouly et al., 2009)] results from the accumulation of translationally impaired or incompetent ribosomes (Davies et al., 2010) that have resulted from the combination of the strong defects in maturation of 16S rRNA 3' terminus and the concomitant loss of the critical 70S ribosome quality control system.
Concluding remarks
Our results provide striking evidence that YbeY is an important RNase involved in late-stage 70S ribosome quality control and in maturation of the 3' terminus of the 16S rRNA. In addition, YbeY might contribute to sRNA regulation (Pandey et al., 2011) by functioning in an as-yet uncharacterized pathway for sRNA-mediated regulation of mRNA. Intriguingly, the mammalian counterpart of YbeY localizes to the mitochondria (Pagliarini et al., 2008), so it will be interesting to determine if YbeY has related roles in mitochondrial RNA metabolism. Finally, the critical physiological roles of the highly conserved YbeY protein make it a potentially interesting target for the development of a new class of antibiotic or an antibiotic adjuvant.
EXPERIMENTAL PROCEDURES
Strains, growth conditions and DNA manipulations
Wild type strain MC4100 (Casadaban and Cohen, 1979) and MC4100 Δ_ybeY_ (Davies et al., 2010) were grown aerobically in Luria-Bertani (LB) at 37°C. Antibiotics were used at the following concentrations, kanamycin 25 μg/ml, kasugamycin 200 μg/ml, rifampicin 400 μg/ml, chloramphenicol 100 μg/ml and ampicillin 100 μg/ml. Allele transfers were done by P1 transduction. DNA manipulations were performed according to the methods of Sambrook (Sambrook and Russell, 2001).
Purification of YbeY
YbeY was purified by affinity, ion exchange and size exclusion chromatography (details in supplement).
In vitro RNA cleavage assays using YbeY, RNase R and RNase A
All YbeY RNase assays were carried out in 50 mM HEPES pH 7.5 in a 20 μl volume and used 0.05 – 500 μM of purified YbeY (wild type or mutant protein) on different RNA substrates as indicated. RNase R (Epicenter) and RNase A (Ambion) were used at a final concentration of 5 μM each. Further information is provided in extended experimental procedures.
Polysome/Ribosome purification and subunit fractionation
Polysomes, 70S ribosomes, 50S and 30S subunits from MC4100, MC4100 Δ_ybeY_, and MC4100 treated with 200 μ/ml of kasugamycin were isolated as described previously (Etchegaray and Inouye, 1999) with minor modifications as described in extended experimental procedures.
rRNA analysis
rRNA was extracted from log phase cultures grown at 37°C, after heat shock at 45°C, and after blocking transcription with rifampicin at 37°C and 45°C at the indicated time points, or from purified ribosome fractions using a Qiagen RNeasy Plus Mini Kit. 16S and 23S rRNAs were separated using Synergel/agarose gel electrophoresis as described (Wachi et al., 1999). A detailed description of the mapping of the 5' and 3' termini of 16S rRNA is provided in extended experimental procedures.
Western blot and Northern blot methods
Both these techniques were done as per standard protocol (Brown et al., 2004; Gallagher et al., 2008). Details are provided in extended experimental procedures.
Supplementary Material
01
Highlights
- YbeY is a previously unrecognized endoribonuclease with unusual characteristics
- YbeY and RNase R together participate in bacterial 70S ribosome quality control
- Defective 30S ribosomal subunit mediates the 70S ribosome quality control mechanism
- YbeY RNase is essential for the maturation of the 3' terminus of 16S rRNA at 45°C
ACKNOWLEDGEMENTS
We thank lab members for their help. This study was supported by grants from National Institute of Health GM31030 and the Deshpande Center to G.C.W., GM17151 to U.L.R., and P30 ES002109 to the MIT Center for Environmental Health Sciences. G.C.W. is an American Cancer Society Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental Information includes six figures and extended experimental procedures.
REFERENCES
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci U S A. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS microbiology reviews. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- Awano N, Rajagopal V, Arbing M, Patel S, Hunt J, Inouye M, Phadtare S. Escherichia coli RNase R has dual activities, helicase and RNase. J Bacteriol. 2010;192:1344–1352. doi: 10.1128/JB.01368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basturea GN, Zundel MA, Deutscher MP. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17:338–345. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Lange H, Gutierrez R, Erhardt M, Stern DB, Gagliardi D. RNR1, a 3'–5' exoribonuclease belonging to the RNR superfamily, catalyzes 3' maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 2005;33:2751–2763. doi: 10.1093/nar/gki576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T, Mackey K, Du T. Analysis of RNA by northern and slot blot hybridization. In: Frederick M, Ausubel, et al., editors. Current protocols in molecular biology. Chapter 4. 2004. Unit 4 9. [DOI] [PubMed] [Google Scholar]
- Cameron V, Uhlenbeck OC. 3'-Phosphatase activity in T4 polynucleotide kinase. Biochemistry. 1977;16:5120–5126. doi: 10.1021/bi00642a027. [DOI] [PubMed] [Google Scholar]
- Campbell TL, Brown ED. Genetic interaction screens with ordered overexpression and deletion clone sets implicate the Escherichia coli GTPase YjeQ in late ribosome biogenesis. J Bacteriol. 2008;190:2537–2545. doi: 10.1128/JB.01744-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Deutscher MP. RNase R is a highly unstable protein regulated by growth phase and stress. RNA. 2010;16:667–672. doi: 10.1261/rna.1981010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Putzer H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002;30:5339–5346. doi: 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Kohrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, RajBhandary UL, Walker GC. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol. 2010;78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Walker GC. A highly conserved protein of unknown function is required by Sinorhizobium meliloti for symbiosis and environmental stress protection. J Bacteriol. 2008;190:1118–1123. doi: 10.1128/JB.01521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Degradation of stable RNA in bacteria. J Biol Chem. 2003;278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–391. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- Gallagher S, Winston SE, Fuller SA, Hurrell JG. Immunoblotting and immunodetection. In: Frederick M, Ausubel, et al., editors. Current protocols in molecular biology. Chapter 10. 2008. Unit 10 18. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci U S A. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Pereto J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiology and molecular biology reviews : MMBR. 2004;68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Alsina J, Chen J, Inouye M. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol Microbiol. 2003;48:1005–1016. doi: 10.1046/j.1365-2958.2003.03475.x. [DOI] [PubMed] [Google Scholar]
- Inoue K, Chen J, Tan Q, Inouye M. Era and RbfA have overlapping function in ribosome biogenesis in Escherichia coli. J Mol Microbiol Biotechnol. 2006;11:41–52. doi: 10.1159/000092818. [DOI] [PubMed] [Google Scholar]
- Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol Cell. 2009;33:227–236. doi: 10.1016/j.molcel.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov AE, Vassieva O, Gelfand MS, Osterman A, Overbeek R. Bioinformatics classification and functional analysis of PhoH homologs. Silico Biol. 2003;3:3–15. [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ. A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell. 2006;24:619–626. doi: 10.1016/j.molcel.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc Natl Acad Sci U S A. 1995;92:6883–6886. doi: 10.1073/pnas.92.15.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5' maturation of 16S ribosomal RNA. The EMBO journal. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA. 1999a;5:139–146. doi: 10.1017/s1355838299981669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5' maturation of 16S ribosomal RNA. The EMBO journal. 1999b;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Deutscher MP. A novel mechanism for ribonuclease regulation: transfer-messenger RNA (tmRNA) and its associated protein SmpB regulate the stability of RNase R. J Biol Chem. 2010;285:29054–29058. doi: 10.1074/jbc.C110.168641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Deutscher MP. Post-translational modification of RNase R is regulated by stress-dependent reduction in the acetylating enzyme Pka (YfiQ) RNA. 2012;18:37–41. doi: 10.1261/rna.030213.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarotti G, Turco E, Ponzetto A, Altruda F. Precursor 16S RNA in active 30S ribosomes. Nature. 1974;247:147–148. doi: 10.1038/247147a0. [DOI] [PubMed] [Google Scholar]
- Muth GW, Ortoleva-Donnelly L, Strobel SA. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science (New York, N.Y.) 2000;289:947–950. doi: 10.1126/science.289.5481.947. [DOI] [PubMed] [Google Scholar]
- Oganesyan V, Busso D, Brandsen J, Chen S, Jancarik J, Kim R, Kim SH. Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus. Acta Crystallogr D Biol Crystallogr. 2003;59:1219–1223. doi: 10.1107/s0907444903011028. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Minesinger BK, Kumar J, Walker GC. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011;39:4691–4708. doi: 10.1093/nar/gkr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penhoat CH, Li Z, Atreya HS, Kim S, Yee A, Xiao R, Murray D, Arrowsmith CH, Szyperski T. NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure. J Struct Funct Genomics. 2005;6:51–62. doi: 10.1007/s10969-005-5277-z. [DOI] [PubMed] [Google Scholar]
- Powers T, Noller HF. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci U S A. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF. Allele-specific structure probing of plasmid-derived 16S ribosomal RNA from Escherichia coli. Gene. 1993;123:75–80. doi: 10.1016/0378-1119(93)90542-b. [DOI] [PubMed] [Google Scholar]
- Purusharth RI, Madhuri B, Ray MK. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3' end processing of 16 and 5 S ribosomal RNA. J Biol Chem. 2007;282:16267–16277. doi: 10.1074/jbc.M605588200. [DOI] [PubMed] [Google Scholar]
- Rasouly A, Davidovich C, Ron EZ. The heat shock protein YbeY is required for optimal activity of the 30S ribosomal subunit. J Bacteriol. 2010;192:4592–4596. doi: 10.1128/JB.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouly A, Schonbrun M, Shenhar Y, Ron EZ. YbeY - a heat shock protein involved in translation in Escherichia coli. J Bacteriol. 2009;191:2649–2655. doi: 10.1128/JB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Chaudhuri B, Kirthi N, Culver GM. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci U S A. 2010;107:4567–4572. doi: 10.1073/pnas.0912305107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbour, N.Y.: 2001. Molecular Cloning. [Google Scholar]
- Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Siibak T, Peil L, Donhofer A, Tats A, Remm M, Wilson DN, Tenson T, Remme J. Antibiotic-induced ribosomal assembly defects result from changes in the synthesis of ribosomal proteins. Mol Microbiol. 2011;80:54–67. doi: 10.1111/j.1365-2958.2011.07555.x. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Thompson J, Kim DF, O'Connor M, Lieberman KR, Bayfield MA, Gregory ST, Green R, Noller HF, Dahlberg AE. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc Natl Acad Sci U S A. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C, Zhou X, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X. Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis. Proc Natl Acad Sci U S A. 2009;106:14843–14848. doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N, Fauvart M, Versees W, Michiels J. The universally conserved prokaryotic GTPases. Microbiology and molecular biology reviews : MMBR. 2011;75:507–542. doi: 10.1128/MMBR.00009-11. second and third pages of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M, Umitsuki G, Shimizu M, Takada A, Nagai K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5' end of 16S rRNA. Biochem Biophys Res Commun. 1999;259:483–488. doi: 10.1006/bbrc.1999.0806. [DOI] [PubMed] [Google Scholar]
- Walter M, Kilian J, Kudla J. PNPase activity determines the efficiency of mRNA 3'-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. The EMBO journal. 2002;21:6905–6914. doi: 10.1093/emboj/cdf686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wireman JW, Sypherd PS. In vitro assembly of 30S ribosomal particles from precursor 16S RNA of Escherichia coli. Nature. 1974;247:552–554. doi: 10.1038/247552a0. [DOI] [PubMed] [Google Scholar]
- Xu Z, O'Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- Zhan C, Fedorov EV, Shi W, Ramagopal UA, Thirumuruhan R, Manjasetty BA, Almo SC, Fiser A, Chance MR, Fedorov AA. The ybeY protein from Escherichia coli is a metalloprotein. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2005;61:959–963. doi: 10.1107/S1744309105031131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01