Mutations causing medullary cystic kidney disease type 1 (MCKD1) lie in a large VNTR in MUC1 missed by massively parallel sequencing (original) (raw)
. Author manuscript; available in PMC: 2014 Jan 24.
Published in final edited form as: Nat Genet. 2013 Feb 10;45(3):299–303. doi: 10.1038/ng.2543
Abstract
While genetic lesions responsible for some Mendelian disorders can be rapidly discovered through massively parallel sequencing (MPS) of whole genomes or exomes, not all diseases readily yield to such efforts. We describe the illustrative case of the simple Mendelian disorder medullary cystic kidney disease type 1 (MCKD1), mapped more than a decade ago to a 2-Mb region on chromosome 1. Ultimately, only by cloning, capillary sequencing, and de novo assembly, we found that each of six MCKD1 families harbors an equivalent, but apparently independently arising, mutation in sequence dramatically underrepresented in MPS data: the insertion of a single C in one copy (but a different copy in each family) of the repeat unit comprising the extremely long (~1.5-5 kb), GC-rich (>80%), coding VNTR in the mucin 1 gene. The results provide a cautionary tale about the challenges in identifying genes responsible for Mendelian, let alone more complex, disorders through MPS.
Medullary cystic kidney disease (MCKD) type 1 (OMIM 174000) is a rare disorder characterized by autosomal dominant inheritance of tubulo-interstitial kidney disease1. Affected individuals variably require dialysis or kidney transplantation in the third to seventh decade of life. Diagnosis of MCKD1 in patients is complicated by the unpredictable progression of kidney disease, the absence of other specific clinical manifestations, and the high frequency of mild kidney disease in the general population2. Nonetheless, the disease has been compellingly and consistently mapped to a single autosomal locus at 1q213-7. Attempts to identify the mutated gene(s), however, have not been successful4.
The advent of massively parallel sequencing (MPS) technologies has made exhaustive sequencing of genomic regions a viable approach to the identification of genes responsible for rare Mendelian diseases caused by high penetrance mutations8,9. Yet, there is also a growing recognition that using MPS to discover disease genes is not always straightforward. Here, we report that MCKD1 is caused by an unusual class of mutations, recalcitrant to detection by MPS. The process of identifying the MCKD1 causal variation is of particular interest for human genetics, because it highlights important challenges in using current MPS for gene discovery.
Linkage analysis was performed on six likely MCKD1 pedigrees (Online Methods, Supplementary Fig. 1 and Supplementary Table 1), and in all families the phenotype showed perfect co-segregation with a single 2-Mb segment of chromosome 1 (Fig. 1). We examined the genotype data for evidence of copy-number variation in the critical interval, but found only two common copy-number polymorphisms, neither of which segregated with disease. Looking at the longest stretches of allelic identity within pairwise comparisons of the pedigrees’ phased risk-haplotypes, we also found no obvious ancestral haplotype shared by a significant fraction of the families (beyond the background LD in the general population). This result suggested that the families carried independently occurring mutations, consistent with the families’ diverse ancestries.
Figure 1. Linkage of six MCKD1 families to chromosome 1.
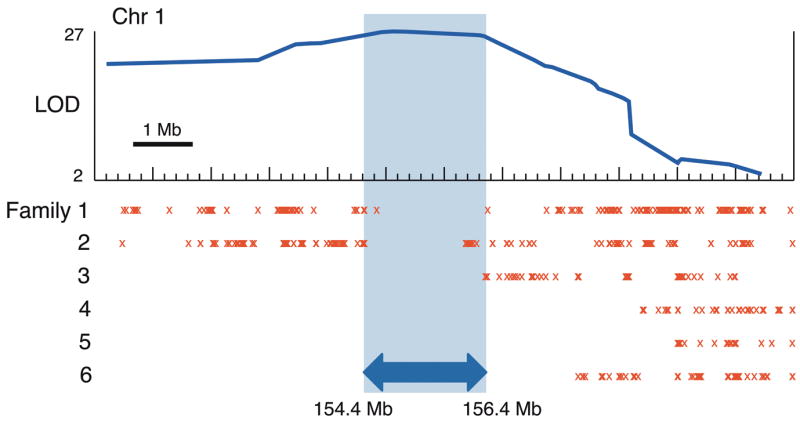
LOD curve shows the combined linkage-score of six MCKD1 pedigrees across 12 Mb of chromosome 1, with the peak score well above the threshold of 3.6 for genome-wide significance17. Red X’s mark the locations of opposite-allele homozygous genotype calls between affected members within each pedigree and highlight regions where affected individuals de facto share no alleles IBD, thereby delineating genomic segments unlikely to harbor causal variation. The shaded region (hg19:chr1:154,370,020–156,439,000) was considered most likely to contain any causal mutations, bounded on each side by recombination breakpoints in two different pedigrees.
To search for mutations, we employed whole exome-, regional-capture- and whole genome sequencing (Online Methods). We selected two affected individuals from each pedigree for sequencing, chosen, where possible, to share only a single haplotype (the risk haplotype) across the linkage region. In protein-coding regions, we found only two rare (<1% in 1000 Genomes Phase I data10), non-silent point variants (SNPs or small indels) shared by both of the affected individuals in any pedigree: each was in a different gene and each in a different pedigree. This finding is consistent with the expected background rate for 75 genes in six independent risk chromosomes given the presence of 100-200 rare coding variants in a typical genome10. In the context of perfect segregation of the phenotype, near-complete coverage of the coding bases in the linked region and the experience with other Mendelian diseases, we had expected to find a gene harboring rare coding variants in multiple families. To our dismay, we found no such evidence.
We next examined the non-coding regions, but found no regional clustering of segregating rare variants. We searched for any large structural variation (hundreds of bases or larger) such as deletions, insertions, duplications and inversions. All variants identified in this manner either failed to segregate with disease or were found at appreciable levels in control populations.
At this point, we concluded that the causal mutation(s) in MCKD1 were either located in a subregion that was recalcitrant to sequencing or represented a novel mutational mechanism. We considered the possibility that MCKD1 might be due to expansions in a coding VNTR sequence, because recurrent mutations at coding VNTRs have been documented as the cause of many genomic disorders11 and because massively parallel sequencing data might not readily reveal such an expansion.
We used SERV (Sequence-based Estimation of minisatellite and microsatellite Repeat Variability)12 to identify highly variable tandem repeats (VNTRs) in or overlapping with coding regions of five genes contained within the disease-linked interval: KCNN3, EFNA3, ASH1L, MEF2D and MUC1. Candidate VNTRs in the first four genes were found either to be non-polymorphic or to show no notable expansion in affected individuals (relative to family members not sharing the risk haplotype and to CEPH family samples), based on direct assays of length by PCR.
The MUC1 VNTR was particularly difficult to assay: it consists of many (20-12513,14) copies of a large repeat unit (60 bases) with very high GC-content (>80%). We ultimately assayed the VNTR by Southern blot and confirmed results with long-range PCR (Online Methods). In our patient samples, VNTR lengths were consistent with published descriptions and were not expanded on risk chromosomes, excluding VNTR length as pathogenic. MUC1 remained particularly interesting as the only gene in the critical region displaying transcripts with kidney-specific expression, based on RNASeq data from an adult control individual (unrelated to this study). MUC1 encodes mucin 1, a transmembrane protein expressed on the apical surface of most epithelial cells, providing (amongst other functions) a protective barrier to prevent pathogens from accessing the cell surface. The protein possesses a heavily glycosylated extracellular domain containing the VNTR and an SEA module with a cleavage site for release of the extracellular domain, which then binds noncovalently to the transmembrane domain17,19 (Fig. 2a).
Figure 2. Discovery of +C insertion within MUC1 coding VNTR.
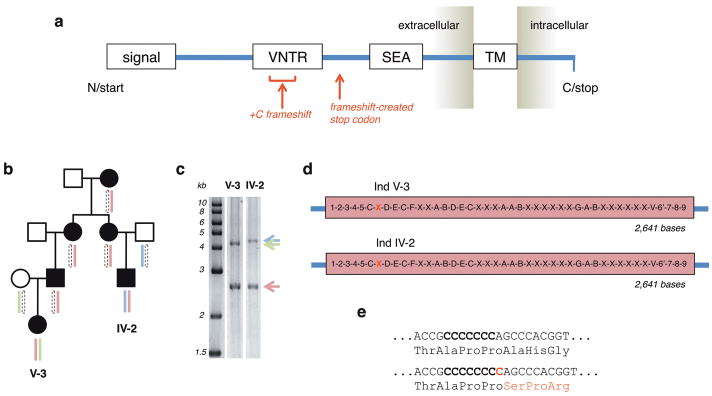
(a) The major domains of the full-length MUC1 precursor protein are shown: N-terminal signal sequence, VNTR, SEA module (where cleavage occurs), transmembrane domain, and C-terminal cytoplasmic domain. Based on fully and unambiguously assembled VNTR alleles, the frameshift caused by insertion of a C in the coding strand (as described in the main text) is expected to introduce a novel stop codon shortly beyond the VNTR domain. (b and c) Where possible, knowledge of segregating phased SNP-marker haplotypes was used to select for de novo VNTR sequencing and assembly of those individuals sharing only a single haplotype across the region, as this aided identification of the VNTR allele segregating with the shared risk haplotype. (d and e) Independent de novo assembly of the shared VNTR allele in two individuals from family 4 shows exactly identical complete sequence, with the seventh 60-base unit (red X) out of 44 containing a +C insertion event. The assembly is oriented relative to the coding strand of MUC1 and covers bases chr1:155,160,963-155,162,030 (hg19). Each unique 60-base repeat segment is represented by a different letter or number (Supplementary Fig. 2). (e) Translational impact of +C frameshift.
We considered the possibility that MCKD1 might be caused by point mutations within the MUC1 VNTR missed due to poor sequence coverage because (i) it was excluded from whole-exome and regional-capture probes due to its low-complexity and extreme sequence composition (and also because it is rarely annotated as coding sequence) and (ii) it was dramatically underrepresented in quality-filtered data from the whole-genome sequence, likely due to its GC-richness and homopolymer content. Because the human reference sequence appeared to significantly underrepresent this region (hg19 predicts a VNTR length far smaller than the published range or that observed in any of our samples, including controls), we undertook to clone and then reconstruct the VNTR alleles of several affected individuals and a CEPH trio; we subcloned, Sanger sequenced and performed de novo assembly for each (Online Methods and e.g. Fig. 2b-d).
We found a number of point variants in the VNTR assemblies, but, with one exception, they either did not segregate with the risk haplotype or were present in the alleles of the CEPH trio and/or unaffected chromosomes. However, we found one variant consistent with disease segregation: the insertion of a single C (relative to the coding strand of MUC1) within a stretch of seven C’s occurring at positions 53-59 in a single copy of the canonical 60-mer repeat (e.g. Fig. 2e). All six families carried such +C insertions, which appear to have arisen independently based on the different overall sizes of the VNTR, different local sequence contexts and different precise repeat units harboring the insertion (Supplementary Figs 2 and 3).
The frameshift caused by the insertion predicts a mutant protein that contains many copies of a novel repeat sequence (obtained by shifted translation of the VNTR) but which lacks, owing to a novel stop codon shortly beyond the VNTR terminus, the downstream SEA self-cleavage module and both transmembrane and intracellular domains characteristic of the normal MUC1 precursor protein (Fig. 2a).
Because discovery of the +C insertion required considerable labor and time, we sought to develop a simple and robust genotyping assay to enable larger population screening. We designed a probe-extension assay (Online Methods and Fig. 3a) capable of distinguishing reference and mutant MUC1 VNTR repeat units, making use of MwoI (which selectively cleaves the reference sequence) to increase the stoichiometric ratio of mutant:reference repeat units.
Figure 3. Detection of MUC1 +C insertion by probe-extension (PE) assay.
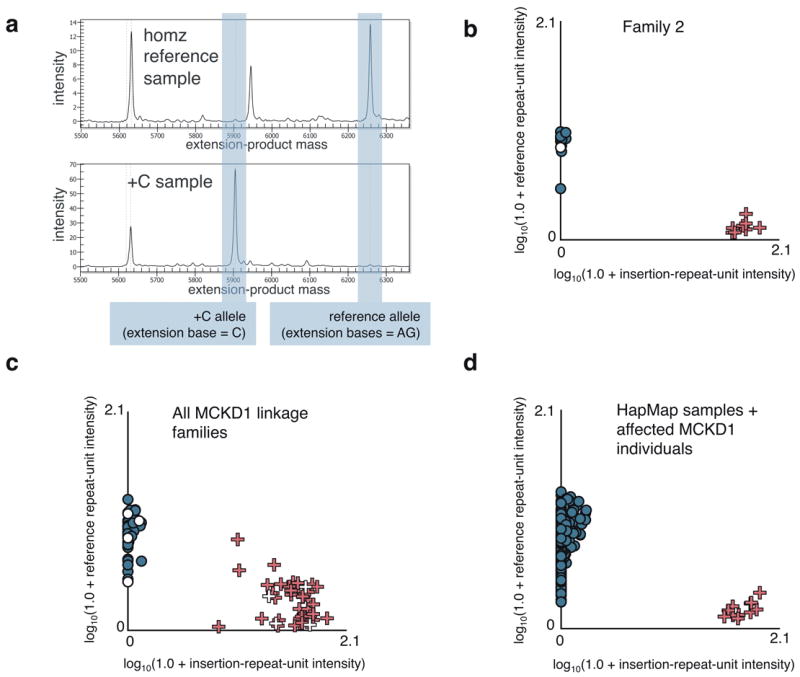
(a) Exemplar electropherograms for the _MUC1-_VNTR +C-insertion PE assay (Online Methods) performed on homozygous reference-allele and heterozygote samples. (b) Allele-intensity scatterplot for large linkage family 2. X-axis values correspond to the detected intensity at the mass of the +C PE product, while Y-axis values reflect that of the reference repeat-unit extension product. Datum coloring reflects MCKD1 diagnosis: blue = unaffected (or HapMap samples), red = affected, white = unknown. Individuals known to carry the linkage-analysis risk haplotype are represented by “+”, while other family members are depicted as dots. (c) Allele-intensity scatterplot for all MCKD1 linkage families. Samples having log-transformed intensities below 0.25 for both alleles were excluded as failed assays. WGA and low DNA-concentration samples were also excluded for underperforming. (d) Allele-intensity scatterplot for HapMap samples together with selected positive controls (MCKD1 individuals known to carry the insertion).
We typed all samples collected from the six MCKD1 families used for linkage analysis, including 62 phenotypically affected and 79 unaffected relatives (Fig. 3b-c), and over 500 control individuals from CEU, Japanese, Chinese, Yoruba and Tuscan HapMap3 populations (Fig. 3d). The genotyping assay was perfectly concordant with sequencing results, and full genotyping of all family members showed that the insertion segregated perfectly with each family’s risk haplotype and yet was not seen in any of the 500 HapMap samples.
Overall, the genotyping results provide strong evidence that the +C insertions are the high-penetrance genetic lesion that leads to development of MCKD1. As a statistical association, the significance of this observation can only be approximated, but it is clearly far less than the reciprocal of the number of bases in the genome (+C seen on 6/6 risk chromosomes vs. 0/1000 HapMap chromosomes). Furthermore, this observation is robust to population structure considerations since the mutations have arisen independently.
To explore the broader impact of MUC1 mutations, we genotyped affected and unaffected individuals from 21 additional small MCKD families screened to be negative for known MCKD mutations (Supplementary Table 1), only one family of which had existing linkage information implicating 1q2115. In 13 of 21 families we found the presence of a +C insertion consistent with being a fully penetrant cause of disease, indicating a substantial role for MUC1 in MCKD1-like phenotypes.
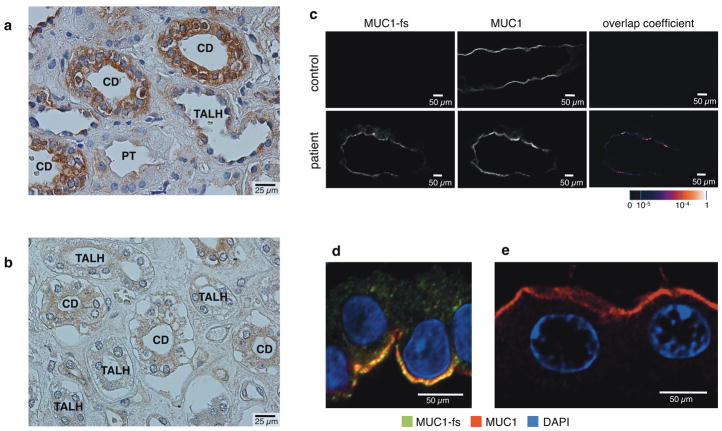
Using antibodies raised against a peptide synthesized based upon the predicted mutant VNTR sequence, we found specific intracellular staining in epithelial cells of Henle’s loop, distal tubule and collecting duct of MCKD1 patients (Fig. 4a), which was absent in control kidney (Fig. 4b). Co-staining of patient and control tissue additionally with antibodies against normal MUC1 demonstrated the specificity of the MUC1-fs (our name for the predicted mutant protein) antibodies for the mutant protein, with diffuse and/or fine granular intracellular localization of the MUC1-fs protein in patient kidney (Fig. 4c), and also patchy co-localization of MUC1-fs and normal MUC1 signals on the apical membrane of collecting duct epithelial cells (Fig. 4c and 4d). Detailed image analysis of patient tissue (Fig. 4d) compared to control tissue (Fig. 4e) detected no intracellular co-localization of MUC1-fs and normal MUC1 proteins in patient tissue, but revealed puncti of colocalization in distinct plasmalemmal subdomains. Antibody to MUC1-fs did not stain normal kidney tissue.
Figure 4. Immunohistochemical and immunofluorescence studies of MUC1-fs protein.
In MCKD1 patients, MUC1-fs is expressed and present in renal epithelial cells of Henle’s loop, distal convoluted tubule, and collecting duct. (a) Strong intracellular staining of MUC1-fs protein in MCKD1 patient, and (b) absence of the specific staining in control; TALH - thick ascending limb of Henle’s loop; CD – collecting duct; PT – proximal tubule. (c) Immunofluorescence analysis showing diffuse and/or fine granular intracellular and membrane staining of MUC1-fs protein, and its partial colocalization with normal MUC1 in collecting duct of an MCKD1 patient. MUC1-fs staining is absent in control, and colocalization with normal MUC1 is therefore not detected. The values of fluorescent signal overlaps are transformed to a pseudo-color scale shown at right bottom in the corresponding lookup table. (d) Immunofluorescence analysis showing different intracellular localizations and partial sub-membrane colocalization of MUC1-fs and normal MUC1 proteins in collecting duct of MCKD1 patient. Note specific staining of both forms in distinct membrane microdomains. (e) Absence of MUC1-fs staining and characteristic membrane localization of normal MUC1 in control.
This study highlights the fact that current MPS technology may not suffice to reveal disease mutations, even when linkage analysis conclusively pinpoints a critical region of a few megabases. Even if the insC event were not dramatically underrepresented in the quality-filtered MPS data and even if the reference genome assembly had been accurate in this region, it still would have been difficult to detect this particular insertion event using typical alignment and variation-detection tools due to (1) the underlying variability of VNTR size within and across individuals, (2) the inability to uniquely place reads within the VNTR, given current MPS read lengths, and (3) the fact that the mutant:reference allelic balance is skewed far from the expected 1:1 of a typical heterozygous variant.
The precise nature of the MCKD1 mutations is notable. Curiously, each independently-arising event is essentially the identical single-base insertion at the same position within one of the repeat units of the VNTR. Yet, insertions at many locations or other events (such as single-base deletions) would also result in out-of-frame translation of MUC1 and/or novel stop codons. Possible explanations for the consistently observed mutation include: (1) this insertion event is strongly favored due to mutational mechanism, (2) other events (eg. delC) are selected against, (3) other events (eg. delC) are benign and not associated with MCKD1, and (4) other MUC1 mutations exist but are undersampled here.
The identified mutation and the associated genotyping assay provide a screening tool for younger members of families in which MCKD1 has been previously diagnosed, as well as a diagnostic tool for sporadic cases. They also alleviate the challenge for living relative kidney donation, as potential donor family members have not known their status as unaffected or (yet-to-be) affected. Much work, however, remains to be done to elucidate the specific mechanism of pathogenesis of the MUC1 mutant protein. We note that knock-out studies indicate that the MUC1 gene is not essential in mice16 and support a possible dominant-negative and/or gain-of-function mode of action for the human MUC1 mutation. Together with the dominant and late-onset nature of the disease, this raises the possibility of preventative or therapeutic approaches based on treatments that decrease expression of the MUC1 gene or splice out its single VNTR-encoding exon.
ONLINE METHODS
Family collection and criteria for diagnosis of affected status
The six analyzed families with autosomal dominant tubulointerstitial kidney disease were among a larger group referred for evaluation. Each showed a clinical phenotype highly suggestive of MCKD1 and lacked UMOD or REN mutations. All had previously demonstrated evidence of linkage to chromosome 1. Written informed consent was obtained from all participants and the study was approved by the Wake Forest School of Medicine Institutional Review Board. Medical records were reviewed and peripheral venous blood samples were obtained for DNA isolation and laboratory determinations. Full diagnostic methods and clinical summaries are described in Supplementary Note.
Linkage and CNV analysis
Family members were genotyped on the Affymetrix 6.0 platform. Whole Affymetrix arrays with genotype call rates < 88% were excluded from analysis, as were samples which yielded low OD measurements (indicating poor sample performance during laboratory steps). Further, markers were excluded for which probe sequences showed excess genomic homology or potential for significant G-quartet formation (those probe sequences for which either allele contained at least three consecutive G’s).
Particularly large pedigrees (>24 bit complexity) were divided into branches where required by computational constraints. LD-independent marker maps were separately created for each pedigree/branch, choosing single, well-typed, informative markers from LD-defined bins of SNPs based on phased, population-specific HapMap data (hapmap.org, release 22). Markers which showed no-call rates > 10% or any Mendelian inheritance errors within a pedigree/branch were excluded from specific pedigree/branch analyses. Additionally, markers were required to be spaced at least 0.1 cM apart according to published sex-averaged recombination positions (affymetrix.com).
All expected intra-pedigree relationships were confirmed from pairwise IBD estimates using PLINK software18 and similarly derived marker sets; however, markers for PLINK were selected agnostic to their being polymorphic within a pedigree/branch so as not to skew IBD calculations. Merlin software19 was used to remove any likely genotyping errors which did not violate Mendelian inheritance rules, and then to perform parametric linkage under a rare, autosomal-dominant model using population-specific allele frequencies (affymetrix.com).
Linkage mapping was performed using the Merlin package under a rare autosomal-dominant model. Scores were combined across pedigrees/branches by summing LOD values, linearly interpolating scores between marker locations as required. The consistency of the alleles carried on the segregating risk haplotype was confirmed across pedigree branches.
The boundaries of the linked region were refined by searching all well-typed markers -- including many that were dropped solely to eliminate markers in LD from the linkage calculations -- for instances where affected members within the same pedigree shared no alleles IBD (by virtue of being homozygous for opposite alleles – for example, one having genotype AA and another CC). Such markers necessarily lie outside the critical linkage interval.
Affymetrix 6.0 intensity data were used by Birdsuite software20 to analyze copy-number variation.
Large-scale sequencing
Because the critical region contains more than 170 separate transcript annotations comprising over 75 RefSeq genes, amplicon-based resequencing of genic regions was initially not considered. Of the 12 sequenced individuals, whole-genome sequencing was performed on 11 of these individuals (~25-fold coverage on average), whole-exome sequencing on 11 individuals (~180-fold coding-sequence coverage on average) and regional-capture sequencing on 5 individuals (~220-fold coverage on average). Sequence processing is described in Supplementary Note. For all but three of the RefSeq genes, at least 99% of the coding bases were covered at ≥10-fold in each pedigree. Further, 98% of non-coding bases were covered at ≥10-fold in each pedigree.
As candidates for being pathogenic MCKD1 mutations, we considered any non-reference allele present in both affected individuals of any pedigree and with a population frequency ≤1%10. Non-coding regions were analyzed similarly.
To discover potential structural variation at the chromosome-1 locus, we ran Genome STRiP21 on the sequenced individuals and on a control population of 32 Finnish genomes sequenced at low coverage by the 1000 Genomes Project10 (Supplementary Note).
_MUC1_-VNTR Southern blot analysis
Genomic DNA (5-8 μg) was digested with 100 u _Hin_fI (NEB). Digests were run on a 0.8% agarose gel, transferred to a BrightStar Plus Nylon membrane (Ambion) and hybridized overnight at 65°C to a quadruply biotinylated synthetic 100mer oligonucleotide probe PS1 (Supplementary Table 3) (IDT) present at 2 ng/ml in SuperHyb hybridization solution (Ambion) supplemented with 100 μg/ml sonicated salmon sperm DNA (Stratagene). After a final high-stringency wash at 65°C in 0.2x SSC and 0.1% SDS, membrane-bound biotin was detected by a BrightStar BioDetect kit (Ambion).
_MUC1_-VNTR long-range PCR
The long-range PCR protocol was adapted from Fowler et al.14. Briefly, 7-μL PCR reactions contained 15 or 30 ng genomic DNA, 1.75 pmol of PS2 and PS3 primers (Supplementary Table 3), 5% DMSO, 625 μM of each dNTP, 1x reaction buffer with 3 mM MgCl2, and 0.25 u DyNAzyme EXT DNA polymerase (Finnzymes). Thermocycling on GeneAmp 9700 instruments (ABI) was as follows: initial denaturation (90 s at 96°C); 22 or 27 cycles (40 s at 96°C, 30 s at 65°C, 6 min at 68°C) and final extension (10 min at 68°C).
_MUC1_-VNTR sequencing and assembly
For selected individuals, we cloned gel-purified long-range-PCR products containing the full-length VNTR. Allele sizes derived from Southern blots and long-range PCR, together with known haplotype sharing between individuals in the same pedigree, in most cases permitted the identification of which MUC1 VNTR allele was part of the segregating risk haplotype (e.g. Fig. 2b and c). In a few cases, the sizes of the risk and non-risk VNTR allele were nearly the same, precluding physical separation of the two alleles prior to molecular cloning. Using transposon hopping and capillary sequencing, we then sequenced clones from each allele (Supplementary Note).
Because the region is exceptionally repetitive and because the read data contain both PCR errors and sequencing errors (exacerbated by the extreme GC content of the repeat), we developed a special assembly algorithm that could distinguish bona fide genomic differences from errors (Supplementary Note). Given the repetitive sequence content, not all assemblies were complete or unambiguous. Instead, some assembly frameworks suggested multiple possible resolutions across areas of uncertainty, forming full networks of possible solutions for a particular allele.
Supplementary Table 2 summarizes the key properties of the assemblies (example shown in Figure 2d), and Supplementary Figures 3 and 4 provide the sequence for those unique alleles (three risk and eight non-risk) where the assembly was fully or almost fully resolved. Supplementary Figure 5 illustrates the notation of graph assembly in a scenario where an allele could not be fully and unambiguously reconciled. We assembled each allele separately and independently. In all situations where two alleles were expected to be identical by haplotype sharing and where the assemblies were fully resolved, the assemblies were indeed identical – thus increasing our confidence that the assemblies were correct.
Genotyping of MUC1 +C insertion event
Genomic DNA was first over-digested using restriction endonuclease MwoI which selectively cleaves the reference repeat-unit sequence (GCCCCCCCAGC), while leaving intact repeat units containing the +C insertion (GCCCCCCC*C*AGC). Tailed primers nested within the 60-bp repeat were then used to PCR amplify the remaining intact VNTR fragments, thus enriching for insertion-containing fragments over reference-sequence background. PCR products were then re-digested with MwoI for a second round of enrichment. A 20-bp probe was then designed just upstream of the insertion site, and probe extension was performed using a high fidelity DNA polymerase and a nucleotide termination mix containing dATP, ddCTP and ddGTP. Following probe extension, reaction products were separated and sized by MALDI-TOF mass-spectrometry using the Sequenom MassArray platform. Spectra were then assessed for the presence of peaks corresponding to the mutant repeat-unit extension-product (at 5,904.83 daltons) and the reference repeat-unit extension-product (at 6258.06 daltons).
Specifically, 100 μg of genomic DNA was digested in a 25-μL reaction volume for 16 hours using 5 units of MwoI restriction endonuclease (New England Biolabs) with supplemental additions of 5 units of enzyme at hours 3 and 15. Digestion reactions were then cleaned using 50 μL AmPure beads according to manufacturers protocol (Agencourt, Beverly, MA), and digested DNA was eluted in 25 μL of nuclease-free water. Remaining intact VNTR fragments were PCR-amplified using 1X HotStart buffer, 1.0 mM MgCl2 (to supplement MgCl2 already in buffers), 0.8 mM dNTPs, 0.8 units of HotStart Taq Plus (Qiagen) and 0.2 μM forward and reverse primers PS6 and PS7 (Supplementary Table 3) in a 25-μL reaction volume. PCR cycling conditions were: one hold at 95°C for 5 min; 45 cycles of 94°C for 30 sec, 67°C for 30 sec, 72°C for 1 min; followed by one hold at 72°C for 10 min. PCR reactions were cleaned using 50 μL AmPure beads, and amplicons were eluted in 25 μL nuclease-free water. A second round of MwoI digestion was performed again for 16 hours with 5 units of enzyme added at hours 0, 3 and 15. Digestion reactions were cleaned using 50 μL AmPure beads and product was eluted in 6.2 μL of nuclease-free water.
Using 5.2 μL of the digested eluate as template, probe extension was performed using 1X HotStart buffer, 0.6 mM MgCl2 (to supplement MgCl2 already in buffers), 1.7 μL SAP buffer (Sequenom, San Diego, CA), 0.2 mM each of nucleotides ddGTP, ddCTP and dATP; 0.7 units of Thermo Sequenase DNA polymerase (Amersham) and 0.6 μM of extension probe PS8 (Supplementary Table 3) in a 10-μL reaction volume. Probe extension was performed on a 384-well ABI GeneAMP 9700 and cycling conditions were: one hold at 94°C for 2 min 55 cycles of 94°C for 5 sec, 52°C for 5 sec, 72°C for 5 sec; followed by one hold at 72°C for 7 min. Reactions were then de-salted by addition of a cation-exchange resin, and ~7 nL of purified extension reaction was spotted onto a SpectroChip (Sequenom) containing matrix 3-hydroxypicoloinic acid. Arrayed reactions were then analyzed by matrix-assisted laser desorbtion/ionization-time of flight (MALDI-TOF) on a Compact mass spectrometer (Sequenom/Bruker).
Assay results were clear enough to assign genotypes based on simple inspection of XY scatterplots depicting log-transformed reference- and mutant-repeat-unit intensities (log10(1.0+peak height)). Samples showing log-transformed intensities < .25 for both alleles were considered failed assays. Similarly, results from whole-genome-amplification samples or samples with low DNA concentrations were typically considered unreliable and discarded.
Antibody generation and kidney immunostaining
Immunodetection of MUC1-fs was performed with custom-prepared rabbit antibodies (PA4 302) raised against the peptide SPRCHLGPGHQAGPGLHRPP, representing the predicted mutant VNTR unit (Open Biosystems, Huntsville, AL; diluted 1:1000 in 5% BSA in PBS). The normal MUC1 protein was detected with monoclonal mouse anti-human Epithelial Membrane Antigen (EMA) mouse monoclonal antibody (DAKO, Glostrup, Denmark; diluted 1:400 in 5% BSA in PBS). Detection of bound primary antibody was achieved using either Dako EnVision + TM Peroxidase Rabbit Kit (Dako) or System-HRP labeled Polymer Anti-mouse (DAKO), for rabbit or mouse antibodies, respectively, with 3,3′-diaminobenzidine as substrate.
Paraformaldehyde-fixed human kidney biopsies were analysed. The specificity of antigen detection was always ascertained by omission of the primary antibody-binding step.
For immunofluorescence analysis, PA4 302 antibody was diluted 1:200 in 5% BSA in PBS and EMA antibody was diluted 1:10 in 5% BSA in PBS. Fluorescence detection used species-specific secondary antibodies. Alexa Fluor® 488 goat-anti rabbit IgG and Alexa Fluor® 568 goat-anti mouse IgG (Molecular Probes, Invitrogen, Paisley, UK). Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). Prepared slides were mounted in Immu-Mount fluorescence mounting medium (Shandon Lipshaw, Pittsburgh, PA) and analyzed by confocal microscopy.
XYZ images sampled according to Nyquist criterion were acquired using a TE2000E C1si laser scanning confocal microscope, Nikon PlanApo objective (40x, N.A.1.30), 488 nm and 543 nm laser lines and 515 +/-15 nm and 590 +/-15 nm band pass filters. Images were deconvolved using the classic maximum likelihood restoration algorithm in Huygens Professional Software (SVI, Hilversum, The Netherlands). Colocalization maps employing single pixel overlap coefficient values ranging from 0-1 were created using Huygens Professional Software. The resulting overlap coefficient values are presented as pseudo-color (scale is shown in corresponding figure lookup tables).
Supplementary Material
1
Acknowledgments
This work was conducted as part of the Slim Initiative for Genomic Medicine, a joint U.S.-Mexico project funded by the Carlos Slim Health Institute. This research was supported in part by the Intramural Research Program of the NIH, NHGRI. S.K., H.H., J.S. and V.B. were funded by Charles University programs PRVOUK-P24/LF1/3 and UNCE 204011, and their work was supported by grants LH12015 and NT13116-4/2012 from the Ministry of Education and Ministry of Health of the Czech Republic. S.A. was supported by NIH DK34854 (The Harvard Digestive Diseases Center). N.P. is a Broad Fellow of the Broad Institute and a postdoctoral research fellow of the Fund for Scientific Research - Flanders (FWO Vlaanderen), Belgium. I.G.V. was supported by HFSP, Alon, the Israeli Centers of Research Excellence (I-CORE), and Edmond J. Safra Center for Bioinformatics at Tel Aviv University. Thanks to T. L. Hatte for reagent use. We thank David Altshuler, Todd Carter, and Johannes Schlondorff for useful discussions, and Maria Cortes, Miguel Ilzarbe, and Miguel Betancourt for helpful project management. We also thank Fran Letendre, Matthew Coole, Robert Paul Frere, Claude Bonnet, Leon Mulrain, Nyima Norbui, and Harindra Arachchi for Sanger sequencing.
Footnotes
AUTHOR CONTRIBUTIONS
A.B., E.L. and M.Daly jointly supervised the research. R.X., M.P. and S.A. provided study design and interpretation advice. C.A., S.J.S., P.S.H. and A.B. performed sample collection. C.Stevens managed the project. C.Sougnez and K.C. provided early genotyping and sequencing support. Linkage analysis was performed by A.K. based on prior work by P.S.H. A.K. and M.Daly developed variation-discovery and analysis methods. A.K., J.R. and R.H. analyzed structural variation. T.G. performed CNV analysis. Supervision of sequencing was by S.G. Custom-capture array design was by S.Sigurdsson and K.L.T. M.P. performed direct PCR of polymorphic-VNTR candidates selected by N.P. A.G. and D.A. performed Southern blot and long-range PCR of the MUC1 VNTR. C.N. supervised the _MUC1_-VNTR sequencing approach. A.G. performed VNTR-allele cloning and generation of sequencing libraries. E.K., R.D., D.P. and S.Steelman performed Sanger sequencing. D.J. assembled and analyzed VNTR Sanger sequencing. M.G. provided RNAseq support. S.K. supervised immunohistochemistry and immunofluorescence work by V.B., H.H., J.S., and P.V. A.K., B.B. and M.DeFelice developed the C-insertion genotype assay. M.Z. provided informatic and sequencing consultation. A.R. provided informatic and analysis consultation. C.Y., J.R., M.C., I.G., R.H. and E.R. provided informatic support. The manuscript was written primarily by A.K., A.G., A.B., E.L. and M.Daly. The supplementary information was prepared mainly by A.K., A.G., D.J., B.B., R.H., S.Sigurdsson, S.K. and A.B.
COMPETING FINANCIAL INTERESTS
Andrew Kirby, Andreas Gnirke, Brendan Blumenstiel and Matthew DeFelice are listed as inventors on the C-insertion genotyping assay under patent review. The other authors declare no competing interests.
References
- 1.Bleyer AJ, Hart PS, Kmoch S. Hereditary interstitial kidney disease. Semin Nephrol. 2010;30:366–373. doi: 10.1016/j.semnephrol.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castro AF, Coresh J. CKD surveillance using laboratory data from the population-based National Health and Nutrition Examination Survey (NHANES) Am J Kidney Dis. 2009;53:S46–55. doi: 10.1053/j.ajkd.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christodoulou K, et al. Chromosome 1 localization of a gene for autosomal dominant medullary cystic kidney disease. Hum Mol Genet. 1998;7:905–911. doi: 10.1093/hmg/7.5.905. [DOI] [PubMed] [Google Scholar]
- 4.Wolf MTF, et al. Medullary cystic kidney disease type 1: mutational analysis in 37 genes based on haplotype sharing. Hum Genet. 2006;119:649–658. doi: 10.1007/s00439-006-0176-3. [DOI] [PubMed] [Google Scholar]
- 5.Serafini-Cessi F, Malagolini N, Cavallone D. Tamm-Horsfall glycoprotein: biology and clinical relevance. Am J Kidney Dis. 2003;42:658–676. doi: 10.1016/s0272-6386(03)00829-1. [DOI] [PubMed] [Google Scholar]
- 6.Vylet’al P, et al. Alterations of uromodulin biology: a common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int. 2006;70:1155–1169. doi: 10.1038/sj.ki.5001728. [DOI] [PubMed] [Google Scholar]
- 7.Scolari F, et al. Uromodulin storage diseases: clinical aspects and mechanisms. Am J Kidney Dis. 2004;44:987–999. doi: 10.1053/j.ajkd.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Romaih KI, et al. Genetic diagnosis in consanguineous families with kidney disease by homozygosity mapping coupled with whole-exome sequencing. Am J Kidney Dis. 2011;58:186–195. doi: 10.1053/j.ajkd.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 12.Legendre M, Pochet N, Pak T, Verstrepen KJ. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007;17:1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horne AW, et al. MUC 1: a genetic susceptibility to infertility? Lancet. 2001;357:1336–1337. doi: 10.1016/s0140-6736(00)04502-5. [DOI] [PubMed] [Google Scholar]
- 14.Fowler JC, Teixeira AS, Vinall LE, Swallow DM. Hypervariability of the membrane-associated mucin and cancer marker MUC1. Hum Genet. 2003;113:473–479. doi: 10.1007/s00439-003-1011-8. [DOI] [PubMed] [Google Scholar]
- 15.Auranen M, Ala-Mello S, Turunen JA, Järvelä I. Further evidence for linkage of autosomal-dominant medullary cystic kidney disease on chromosome 1q21. Kidney Int. 2001;60:1225–1232. doi: 10.1046/j.1523-1755.2001.00931.x. [DOI] [PubMed] [Google Scholar]
- 16.Spicer AP, Rowse GJ, Lidner TK, Gendler SJ. Delayed mammary tumor progression in Muc-1 null mice. J Biol Chem. 1995;270:30093–30101. doi: 10.1074/jbc.270.50.30093. [DOI] [PubMed] [Google Scholar]
- 17.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 20.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 2011;43:269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1