Clonal development and organization of the adult Drosophila central brain (original) (raw)
. Author manuscript; available in PMC: 2014 Apr 22.
Published in final edited form as: Curr Biol. 2013 Mar 28;23(8):633–643. doi: 10.1016/j.cub.2013.02.057
Summary
Background
The insect brain can be divided into neuropils that are formed by neurites of both local and remote origin. The complexity of the interconnections obscures how these neuropils are established and interconnected through development. The Drosophila central brain develops from a fixed number of neuroblasts (NBs) that deposit neurons in regional clusters.
Results
By determining individual NB clones and pursuing their projections into specific neuropils we unravel the regional development of the brain neural network. Exhaustive clonal analysis revealed 95 stereotyped neuronal lineages with characteristic cell body locations and neurite trajectories. Most clones show complex projection patterns, but despite the complexity, neighboring clones often co-innervate the same local neuropil(s) and further target a restricted set of distant neuropils.
Conclusions
These observations argue for regional clonal development of both neuropils and neuropil connectivity throughout the Drosophila central brain.
Introduction
In the adult brain of Drosophila melanogaster neuropils are defined as distinct synapse-dense areas arising due to denser local interconnectivity between neurites within one region compared to the adjacent region. These anatomical features are thereby a convenient anatomical proxy for decomposing brain circuitry into distinct subcircuits. The sets of neurons derived from the same neural stem cell progenitor, or neuroblast (NB), are one of the few levels of organization operating at this same scale between individual neurons and gross anatomy motivating the analysis of how NBs generate neuropils and wire them together. Given the lack of active migration of neurons outside the optic lobes (OLs), the NB lineages are expected to build regional neuropils through a series of clonal units. One common convention is to classify neurons relative to the neuropil they innervate, with a primary distinction between local interneurons (LNs), which elaborate solely within a single neuropil, and projection neurons (PNs), which project between neuropils thereby connecting them together.
The most studied neuropils in the Drosophila central brain are those with a striking morphology and clear boundaries. These include the antennal lobe (AL), the mushroom body (MB), and the components of the central complex (CX), which include the protocerebral bridge (PB), the fan-shape body (FB), the ellipsoid body (EB), and the paired noduli (NO). All of these neuropils are composed of anatomically distinct subregions, such as the glomeruli of the AL [1], the input calyx and output lobes of the MB [2], as well as array-like structures within the components of the CX [3]. These cases make clear that the subdivisibility of the brain into neuropils represents a convenient idealization, but the substructure within neuropils and superstructures that span them indicate that the level to draw the division is somewhat arbitrary. Recently the Insect Brain Name Working Group has generated a standardized set of 33 neuropils building off of previous efforts at generating a standard brain [4, 5](Ito et al., submitted). Although the choice of neuropil boundaries that best reflect the underlying circuitry can be debated, the effort at standardization makes them a good set to analyze based on current knowledge and common terminology.
Most NBs in the CNS have similar proliferation patterns where repeated asymmetric divisions generate a series of ganglion mother cells (GMCs), which divide once to produce a Notch-high A sibling and Notch-low B sibling [6, 7]. Serially produced neurons that share an A or B fate tend to be of the same neuronal class, such as LN vs. PN, and has led to the concept of “hemilineages” [8]. More recently the posterior asense-negative (PAN) or type II NBs [9, 10] have been found, which generate a series of intermediate neural progenitors (INPs) through asymmetric divisions that then produce a relatively short series of GMCs. Most NBs undergo two periods of proliferation, one during embryogenesis that generates the larval nervous system and a second during larval development that generates the adult nervous system [11-13]. The only exceptions are the MB NBs and the lateral lineage of the AL (lAL), which skip the quiescent period beginning in late embryogenesis to generate many more neurons than most lineages [11]. Another exception to this pattern is the NB precursors of the OL, which form a separate neuroepithelium that proliferates to generate many NBs before producing migratory neurons that do not maintain cell body clustering [14].
Technau and co-workers have identified 106 uniquely identifiable NBs that delaminate in a stereotyped spatiotemporal pattern within the procephalic neurogenic region of early Drosophila embryos [15, 16]. The procephalic region plus the OL primordium form the supraesophageal ganglion (SPG), which is fused with the subesophageal ganglion (SEG) that together form the adult fly brain. As the OL has a more complex clonal structure, we focus on the central portion of the SPG, which is termed the cerebrum (Ito et al. submitted), to analyze the relationship between NBs and brain anatomy. In both the brain and thoracic ganglion of larvae, NB clones containing immature larval born neurons generate just one or two well-defined tracts [12, 17], but it is unclear if this simplicity persists in the adult brain.
Through clonal analysis of larval born neurons labeled with ubiquitous drivers, we identified 95 stereotyped neuronal lineages in the adult Drosophila cerebrum. The clones show lineage-characteristic features in clone size, cell body distribution, neurite projection, and neuropil innervation. Most clones show immediate neurite elaborations in specific local neuropil(s) before targeting other brain regions. Using these clones aligned to a pre-selected brain [4, 18], we outline the strongest clonal trajectories between neuropils. In sum, this exhaustive clonal study has uncovered the majority of the developmental building blocks of the adult Drosophila cerebrum, paving the way for single cell lineage mapping of brain development and circuitry to ultimately build a complete cellular and developmental fly brain map.
Results
Identification of 95 NB lineages
In order to clearly visualize all the progeny present in one NB clone it is necessary to generate sparse clones with ubiquitous drivers. However, the lack of cell type specificity in ubiquitous drivers requires a genetic strategy that can label one to two NBs out of the hundreds of NBs and even larger pool of GMCs dividing during larval development. In order to achieve the necessary specificity we utilized ‘flip-out MARCM,’ [19] a composite strategy where mitotic MARCM clones [20] and flip-out of a stop cassette blocking GAL4 expression were both under control of the FLP recombinase driven by a heat shock inducible promoter. Notably, flip-out MARCM significantly increased the specificity of clone induction as evidenced by a drastic reduction of background clones presumably due to the requirement of two distinct FLP-mediated events.
Using flip-out MARCM, we collected confocal stacks for over 1,500 of our most sparsely labeled NB clones in the cerebrum generated upon larval hatching. Such NB clones lack embryonic-born primary neurons. We grouped clones based on cell body cluster position followed by neurite projection pattern and found stereotypy in cell body distribution and neurite elaboration among members of the same group (e.g. Figure 1, Table S1). This stereotypy facilitated identification of neuronal lineages and is consistent with previous work where individual precursors generate defined sets of progeny neurons. For example, clones of the SLPa&l1 lineage (see below for the naming convention) consistently contain two cell body clusters located near the anterior dorsolateral corner of the cerebrum (Figures 1A & B). Their neurites selectively innervate the lateral domain of the SLP, the Clamp (SCL/ICL) surrounding the MB peduncle, the LAL, the WED, and the LO in the optic lobe (see Fig. 3 for a neuropil schematic). Distinct lineages exhibit different characteristic morphologies, such as the neurite entry bundle where the primary neurites from a cell body cluster enter the neuropil, allowing unambiguous separation of nearby clones into different lineage groups. For instance, the neighboring LHl1 lineage carries only one cell body cluster and specifically targets the LH, SLP, and SIP plus the laterally located AVLP and PVLP in addition to the LO of the optic lobe (Figures 1C & D).
Figure 1. Representative NB clones with stereotyped morphologies.
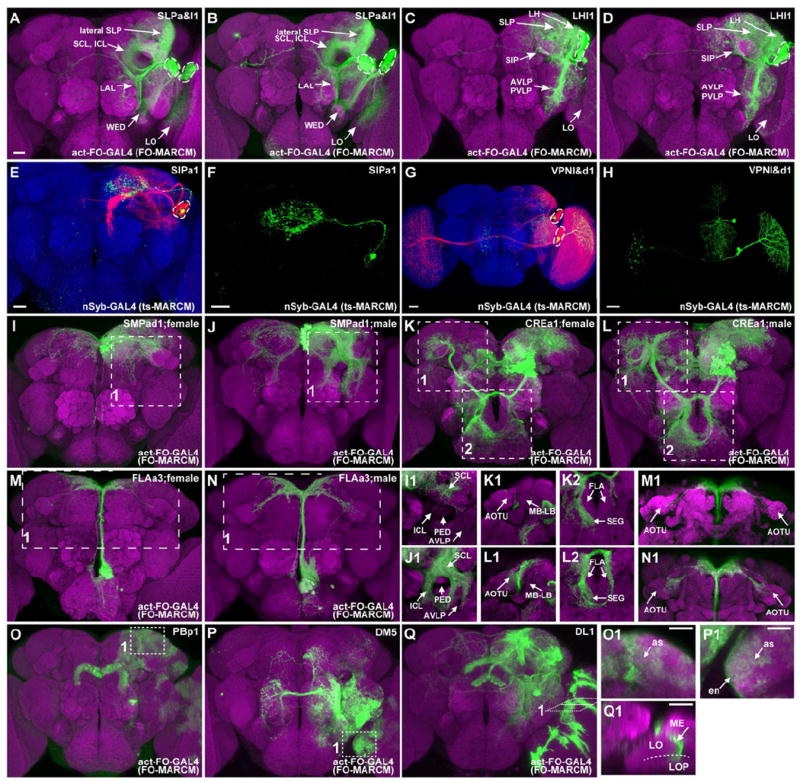
(A-D) Merged confocal images of MARCM clones (green) in nc82-counterstained adult Drosophila brains (magenta). The same NB clone (SLPa&l1) was hit in [A] and [B]; a neighboring but distinct clone (LHl1) were identified in [C] and [D]. Note their possession of two versus one cluster of cell bodies (dashed circles) and the innervation of distinct sets of neuropils (arrows).
(E-H) Adult SIPa1 (E, F) and VPNl&d1 (G, H) clones, induced shortly after larval hatching, were labeled with twin-spot MARCM. As revealed from the paired GMC clones (green), the first larval-born GMC yielded only one viable neuron in the SIPa1 lineage but produced two neurons in the VPNl&d1 lineage. Note the segregation of the twin VPNl&d1 neurons with distinct projections (H) into each of the two cell body clusters (dashed circles) present in the NB clone (G).
(I-N) SMPad1, CREa2, and FLAa3 clones exhibit gender-specific neurite elaborations, as pointed out with arrows in the subpanels of [I] - [N].
(O-Q) PBp1, DM5, and DL1 clones show glia-like elaborations. Close-up views (insets) reveal astrocyte-like (as) glia in the PBp1 clone, both ensheathing (en) and astrocyte-like (as) glia in the DM5 clone, and optic lobe glia separating the medulla (ME) and lobule/lobule plate (LO/LOP) in the DL1 clone.
Scale bars, 20 μm. Spatially segregated background clones were removed in some cases.
Figure 3. Cell body distribution of neuropil-characteristic neuronal lineages.
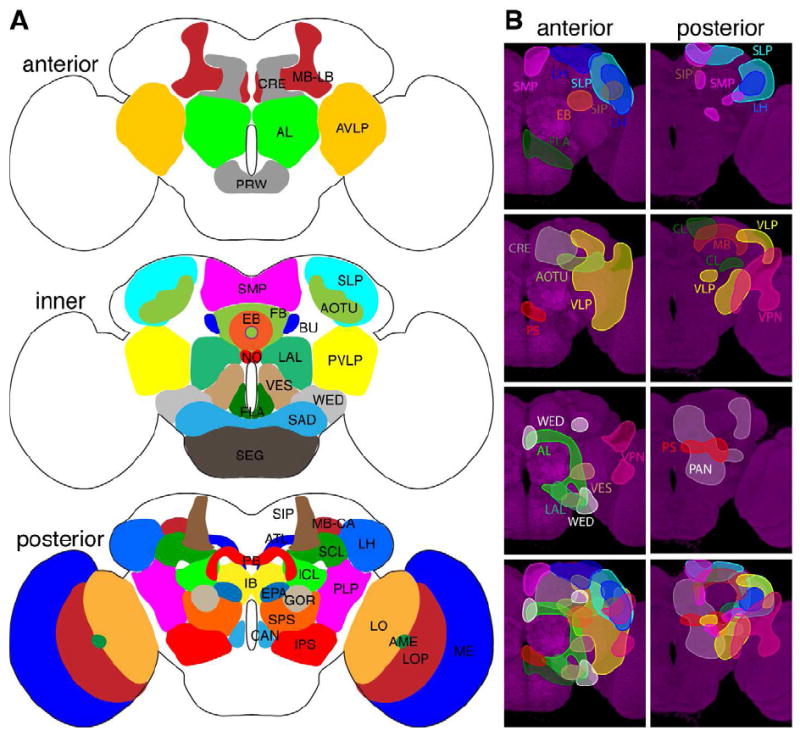
(A) Illustration of neuropils that have been arbitrarily assigned to three cross sections in the adult Drosophila brain. Anterior neuropils, AL: antennal lobe, AVLP: anterior ventrolateral protocerebrum, CRE: crepine, MB-LB: mushroom body lobe, PRW: prow. Inner neuropils, AOTU: anterior optic tubercle, BU: bulb, EB: ellipsoid body, FB: fan shaped body, FLA: flange, LAL: lateral accessory lobe, NO: Noduli, PVLP: posterior ventrolateral protocerebrum, SAD: saddle, SEG: subesophageal ganglion, SLP: superior lateral protocerebrum, SMP: superior medial protocerebrum, VES: vest, WED: wedge. Posterior neuropils, AME: accessory medulla, ATL: antler, CAN: cantle, EPA: epaulette, IB: inferior bridge, ICL: inferior clamp, IPS: inferior posterior slope, LH: lateral horn, LO: lobula, LOP: lobula plate, MB-CA: mushroom body calyx, ME: medulla, PB: protocerebral bridge, PLP: posterior lateral protocerebrum, SCL: superior clamp, SIP: superior intermediate protocerebrum, SPS: superior posterior slope.
(B) Illustration of neuropil-characteristic clonal cell body distributions on the anterior or posterior brain surface. Clonal cell body loci for various neuropils shown in different colors in the top panels are superimposed in the bottom panel, to reveal the overall coverage by the identified clones.
Through manual annotation of the detailed neuropil innervation patterns, we established the presence of 92 stereotyped clones (Table S1; Figure 2; See Figure S2 for larger images of the clones). Given four ‘equivalent’ copies of the MB lineage that yield undistinguishable neurons during post-embryonic development [21], we have identified 95 neuronal lineages per cerebral hemisphere. The difficulty in distinguishing the four MB lineages based on adult clone morphology suggests our catalog of clones may actually cover more than 95 distinct NB lineages. Given the large range in clone frequency (Table S1), it is not possible to use this sort of information to guess if certain NBs are indistinguishable duplicates as in the MB case. The fusion of segments in the brain and integration of adjacent neuropils complicates assignment of NB clones bordering the OL and SEG such that a few may have inadvertently been missorted. As there is currently no suitable technique for reliably tracking NBs between the embryo and adult, position and lack of neuronal migration were used as criteria to focus on the cerebral NB clones.
Figure 2. Catalog of cerebral NB clones.
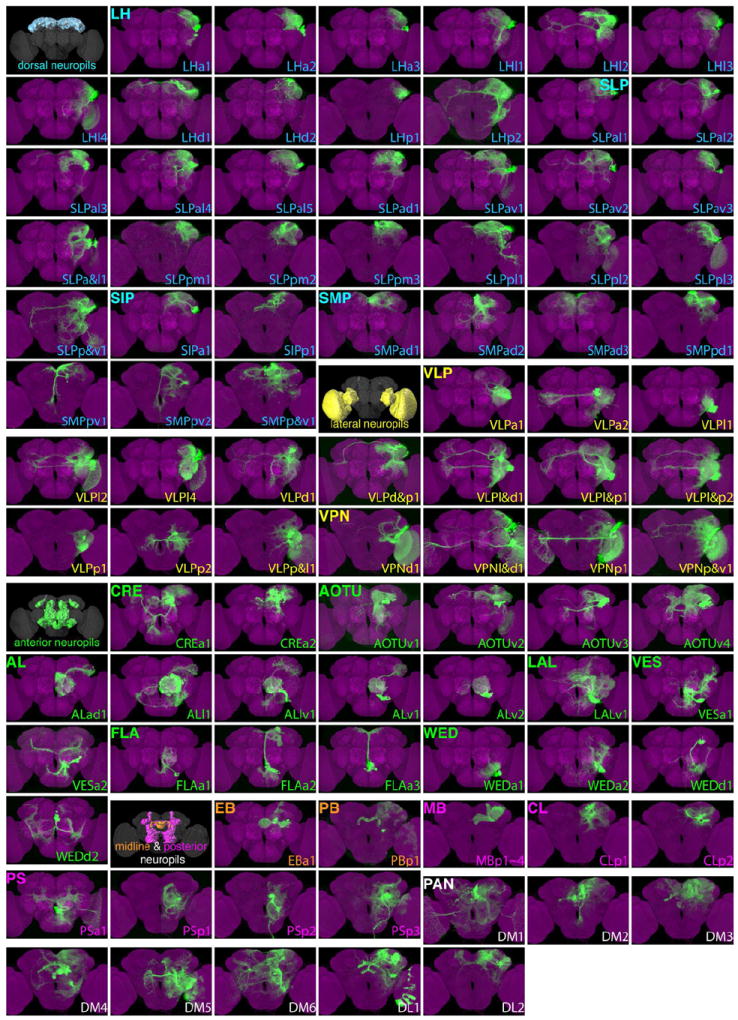
Ninety-two stereotyped NB clones (green) shown individually after warping into an nc82-counterstained adult Drosophila brain (magenta). They are named according to their primary immediate neuropil targets, referred to as home neuropils, and cataloged by grouping home neuropils into dorsal, lateral, anterior, and midline/posterior neuropil sets. Note the brains are shown with the anterior or posterior surface up depending on the location of the clone cell bodies. Spatially segregated background clones were removed in some cases. Note that the PSa1 clone originates from anterior brain surface. Please find the same images at higher resolution in supplemental Figure S2 and the raw 3D images in Virtual Fly Brain: www.virtualflybrain.org.
Lineage-characteristic clone sizes and morphologies
Distinct lineages differ greatly in clone size and some lineages carry two clusters of cell bodies with independent neurite tracts. Counts of cell bodies demonstrated that each lineage produced a characteristic number of offspring that survived into the adult stage (Table S1). Although most lineages generated 30-150 neurons, the MB, ALl, and PAN lineages generate ~200 or more neurons due to their special proliferation patterns. There were also two notably small lineages FLAa1 and PBp1 lineages that contain only nine neurons (Table S1 & Figure S1A). Such ultra-short lineages might result from premature NB death [22].
To assess the fates of the first larval neurons derived from a given NB, we repeated the clonal analysis with twin-spot MARCM, which allows differential labeling of sister clones and thus the detection of NB clones with their paired GMC (or INP) progeny (Figures 1E-H) [23]. We obtained NB clones for 73 of the 95 cerebral lineages and saw that each had a characteristic first progeny. Distinct NB clones associated with different numbers of post-mitotic neurons (Table S1). First, we validated seven PAN lineages as their NB clones consistently pair with 6 to 9 mature neurons. However, the much smaller eighth PAN NB clone (the DL2 lineage) paired with only one adult neuron, requiring other strategies to confirm its PAN NB origin (manuscript in preparation). The six dorsomedial PAN NB clones correspond to the known larval DM1-6 lineages, while the two dorsolateral PAN NB clones are referred to as the DL1 and DL2 lineages respectively. Second, other NB clones paired with no more than two neurons, consistent with their generating GMCs as in other lineages. In cases where NB clones have two cell body clusters and paired with two viable neurons, we consistently observed presence of one first larval-born neuron in each cell body assembly (Figures 1G & H) [17]. As implicated from the ensembles’ morphologies, these sister neurons show distinct cluster-specific neurite projections indicating a sister hemilineage relationship between the separate ensembles from the same lineage. In addition, we have detected in different individuals not only the same cell number (Table S1), but also undistinguishable morphology for the first born larval offspring, supporting the notion that diverse neurons arise from each neural progenitor in a stereotyped sequence.
Notably, two thirds of single-cell-paired NB clones carry fewer than 80 neurons. By contrast, two thirds of two-cell-paired NB clones contain more than 100 neurons. These observations support the interpretation that hemilineage-dependent cell death plays an important role in NB clone size differences (Figure S1B). In clones with two distinct cell clusters, the smaller cluster contains roughly 40% of the neurons in the larger cluster (Table S1), suggesting a significant amount of temporally controlled cell death may also occur. The LHa1 and WEDd2 lineages exhibit an extreme example of this phenomenon with one separate neuron with a unique projection paired with the main cell body cluster (Figure 2). Differential apoptosis may also explain why the DM5 and DL2 PAN NBs produce many fewer neurons than the other PAN clones [24].
Few NB clones show obvious sexual dimorphism or glia-like elaboration
Three lineages, including SMPad1, CREa1, and FLAa3, exhibit obvious gender-dependent dimorphic phenotypes (Figures 1I-N, Table S1). Notably, the male clones of all three lineages carry more cells and elaborate more exuberantly than their female counterparts. The SMPad1 lineage shows male-specific elaborations around the MB peduncle in both brain hemispheres, especially within the ipsilateral SCL, ICL, and AVLP (Figures 1I & J). The CREa1 lineage differentially innervates the contralateral AOTU/SLP/MB-LB and the ipsilateral FLA/SEG in male versus female brains (Figures 1K & L). The FLAa3 lineage, by contrast, acquires bilaterally symmetric neurite trajectories that prominently extend into the AOTUs in the male brain only (Figures 1M & N). These sexually dimorphic cerebral lineages probably contain the aSP-a/aSP2, aDT-b/mAL/aDT2, and aDT-h/mcALa/aDT6 clusters of Fru-expressing neurons, respectively [25-27]. The sexual dimorphism of the CREa1 clones has been shown to result from programmed cell death of neurons with malecharacteristic projections in the female brain lacking male-specific Fru, again highlighting cell death in shaping lineage composition [28].
Three lineages, including PBp1 and two PAN lineages, make glia-like progenies occupying large brain territories (Figures 1O-Q). The PB lineage yields about nine neurons that exclusively innervate the PB, but produces many astrocyte-like cells that scatter in the dorsal and lateral cerebrum and further into the optic lobe (OL). By contrast, the DM5 PAN lineage deposits heterogeneous populations of neurons and glia. We detected ensheathing as well as astrocyte-like glia in DM5 NB clones. Notably, the astrocyte-like glia of the PB and DM5 clones show a complementary distribution. In both the cerebrum and OL, the glial cells of PBp1 lineage populate more dorsolateral areas than the analogous DM5-derived glia. The origin-dependent production of specific glial subsets is also evident in the DL1 PAN lineage, which makes elongated OL glia that align at the interface between LO/LOP and ME.
There should be additional sexually dimorphic lineages, including possibly male-only lineages, that carry Fru-positive neurons [25-27]; and most, if not all, of the PAN lineages yield neurons plus Repo-positive glial offspring [29, 30]. We tentatively mapped major Fru-positive clusters onto those NB clones whose sexual dimorphism could be masked by the gross complexity (Table S1), but it is difficult to map fru clones containing only a few neurons. To unveil all sexually dimorphic or glia-producing neuronal lineages requires more detailed morphological analysis of not only entire clones but also their specific constituents.
Regional clustering of neuropil-characteristic neuronal lineages
Notably, almost all cerebral clones have an entry bundle that leads to one or more local neuropils, suggesting regional establishment of neuropils may be a general phenomenon. Our clones further group well based on the defined neuropil regions and roughly support the current neuropil definitions. We therefore use a regional naming convention based on the main adjacent neuropil except for the eight PAN NBs, which have complex projection patterns and are more simply designated by cell body locations.
Eighty-four stereotyped clones were individually assigned a ‘home’ neuropil based on prominent proximal neurite elaboration and subsequently grouped into 18 neuropil-oriented families. They were then named following the pattern NPp#, where NP is the neuropil, p is the cell body position, and # is the number that together yield a unique name. Our collection and the independent effort by Ito et al. have been coordinated to follow the same naming convention and clone assignment. For example, the two collections have jointly uncovered 12 non-PAN clones that show prominent immediate neurite elaboration in the LH and are thus placed into the LH lineage family. The cell bodies of these LH clones lie anterior (a), lateral (l), dorsal (d), or posterior (p) to the LH and are therefore called LHa1-4, LHl1-4, LHd1- 2, and LHp1-2, respectively. 10 of the 12 LH lineages, excluding LHa4 (unique in the Tokyo collection) and LHd2 (unique in the Janelia collection), were dually identified (Table S1). For known lineages, such as adPN or lAL, we renamed them ALad1 and ALl1, which essentially maintains the original naming. In common use we expect people to give the full name prior to simplifying to the minimal unique length such as ALad. In this way if additional clones are found in a group uniqueness will be maintained, but shorter names can be used in practice. In addition to the eight PAN lineages, we identified 11 LH lineages, 17 SLP lineages, 2 SIP lineages, 7 SMP lineages, 13 VLP lineages, 4 VPN (Visual Projection Neuron) lineages (with the OL as the home neuropil), 2 CRE lineages, 4 AOTU lineages, 5 AL lineages, 1 LAL lineage, 2 VES lineage, 3 FLA lineages, 4 WED lineages, 2 CL lineages, 4 PS lineages, 1 PB lineage, 1 EB lineage and 4 MB lineages (Figure 2).
Some neuropils host many distinct lineages while certain neuropils are pioneered by just a few lineages. For instance, the 17 SLP lineages occupy different subregions within the SLP and further relay the zone-specific information to distinct brain domains. This could reflect the relatively large size or substructural complexity of the SLP and its possible function as a regional hub in the neural circuitry of the cerebrum. By contrast, the CRE is mainly patterned by two lineages with analogous CRE-related neurite elaboration and projection patterns. Interestingly, the most obvious difference between the CREa1 and CREa2 clones is the extra sexually dimorphic neurite tract system that primarily innervates regions outside the CRE. We did not find lineages selectively dedicated to 8 neuropils: the anterior PRW/SAD, the central FB/NO, and the posterior ATL/IB/EPA/GOR/CAN neuropils. The lack of obvious neuropil-specific lineages could result from derivation from the SEG (e.g. PRW, SAD, CAN), the PAN lineages (e.g. FB, NO, ATL, IB), or assignment of the clone to adjacent neuropils where the NB elaborates more extensively (e.g. SAD, ATL, EPA, GOR). The ALl lineage produces non-AL neurons based on a fate switch in the PN hemilineage leading to innervation of other target neuropils including the SAD and SEG [31]. This indicates anatomical neuropils and developmental origin are related, but not necessarily in a one to one fashion.
Given the minimal migration of post-mitotic neurons, the clonal cell body positions should roughly reflect the responsible progenitors’ relative developmental loci in the brain primordium. Mapping the cell body distribution for clones that coinnervate a given neuropil should therefore hint where the neuropil arises. Notably, the clonal-unit families of dorsal neuropils (LH/SLP/SIP/SMP) as well as lateral neuropils (VLP/OL) consist of clones whose cell bodies may reside anterior, posterior, dorsal or lateral to the involved neuropils that jointly cover the dorsal and lateral periphery of the cerebrum (Figure 3B). By contrast, centrally located neuropils originate largely from either anterior or posterior situated clones (Figure 3B).
Neuropil connectivity deduced from clonal innervation
It has been proposed that the neural circuits of the Drosophila brain are generated in a modular fashion from a series of clonal units [12, 17, 32]. However, only seven lineages have restricted elaboration in five or fewer neuropils; and over 40 lineages target 15 or more brain compartments indicating the situation is more complex (Figure S1C). The innervation of multiple neuropils by a given clone is likely to reflect neuronal diversity with overlapping projection patterns complicating determination of neuropil connectivity. Given that our clones cover 90% of the neurons in the cerebrum and there are clear trends in neuropil connectivity, we combined manual analysis of projection patterns in neuropils generated by two or fewer clones with computational analysis of preferred distal targets shared by the majority of clones in the larger families. By analyzing “anatomical connectivity” between neuropils rather than connectivity at the neuronal level we avoid the difficulties of determining neuronal connections at the light level. Although this approach may not be optimal, it is straightforward and provides a map of the major NB based connections that can be refined by future single cell lineage studies.
Toward this aim, we first aligned 95 clean clones (representing 92 distinct lineages plus three male clones showing sexual dimorphism) with a fly brain template containing 33 segmented neuropil regions in each hemisphere [4, 18]. This allowed us to compute the fractional innervation of each neuropil by each NB, which can be conveniently visualized as a heat map (Figure 4). To use the clonal innervation to generate directional connectivity we made the simplifying assumption that the proximal home neuropil was input and any other neuropil with 20% or higher voxel coverage by more than half family members was a target neuropil or output neuropil (Figure 5). The threshold of 20% was simply set to minimize issues with alignment, neuropil boundaries, and emphasize strong connections. Although the approach may not be optimal, the results are reasonable upon inspection and it fulfills the goal of providing a rough guide to connectivity without attempting to over interpret our data. Due to their complex innervation patterns and lack of home neuropil, the PAN lineages were not included in this analysis. For families that consist of no more than two lineages, we identified the distal targets by manually following any neurite fascicles that could be reliably tracked in individual clones. In addition, we included the FLA-to-SMP and the SMP-to-contralateral FLA projections, as evidenced in the relatively simple FLAa3 and SMPpv1 clones, which create some circular connections among the bilateral FLAs and SMPs. We combined these multiple lines of information to derive a neuropil connectivity matrix between the identified ‘home’ neuropils and the entire set of brain neuropils (Figure 5B). One can thus predict the inputs as well as outputs for most neuropils. However, we provide no insight into the output from neuropils that lack obvious founding lineages (e.g. SAD) or are founded by lineages that do not project out of the neuropil (e.g. MB and PB). The complexity of the PAN lineages severely limits inference of connectivity, but may relate to the integrated, but modular composition of the CX. Together with other data, this connectivity will be analyzed elsewhere (Yang et al. submitted).
Figure 4. Heat map of neuropil innervations by various NB clones.
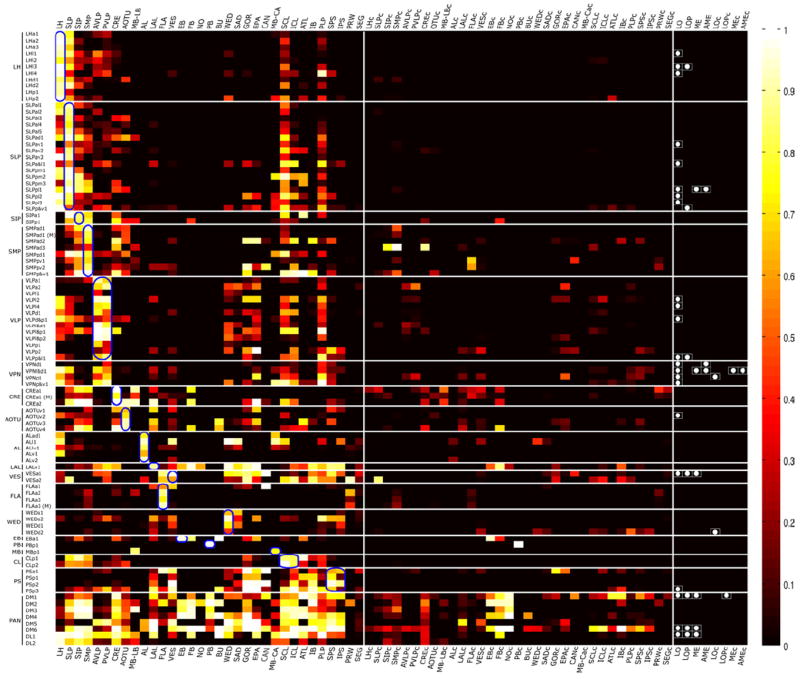
Degrees of voxel coverage in distinct neuropils in the ipsilateral as well as contralateral hemisphere by 95 representative NB clones, including one female sample from each of the 92 stereotyped lineages plus three male clones showing obvious sexual dimorphism. Note the neurite elaborations in various optic lobe neuropils are simply indicated with white dots. Blue circles indicate home neuropils.
Figure 5. Putative clonal-level neuropil connectome.
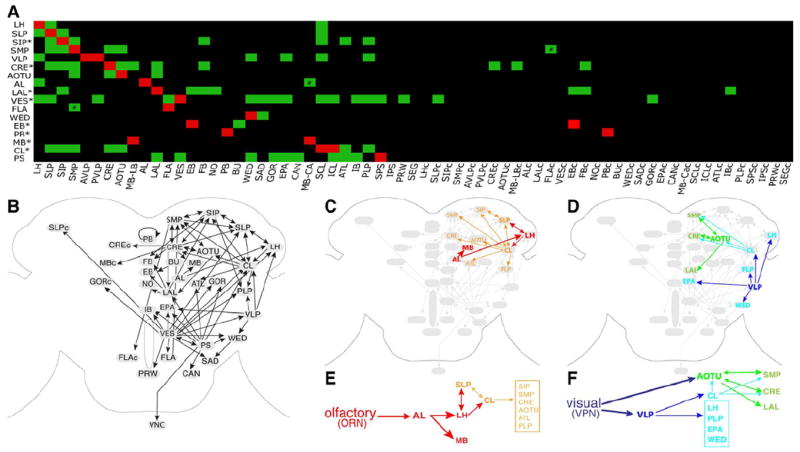
(A) The likely connections between various home neuropils (listed on the Y-axis) and other neuropil regions (arranged along the X-axis) are indicated with green boxes. This putative neuropil connectome was deduced primarily through the determination of the major distal targets shared by most of the NB clones cofounding a home neuropil, as revealed from the heat map of clonal neuropil innervation patterns (Figure 4). For those home neuropils founded by no more than two lineages (*), their possible distal targets were identified by manually tracking the readily traceable neurite fascicles in individual clones. The reciprocal connections between the SMP and FLA (#) are evident in the relatively simple FLPa3, and SMPpv1 clones; the AL-to-MB-CA connection (#) is known.
(B) Diagrams of possible information flow across distinct neuropils in the Drosophila cerebrum, as judged from the neuropil connection matrix shown in [A]. And neural activities presumably propagate from home neuropils to their connected neuropils, except that the EB lies distal to the BU in the EBa1 clone, the sole EB founding lineage. The putative sub-networks that process olfactory or visual information are illustrated in additional diagrams.
The connectivity matrix correctly identified the LH and MB calyx as the main targets of the AL lineage family, which process the olfactory information downstream of the AL. This motivated us to use the neuropil connectivity matrix to build a rough map of anatomical connectivity for less studied brain regions (Figure 5B). Tracing the circuit downstream of the AL reveals that the olfactory information processed by the LH [33, 34] may enter the SLP and SCL, which can further communicate with various neuropils, including the SIP, SMP, CRE, AOTU, ATL, and PLP (Figures 5C & E). By contrast, the visual information can be processed through the AOTU [35] into the SMP, CRE and LAL (the CX input/output center) or computed via the VLP [35, 36] into the SCL/ICL (CL), LH, PLP, EPA, and WED (Figures 5D & F). Notably, the CL may relay the VLP-processed visual information back to the AOTU. In addition, reciprocal connections exist broadly among higher brain centers (bidirectional arrows in Figure 5B). For example, extensive mutual innervations are evident among neighboring neuropils in the dorsal brain domain and between the dorsal neuropils and CRE/CL.
In our analysis the primary cerebral output appears to pass through the posterior slope (SPS/IPS) to the ventral ganglion. Interestingly, this neuropil has projections to a number of neuropils including the LAL, VES, WED, GOR, EPA, CAN, ATL, CL, and IB. This may suggest that the current output is relayed throughout the brain. Interestingly all PAN lineages innervate either the SPS or IPS except for DL2. Given the role of the PAN lineages in establishing the CX and the role of the CX in navigation and locomotion, just such a connection from the CX to an output region like the PS would be expected. We did not uncover the SEG as a major distal target for any cerebral home neuropil, although its intermediate position makes it a potential output region. However, some neuropils such as the FLA, PRW, and CAN are not well separated from the SEG in neuropil staining and may play such an intermediate role through clones like FLAa2 and PSp3 that elaborate rather broadly in the SEG.
Discussion
The stereotyped nature of NB clones indicates extensive lineage-intrinsic neural development where the unique fate of each NB at delamination programs it to make a specific set of distinct neurons [37]. Distinct siblings probably arise in an invariant sequence, as the first larval-born GMCs reproducibly generate clones with the same morphology. Such birth order/time-dependent neuron fate specification is evident even in the complex PAN lineages that consistently produce the same sets of offspring from the same INPs (manuscript in preparation). Although the length of lineages and the rate of NB divisions could affect final clone sizes, patterned apoptosis governed by hemilineage identity and neuronal temporal fate seems to account for much of the difference in lineage cell number [6, 17, 38].
Likely coverage of the 106 NBs of the procephalic origin
Urbach et al. have identified 106 unique NBs that are formed in a stereotyped spatiotemporal pattern on either side of the procephalic neurogenic region of early Drosophila embryos that underlie the formation of the Drosophila cerebrum [15, 16]. Given the increased interest in Drosophila behavior and brain anatomy, we have determined the morphology of 95 NB clones, about 90% of the expected 106, and examined how they contribute to diverse neuropils. We believe that a similar number of NBs generate the adult nervous system based on counts of large Dpn-positive cells during larval neurogenesis (data not shown). The 95 lineages we outline here should generate roughly 10,100 neurons with an average (not including the PAN, MB, or ALl lineages) of about 70 neurons surviving in adult, suggesting all 106 lineages should generate about 10,800 larval born neurons. These numbers begin to provide increasingly detailed target information for refining our knowledge of the composition of the Drosophila brain. Most of the uncertainty in determining our coverage is that the extensive fusion of neuropils clouds the developmental origin of clones at the boundary of our region of focus. However, the difficulty in comprehensively tracing NBs from embryonic to adult stages makes this an unavoidable issue. Given this fusion, our coverage of NBs contributing to the cerebrum is fairly robust and largely a developmental accounting issue from an anatomical standpoint. In our opinion, the greater issue is developing future strategies that permit comprehensive analysis of the SEG and OL NBs.
Independent efforts by Ito et al. at Tokyo University have recovered a comparable number of NB clones in the adult Drosophila cerebrum. Cross-comparison based on 3-D images revealed 77 clones present in both collections, leaving 19 Tokyo clones (including two two-cell clones with unclear lineage identity) and 18 Janelia clones unmatched. This yields a combined set of 114 NB clones. There are 13 VPN clones at the cerebrum/OL border, including nine Tokyo-unique VPN clones. Some of them may derive from the OL and had been excluded from our collection because of variation in twin clone size suggesting a more complex proliferation pattern (data not shown). Together the two collections jointly cover 114 lineages and are likely to cover almost all of the 106 NB clones of the procephalic origin plus some OL-derived VPN clones.
Complex interrelation between lineages and neuropils
The adult Drosophila cerebrum appears as an indivisible structure, showing no anatomical evidence for its development from three procephalic neuromeres, namely the tritocerebrum, deutocerebrum, and protocerebrum. Similarly, the fusion of the SEG with the central brain is another example of the trend towards greater integration of distinct segmental circuitry over the course of evolution. This trend from local circuitry towards more integrated circuits has occurred throughout the various metazoan kingdoms [39]. The complex interrelation between lineages and neuropils could partially result from the fusion of segments and other neuropil units to integrate information across the brain to generate coherent behavior from diverse circuitry.
However, despite omitting highly diverse primary neurons, many clones have extended neurites along discrete tracts and apparently contributed to distinct circuits. The existence of clones innervating multiple separate neuropils may be an indication that they contain greater diversity in neuronal class than the hemilineage based fating mechanisms inherent to the GMC program. In support of this interpretation, the derivation of PN classes of various sensory modalities from a single lAL hemilineage has recently be shown to involve Notch signaling to accomplish a binary fate switch within a hemilineage that is analogous to GMC sister fating [31]. This blurs the distinction of neuron class specification based on hemilineage origin versus neural type specification through GMC birth order.
NB lineages as modules for evolution
The existence of 4 highly similar yet distinct MB lineages [40] suggests that expansion of brain anatomy during insect evolution has involved reuse of existing NB fating programs [41]. Such mechanisms may explain the derivation of neurons of the same class from hemilineages of distinct NB origin such as uniglomerular PNs from the ALl and ALad lineages. If the set of NB lineages laid out a simple set of modular evolutionary building blocks their determination would lay out a clear road map of brain anatomy, but this is unfortunately not the case. The NB mode of specification appears to have arisen within crustacea and likely involved packaging of preexisting neuronal cell types into a modular developmental unit derived from a single precursor [42]. The complexity of the sets of neurons in a NB suggests that there may be just such a lack of modularity in the ancestral set of neurons. If selection acted to expand a certain neural class through duplication of a NB, the differential requirement for distinct subtypes might be expected to lead to selection for developmentally programmed cell death to remove unnecessary neurons – a prominent feature in our data.
Anatomical brain connectivity
As to the detailed neural network, the existence of few clones with simple tracts and obvious polarity limits our ability to interpret this information. The LH and VLP/AOTU family of clones suggest convergence of olfactory and visual information in the SCL and CRE (Figure 5). For those interested in inputs into a specific neuropil, our map provides potential clues as to input pathways. Notably, the neuropils (e.g. VLP and SAD) already implicated in specific sensory modalities receive inputs from limited sources whereas most deep neuropils are targeted by diverse clones and possibly integrate diverse information. By analyzing the inputs and outputs together certain neuropils appear to cluster into functional networks such as the SMP, SIP, and CRE or the SIP, SLP, and CL.
However, single-cell analysis is essential to validate these hypothetical circuits and refine our understanding of anatomical connectivity. In contrast to the existing fly circuit map [43] that generated single neuron clones from broad neuronal drivers, our study lacks the same resolution. However, our approach allows determination of coverage, giving significant advantages in gauging whether important anatomical connections are missing based on driver choice or other technical considerations. While Chiang et al. [43] emphasize the importance of single cell patterns in defining neuropil boundaries, it is difficult to judge the meaning of neuropil units defined from incomplete single neuron sampling. Until a more agreed upon set of neuropil boundaries is determined, lack of a standard neuropil set makes cross comparison difficult. However, their study found few connections with the CMP, which corresponds to the PS region, and appears to be an important output neuropil suggesting their sampling may miss important regions. Although our NB connectivity likely covers 90% of the neurons in the cerebrum, our strategy to filter out the strongest connections reduces our sensitivity to detect minor connections. We plan to utilize and extend our NB atlas to systematically map the lineages of the Drosophila brain at single cell resolution to achieve the best coverage possible.
Experimental Procedures
Induction of Drosophila NB clones
Flip-out MARCM clones were generated by heat-shocking newly hatched larvae with the following genotype: hs-FLP1/+; FRTG13, UAS-mCD8∷GFP/FRTG13, tubP-GAL80; actin5cP-FRT-stop-FRT-GAL4/+ or hs-FLP1/ actin5cP-FRT-stop-FRT-GAL4; FRTG13, UAS-mCD8∷GFP/FRTG13, tubP-GAL80; + for 15-25 minutes at 38°C. We utilized independent actin5cP-FRT-stop-FRT-GAL4 transgenes to exclude positional effects on the clone coverage, though only female brains carried clones during use of the X-chromosome driver resulting in a 3:1 female-dominant ratio. Twin-spot MARCM clones were induced by heat-shocking newly hatched larvae with the following genotype: hs-FLP1/+; FRT40A, UAS-mCD8∷GFP, UAS-rCD2-Mir/FRT40A, UAS-rCD2∷RFP, UAS-gfp-Mir; nSyb-GAL4/+, for 10 minutes at 38°C. The clones of six lineages, including LHd2, SLPpm1, SIPp1, SMPad3, SMPpv2, WEDd1, were obtained via embryonic NB-specific stochastic excision of the STOP cassette from actin5cP-loxPstop-loxP-lexA∷p65 (details will be described elsewhere).
Immunostaining and confocal imaging
Brains were dissected, fixed, and processed as described previously [20, 23]. Antibodies used in this study include rabbit anti-GFP (1:1000, Invitrogen, Carlsbad, CA), rat monoclonal anti-mCD8 (1:100, Invitrogen), rabbit anti-RFP (1:1000, Clontech), mouse monoclonal anti-Bruchpilot, nc82 (1:50, DSHB; Developmental Studies Hybridoma Bank, Iowa City, IA), Alexa-488 (Invitrogen), Cy3 (JIR; Jackson Immuno Research, West Grobe, PA) or Cy5 (JIR) conjugated anti-mouse, rabbit and rat antibody (1:300). Fluorescent signals of whole-mount adult fly brains and cell bodies for counting cell number of each lineage were collected by confocal serial scanning at 0.8 or 1.0 μm intervals using LSM710 microscope (Carl Zeiss, Oberkochen, Germany).
Manual analysis of neuropil innervation patterns and clone sizes
For manual analysis of neuropil innervation patterns, a maximum intensity projection of each confocal Z-stack was generated. Similar patterns of 2-D confocal images of NB clones were initially sorted into same groups. Confocal image stacks of all collected NB clones were then carefully re-examined to refine grouping accuracy. Neuropil innervation patterns were annotated based on clear staining within nc82-identifiable neuropils. The three highest quality samples of each lineage were then used to count cell number with a Fiji [44] macro written by Arnim Jenett (available at http://janelia.org/janelia-technology/available-technology).
Heatmap Generation
Confocal stacks of each clonal lineage were registered using BrainAligner [18] to a pre-selected brain with predefined neuropil boundaries [4, 18]. For each aligned stack, we extracted GFP signals using adaptive thresholding and visually confirmed the threshold appropriately detected the signal from the clone. The fraction of voxels above the threshold was computed for each neuropil with cases of extremely low voxel coverage not included. The manual annotation of neuropil innervation was compared against the automated annotation and discrepancies were resolved (e.g. confirmation of missed neuropils and removal of false positives adjacent to the strong signal in cell body regions).
Supplementary Material
01
Highlights.
- Determination of 90% of the Drosophila cerebral neural progenitors.
- Demonstration that these units have more neuronal diversity than expected.
- Estimation of neural number and cell death and description of developmental trends.
- Generation of an anatomical map of neural circuits of the Drosophila cerebrum.
Acknowledgments
We thank Janelia Farm fly core and fly light team for various technical supports in the data production. We thank Arnim Jenett for sharing the fly brain neuropil masks and the Fiji macro for counting cell bodies, Julie Simpson for nSyb-GAL4, Barret Pfeifer for advice on the construction of flip out constructs, Jim Truman for helpful discussions, and Crystal Sullivan for administrative support. This work was supported by Howard Hughes Medical Institute and National Institute of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stocker RF, Lienhard MC, Borst A, Fischbach KF. Neuronal architecture of the antennal lobe in Drosophila melanogaster. Cell Tissue Res. 1990;262:9–34. doi: 10.1007/BF00327741. [DOI] [PubMed] [Google Scholar]
- 2.Leiss F, Groh C, Butcher NJ, Meinertzhagen IA, Tavosanis G. Synaptic organization in the adult Drosophila mushroom body calyx. J Comp Neurol. 2009;517:808–824. doi: 10.1002/cne.22184. [DOI] [PubMed] [Google Scholar]
- 3.Young JM, Armstrong JD. Structure of the adult central complex in Drosophila: organization of distinct neuronal subsets. J Comp Neurol. 2010;518:1500–1524. doi: 10.1002/cne.22284. [DOI] [PubMed] [Google Scholar]
- 4.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rein K, Zockler M, Mader MT, Grubel C, Heisenberg M. The Drosophila standard brain. Curr Biol. 2002;12:227–231. doi: 10.1016/s0960-9822(02)00656-5. [DOI] [PubMed] [Google Scholar]
- 6.Lin S, Lai SL, Yu HH, Chihara T, Luo L, Lee T. Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain. Development. 2010;137:43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skeath JB, Doe CQ. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 1998;125:1857–1865. doi: 10.1242/dev.125.10.1857. [DOI] [PubMed] [Google Scholar]
- 8.Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- 12.Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa E, Kitada Y, Kaido M, Takayama R, Awasaki T, Tabata T, Sato M. Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development. 2011;138:983–993. doi: 10.1242/dev.058370. [DOI] [PubMed] [Google Scholar]
- 15.Urbach R, Schnabel R, Technau GM. The pattern of neuroblast formation, mitotic domains and proneural gene expression during early brain development in Drosophila. Development. 2003;130:3589–3606. doi: 10.1242/dev.00528. [DOI] [PubMed] [Google Scholar]
- 16.Urbach R, Technau GM. Neuroblast formation and patterning during early brain development in Drosophila. BioEssays : news and reviews in molecular, cellular and developmental biology. 2004;26:739–751. doi: 10.1002/bies.20062. [DOI] [PubMed] [Google Scholar]
- 17.Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- 18.Peng H, Chung P, Long F, Qu L, Jenett A, Seeds AM, Myers EW, Simpson JH. BrainAligner: 3D registration atlases of Drosophila brains. Nature methods. 2011;8:493–500. doi: 10.1038/nmeth.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 22.Kuert PA, Bello BC, Reichert H. The labial gene is required to terminate proliferation of identified neuroblasts in postembryonic development of the Drosophila brain. Biology open. 2012;1:1006–1015. doi: 10.1242/bio.20121966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 2009;12:947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Reichert H. Programmed cell death in type II neuroblast lineages is required for central complex development in the Drosophila brain. Neural Dev. 2012;7:3. doi: 10.1186/1749-8104-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GS. Sexual dimorphism in the fly brain. Curr Biol. 2010;20:1589–1601. doi: 10.1016/j.cub.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59:759–769. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Yu JY, Kanai MI, Demir E, Jefferis GS, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 28.Kimura K, Ote M, Tazawa T, Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438:229–233. doi: 10.1038/nature04229. [DOI] [PubMed] [Google Scholar]
- 29.Izergina N, Balmer J, Bello B, Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 2009;4:44. doi: 10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viktorin G, Riebli N, Popkova A, Giangrande A, Reichert H. Multipotent neural stem cells generate glial cells of the central complex through transit amplifying intermediate progenitors in Drosophila brain development. Dev Biol. 2011;356:553–565. doi: 10.1016/j.ydbio.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Lin S, Kao CF, Yu HH, Huang Y, Lee T. Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions. PLoS biology. 2012;10:e1001425. doi: 10.1371/journal.pbio.1001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito K, Awasaki T. Clonal unit architecture of the adult fly brain. In: Technau G, editor. Brain Development in Drosophila. Landes Bioscience/Springer; Austin: 2007. pp. 137–158. [Google Scholar]
- 33.Marin EC, Jefferis GS, Komiyama T, Zhu H, Luo L. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 2002;109:243–255. doi: 10.1016/s0092-8674(02)00700-6. [DOI] [PubMed] [Google Scholar]
- 34.Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 35.Otsuna H, Ito K. Systematic analysis of the visual projection neurons of Drosophila melanogaster. I. Lobula-specific pathways. J Comp Neurol. 2006;497:928–958. doi: 10.1002/cne.21015. [DOI] [PubMed] [Google Scholar]
- 36.Mu L, Ito K, Bacon JP, Strausfeld NJ. Optic glomeruli and their inputs in Drosophila share an organizational ground pattern with the antennal lobes. J Neurosci. 2012;32:6061–6071. doi: 10.1523/JNEUROSCI.0221-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Technau GM, Berger C, Urbach R. Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev Dyn. 2006;235:861–869. doi: 10.1002/dvdy.20566. [DOI] [PubMed] [Google Scholar]
- 38.Yu HH, Kao CF, He Y, Ding P, Kao JC, Lee T. A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM. PLoS biology. 2010;8 doi: 10.1371/journal.pbio.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bullock TH, Horridge GA. Structure and Function in the Nervous System of Invertebrates. Netherlands: W. H. Freeman; 1965. [Google Scholar]
- 40.Kunz T, Kraft KF, Technau GM, Urbach R. Origin of Drosophila mushroom body neuroblasts and generation of divergent embryonic lineages. Development. 2012;139:2510–2522. doi: 10.1242/dev.077883. [DOI] [PubMed] [Google Scholar]
- 41.Urbach R, Technau GM. Early steps in building the insect brain: neuroblast formation and segmental patterning in the developing brain of different insect species. Arthropod Struct Dev. 2003;32:103–123. doi: 10.1016/S1467-8039(03)00042-2. [DOI] [PubMed] [Google Scholar]
- 42.Harzsch S. Ontogeny of the ventral nerve cord in malacostracan crustaceans: a common plan for neuronal development in Crustacea, Hexapoda and other Arthropoda? Arthropod Struct Dev. 2003;32:17–37. doi: 10.1016/S1467-8039(03)00008-2. [DOI] [PubMed] [Google Scholar]
- 43.Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 44.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01