Signatures of mutational processes in human cancer (original) (raw)
. Author manuscript; available in PMC: 2014 Feb 22.
Published in final edited form as: Nature. 2013 Aug 14;500(7463):415–421. doi: 10.1038/nature12477
Abstract
All cancers are caused by somatic mutations. However, understanding of the biological processes generating these mutations is limited. The catalogue of somatic mutations from a cancer genome bears the signatures of the mutational processes that have been operative. Here, we analysed 4,938,362 mutations from 7,042 cancers and extracted more than 20 distinct mutational signatures. Some are present in many cancer types, notably a signature attributed to the APOBEC family of cytidine deaminases, whereas others are confined to a single class. Certain signatures are associated with age of the patient at cancer diagnosis, known mutagenic exposures or defects in DNA maintenance, but many are of cryptic origin. In addition to these genome-wide mutational signatures, hypermutation localized to small genomic regions, kataegis, is found in many cancer types. The results reveal the diversity of mutational processes underlying the development of cancer with potential implications for understanding of cancer etiology, prevention and therapy.
INTRODUCTION
Somatic mutations found in cancer genomes1 may be the consequence of the intrinsic slight infidelity of the DNA replication machinery, exogenous or endogenous mutagen exposures, enzymatic modification of DNA or defective DNA repair. In some cancer types, a substantial proportion of somatic mutations is known to be generated by exposures, e.g., tobacco smoking in lung and ultraviolet (UV) light in skin cancers2, or by abnormalities of DNA maintenance, e.g., defective DNA mismatch repair in some colorectal cancers3. However, our understanding of the mutational processes that cause somatic mutations in most cancer classes is remarkably limited.
Different mutational processes often generate different combinations of mutation types, termed “signatures.” Until recently, mutational signatures in human cancer have been explored through a small number of frequently mutated cancer genes, notably TP534. Although informative, these studies have limitations. To generate a mutational signature, a single mutation from each cancer sample is entered into a mutation set aggregated from several cases of a particular cancer type. A signature that contributes the large majority of somatic mutations in the tumor class is accurately reported. However, if multiple mutational processes are operative, a jumbled composite signature is generated. Furthermore, because such studies are based on “driver” mutations1, signatures of selection are superimposed on the signatures of mutational processes.
Recent advances in sequencing technology have overcome past limitations of scale1. Thousands of somatic mutations can now be identified in a single cancer sample, offering the possibility of deciphering mutational signatures even when several mutational processes are operative. Moreover, since most mutations in cancer genomes are “passengers”1 they do not bear strong imprints of selection.
We recently developed an algorithm to extract mutational signatures from catalogues of somatic mutations and applied it to 21 breast cancer whole genome sequences5,6. Novel and known signatures were revealed, with the contribution of each signature to each cancer sample and the timing of its activity estimated6,7. Further studies have demonstrated that the approach can also be applied, albeit with less power, to mutational catalogues from sequences of all coding exons (“exomes”)5. Global sequencing initiatives are now yielding catalogues of somatic mutations from thousands of cancers8. We have therefore applied this method to survey the repertoire of mutational signatures and processes operating across the spectrum of human neoplasia.
RESULTS
Mutational catalogues
We compiled 4,938,362 somatic substitutions and small insertions/deletions (indels) from the mutational catalogues of 7,042 primary cancers of 30 different classes (507 from whole genome and 6,535 from exome sequences) (Supplementary Fig. 1). In all cases, normal DNA from the same individuals had been sequenced to establish the somatic origin of variants.
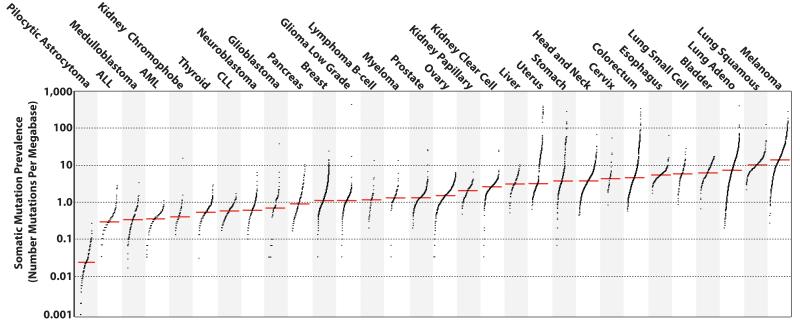
The prevalence of somatic mutations was highly variable between and within cancer classes, ranging from about 0.001/Mb to more than 400/Mb (Fig. 1). Certain childhood cancers carried fewest mutations whereas cancers related to chronic mutagenic exposures such as lung (tobacco) and malignant melanoma (UV) exhibited the highest prevalence. This variation in mutation prevalence is attributable to differences between cancers in the duration of the cellular lineage between the fertilized egg and the sequenced cancer cell and/or to differences in somatic mutation rates during the whole or parts of that cellular lineage1.
Figure 1. The prevalence of somatic mutations across human cancer types.
Every dot represents a sample while the red horizontal lines are the median numbers of mutations in the respective cancer types. The vertical axis (log scaled) shows the number of mutations per megabase while the different cancer types are ordered on the horizontal axis based on their median numbers of somatic mutations. We would like to thank Gad Getz and colleagues for the design of this figure26.
The landscape of mutational signatures
In principle, all classes of mutation (substitutions, indels, rearrangements, etc.) and any accessory mutation characteristic, e.g., the sequence context of the mutation or the transcriptional strand on which it occurs, can be incorporated into the set of features by which a mutational signature is defined. In the first instance, we extracted mutational signatures using base substitutions and additionally included information on the sequence context of each mutation. Since there are six classes of base substitution C>A, C>G, C>T, T>A, T>C, T>G (all substitutions are referred to by the pyrimidine of the mutated Watson-Crick base pair) and since we incorporated information on the bases immediately 5′ and 3′ to each mutated base, there are 96 possible mutations in this classification. This 96 substitution classification is particularly useful for distinguishing mutational signatures which cause the same substitutions but in different sequence contexts.
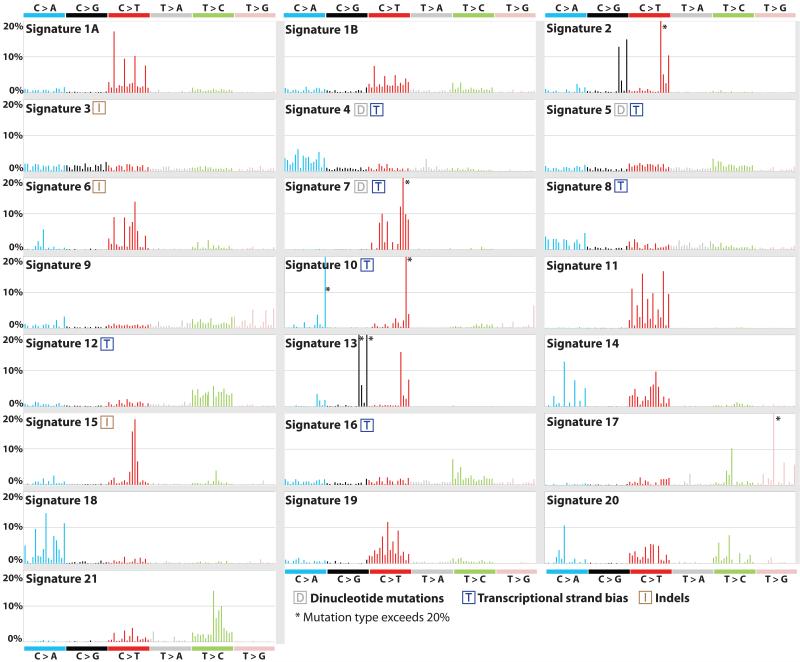
Applying this approach to the 30 cancer types revealed 21 distinct validated mutational signatures (Supplementary Table 1, Supplementary Figs 2 to 28). These show substantial diversity (Fig. 2 and Supplementary Figs 2 to 23). There are signatures characterized by prominence of only one or two of the 96 possible substitution mutations, indicating remarkable specificity of mutation type and sequence context (Signature 10). By contrast, others exhibit a more-or-less equal representation of all 96 mutations (Signature 3). There are signatures characterized predominantly by C>T (1A/B, 6, 7, 11, 15, 19), C>A (4, 8, 18), T>C (5, 12, 16, 21) and T>G mutations (9, 17), with others showing distinctive combinations of mutation classes (2, 13, 14).
Figure 2. Validated mutational signatures found in human cancer.
Each signature is displayed according to the 96 substitution classification defined by the substitution class and sequence context immediately 3′ and 5′ to the mutated base. The probability bars for the six types of substitutions are displayed in different colors. The mutation types are on the horizontal axes, while vertical axes depict the percentage of mutations attributed to a specific mutation type. All mutational signatures are displayed based on the trinucleotide frequency of the human genome. A higher resolution of each panel is found respectively in Supplementary Figures 2 to 23.
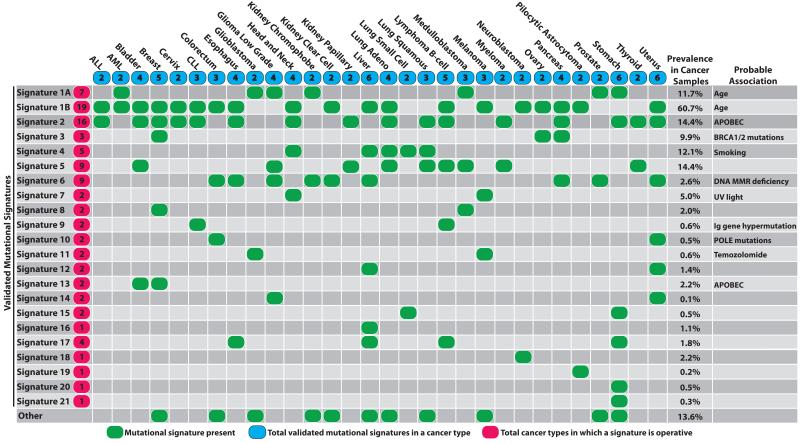
Signatures 1A and 1B were observed in 25/30 cancer classes (Fig. 3). Both are characterized by prominence of C>T substitutions at NpCpG trinucleotides. Since they are almost mutually exclusive among tumor types they probably represent the same underlying process, with Signature 1B representing less efficient separation from other signatures in some cancer types. Signature 1A/B is likely related to the relatively elevated rate of spontaneous deamination of 5-methyl-cytosine which results in C>T transitions and which predominantly occurs at NpCpG trinucleotides9. This mutational process operates in the germline, where it has resulted in substantial depletion of NpCpG sequences, and in normal somatic cells10.
Figure 3. The presence of mutational signatures across human cancer types.
Cancer types are ordered alphabetically as columns while mutational signatures are displayed as rows. “Other” indicates mutational signatures for which we were not able to perform validation or for which validation failed (Supplementary Figs 24 to 28). Prevalence in cancer samples indicates the percentage of samples from our dataset of 7,042 cancers in which the signature contributed significant number of somatic mutations. For most signatures, significant number of mutations in a sample is defined as more than 100 substitutions or more than 25% of all mutations in that sample.
Signature 2 is characterized primarily by C>T and C>G mutations at TpCpN trinucleotides and was found in 16/30 cancer types (Fig. 3). On the basis of similarities in mutation type and sequence context we previously proposed that Signature 2 is due to over-activity of members of the APOBEC family of cytidine deaminases, which convert cytidine to uracil, coupled to activity of the base excision repair and DNA replication machineries6,11.
In most cancer classes at least two mutational signatures were observed, with a maximum of six in cancers of the liver, uterus and stomach. Although these differences may, in part, be attributable to differences in the power to extract signatures, it seems likely that some cancers have a more complex repertoire of mutational processes than others.
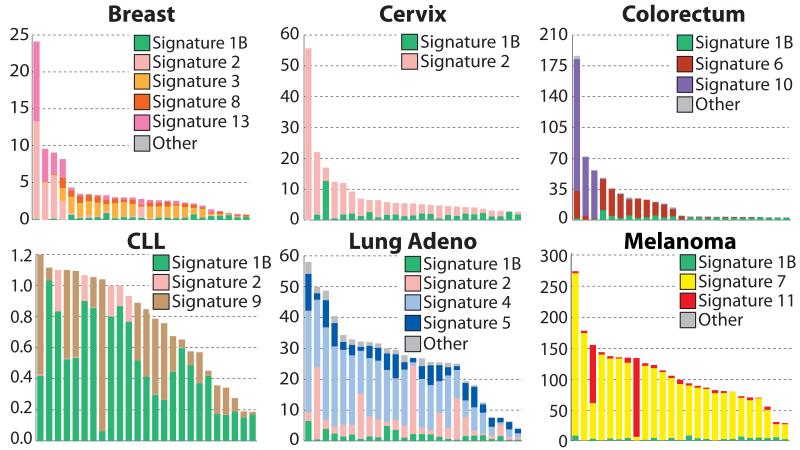
Most individual cancer genomes exhibit more than one mutational signature and many different combinations of signatures were observed (Fig. 4 and Supplementary Figs 29 to 88). The patterns of contribution to individual cancer samples vary markedly between signatures. Signature 1A/B contributes relatively similar numbers of mutations to most cancer cases whereas other signatures contribute overwhelming numbers of mutations to some cancer samples but very few to others of the same cancer class, for example Signatures 2, 3, 4, 6, 7, 9, 10, 11, 13 (Fig. 4).
Figure 4. The contributions of mutational signatures to individual cancers of selected cancer types.
Each bar represents a typical selected sample from the respective cancer type and the vertical axis denotes the number of mutations per megabase. Contributions across all cancer samples could be found in Supplementary Figures 29 to 58. Summary of the total contributions for all operative mutational processes in a cancer type could be found in Supplementary Figures 59 to 88. “Other” indicates mutational signatures for which we were not able to perform validation or for which validation failed (Supplementary Figs 24 to 28).
Mutational signatures and age of cancer diagnosis
We examined each cancer type for correlations between age of diagnosis and the number of mutations attributable to each signature in each sample. Signature 1A/B exhibited strong positive correlations with age in the majority of cancer types of childhood and adulthood (Supplementary Table 2). No other mutational signature showed a consistent correlation with age of diagnosis.
The mutations in a cancer genome may be acquired at any stage in the cellular lineage from the fertilized egg to the sequenced cancer cell. The correlation with age of diagnosis is consistent with the hypothesis that a substantial proportion of Signature 1A/B mutations in cancer genomes have been acquired over the lifetime of the cancer patient, at a relatively constant rate which is similar in different people, probably in normal somatic tissues. The absence of consistent correlation of all other signatures with age suggests that mutations associated with these have been generated at different rates in different people, possibly as a consequence of differing carcinogen exposures or after neoplastic change has been initiated.
Mutational signatures with transcriptional strand bias
The efficiency of DNA damage and DNA maintenance processes can differ between the transcribed and untranscribed strands of genes. The most celebrated cause of this phenomenon is transcription-coupled nucleotide excision repair (NER) that operates predominantly on the transcribed strand of genes and is recruited by RNA polymerase II when it encounters bulky DNA helix-distorting lesions12.
We re-extracted substitution mutational signatures incorporating the transcriptional strand on which each mutation has taken place. Since a mutation in a transcribed genomic region may be either on the transcribed or the untranscribed strand, this generates a classification with 192 mutation subclasses.
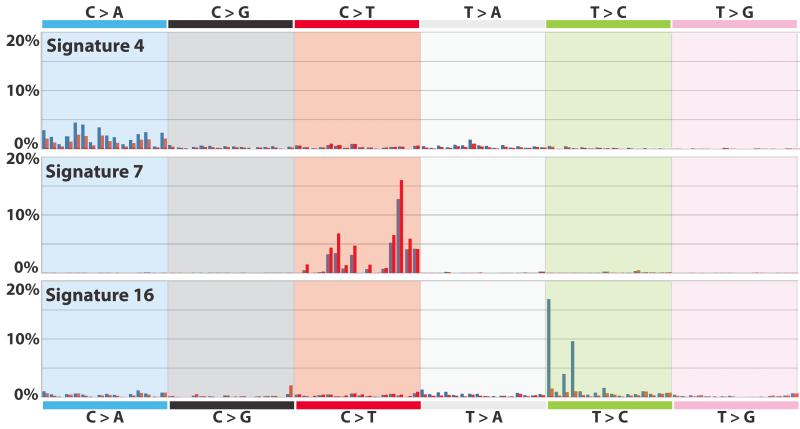
Several signatures showed substantial differences in mutation prevalence between transcribed and untranscribed strands (known as transcriptional strand bias) (Fig. 5 and Supplementary Figs 89 to 95). For example, Signature 4 shows transcriptional strand bias for C>A mutations (Fig. 5). Signature 4 is observed in lung adeno, squamous and small cell carcinomas, head and neck squamous, and liver cancers (Fig. 3), most of which are known to be caused by tobacco smoking. Therefore, Signature 4 is probably an imprint of the bulky DNA adducts generated by polycyclic hydrocarbons found in tobacco smoke and their removal by transcription-coupled NER13. The higher prevalence of C>A mutations on transcribed compared to untranscribed strands is consistent with the propensity of many tobacco carcinogens to form adducts on guanine.
Figure 5. Mutational signatures with strong transcriptional strand bias.
Mutations are shown according to the 192 mutation classification incorporating the substitution type, the sequence context immediately 5′ and 3′ to the mutated base and whether the mutated pyrimidine is on the transcribed or untranscribed strand. The mutation types are displayed on the horizontal axis, while the vertical axis depicts the percentage of mutations attributed to a specific mutation type. A higher resolution version of each panel is found respectively in Supplementary Figures 89 to 95.
Similarly, Signature 7, mainly found in malignant melanoma, shows a higher prevalence of C>T mutations on the untranscribed compared to the transcribed strands consistent with the formation, through UV exposure, of pyrimidine dimers and other lesions which are known to be repaired by transcription-coupled NER14.
Beyond these known examples of DNA damage processed by transcription-coupled NER, other signatures show strong transcriptional strand bias (5, 8, 10, 12, 16). Notably, Signature 16, which is characterized by T>C mutations at ApTpA, ApTpG and ApTpT trinucleotides and is observed in hepatocellular carcinomas, shows the strongest transcriptional strand bias of any signature, with T>C mutations occurring almost exclusively on the transcribed strand (Fig. 5). Similarly, Signature 12, which features T>C mutations at NpTpN trinucleotides, also found in hepatocellular carcinomas, shows strong transcriptional strand bias with more T>C mutations on the transcribed than untranscribed strands (Supplementary Fig. 94). On the assumption that the transcriptional strand biases in Signatures 12 and 16 are introduced by transcription coupled NER, these currently unexplained signatures may be the result of bulky DNA helix-distorting adducts on adenine. However, there is no prior basis for invoking transcription-coupled NER in the genesis of these signatures and other causes of transcriptional strand bias may exist.
Mutational signatures with insertions and deletions
We re-extracted the mutational signatures including, in addition to the 96 substitution types, two further classes of mutation: indels at short nucleotide repeats and indels with overlapping microhomology at breakpoint junctions. Three of the 21 base substitution signatures associated with large numbers of indels. Signature 6, which is characterized predominantly by C>T at NpCpG mutations, but is distinct from Signature 1A/B, contributes very large numbers of substitutions and small indels (mostly of 1bp) at nucleotide repeats to subsets of colorectal, uterine, liver, kidney, prostate, esophageal and pancreatic cancers. This pattern of indels, often termed “microsatellite instability”, is characteristic of cancers with defective DNA mismatch repair15. Consistent with this explanation, the presence of Signature 6 was strongly associated with the inactivation of DNA mismatch repair (MMR) genes in colorectal cancer (P = 3.3 × 10−5).
Signature 15 also contributes very large numbers of substitutions and small indels at nucleotide repeats but, compared to Signature 6, exhibits greater prominence of C>T at GpCpN trinucleotides. Signature 15 was found in several samples of lung and stomach cancer and its origin is currently unknown.
By contrast, substantial numbers of larger deletions (up to 50 bp) with overlapping microhomology at breakpoint junctions were found in breast, ovarian and pancreatic cancer cases with major contributions from Signature 3. A subset of cancer cases of these three classes is known to be due to inactivating mutations in BRCA1 and BRCA2 and the presence of Signature 3 was strongly associated with BRCA1/2 mutations within the individual cancer types (P = 1.6 × 10−8 for breast cancer and P = 0.02 for pancreatic cancer)6. Indeed, almost all cases with BRCA1/2 mutations showed a large contribution from Signature 3. However, some cases with a substantial contribution from Signature 3 did not have BRCA1/2 mutations suggesting that other mechanisms of BRCA1/2 inactivation or abnormalities of other genes may also generate it.
BRCA1 and BRCA2 are implicated in homologous recombination based DNA double strand break repair16. Abrogation of their functions results in non-homologous end-joining (NHEJ) mechanisms, which can utilize microhomology at rearrangement junctions to rejoin double strand breaks, taking over DNA double strand break repair. The results show that, in addition to the genomic structural instability conferred by defective double strand break repair, a base substitution mutational signature is associated with BRCA1/2 deficiency.
Associating cancer etiology and mutational signatures
Each mutational signature is the imprint left on the cancer genome by a mutational process that may include one or more DNA damage and/or DNA maintenance mechanisms, with the latter either functioning normally or abnormally. Here, we consider likely mechanisms or underlying causes by comparing signatures with mutation patterns of known causation in the scientific literature or by associating them with epidemiological and biological features of particular cancer types.
Signature 1A/B is probably due to the endogenous mutational process present in most normal and neoplastic cells that is initiated by deamination of 5-methyl-cytosine9. Other signatures are likely attributable to exogenous mutagenic exposures. Signature 7 is observed in malignant melanoma and squamous carcinoma of the head and neck and has the known features of UV induced mutations. Signature 4 is found in cancers associated with tobacco smoking (Fig. 3) and has the mutational features associated with tobacco carcinogens13. The causal relationship between tobacco smoking and Signature 4 is supported by a strong positive association between smoking history and the contributions of Signature 4 to individual cancers (P = 1.1 × 10−7, Supplementary Figs 44 to 46, 74 to 76, 96).
Cigarette smoke contains over 60 carcinogens13 and it is possible that this complex mixture may initiate other mutational processes. Signatures 1A/B, 2 and 5 were also found in lung adenocarcinoma. Signature 5, but not Signatures 1A/B and 2, also showed a positive correlation between smoking history and mutation contribution (P = 8.0 × 10−3, Supplementary Fig. 96). Thus, in lung cancer, Signature 5, which is characterized predominantly by C>T and T>C mutations, may also be due to tobacco carcinogens. However, it is also present in nine other cancer types, most of which are not strongly associated with tobacco consumption, and therefore its etiology overall is unclear (Fig. 3).
Some anticancer drugs are mutagens17. Signature 11 is found in malignant melanomas and glioblastomas multiforme pretreated with the alkylating agent temozolomide (P = 4.0 × 10−3) and has mutational features very similar to those previously reported in experimental studies of alkylating agents18.
Abnormalities in DNA maintenance may also be responsible for mutational signatures and the roles of defective DNA mismatch repair (Signature 6) and defective homologous recombination based DNA double strand break repair (Signature 3) have been discussed above. Other signatures may result from abnormal activity of enzymes that modify DNA or of error-prone polymerases. Signatures 2 and 13 have been attributed to the AID/APOBEC family of cytidine deaminases6. On the basis of similarities in the sequence context of cytosine mutations caused by APOBEC enzymes in experimental systems, a role for APOBEC1, APOBEC3A and/or APOBEC3B in human cancer appears more likely than for other members of the family19-21. However, the reason for the extreme activation of this mutational process in some cancers is unknown. Since APOBEC activation constitutes part of the innate immune response to viruses and retrotransposons22 it may be that these mutational signatures represent collateral damage on the human genome from a response originally directed at retrotransposing DNA elements or exogenous viruses. Confirmation of this hypothesis would establish an important new mechanism for initiation of human carcinogenesis.
Signature 9, observed in chronic lymphocytic leukaemia and malignant B-cell lymphomas is characterized by T>G transversions at ApTpN and TpTpN trinucleotides, and is restricted to cancers that have undergone somatic immunoglobulin gene hypermutation (IGHV-mutated) associated with Activation Induced Deaminase (AID) (P = 2.5 × 10−4 in CLL). Signature 9 does not, however, have the known mutational features of AID20, and has been proposed to be due to POL η, an error prone polymerase involved in processing AID induced cytidine deamination11,23. Similarly, Signature 10, which generates huge numbers of mutations in subsets of colorectal and uterine cancer, has been previously associated with altered activity of the error-prone polymerase POL ε (POLE) consequent on mutations in the gene24,25.
Many mutational signatures do not, however, have an established or proposed, underlying mutational process or etiology. Some, for example Signatures 8, 12 and 16, show strong transcriptional strand bias (Fig. 5) and possibly reflect the involvement of transcription coupled nucleotide excision repair acting on bulky DNA adducts due to exogenous carcinogens. Others, for example Signatures 14, 15, and 21, show overwhelming activity in a small number of cancer cases (respectively Supplementary Figs 38, 45, and 56) and are perhaps more likely to be due to currently uncharacterized defects in DNA maintenance.
Localized hypermutation: kataegis
Foci of localized substitution hypermutation, termed kataegis after the Greek for thunderstorm, were recently described in breast cancer6. Kataegis is characterized by clusters of C>T and/or C>G mutations which are substantially enriched at TpCpN trinucleotides and on the same DNA strand. Foci of kataegis include from a few to several thousand mutations and are often found in the vicinity of genomic rearrangements. The genomic regions affected are different in different cancers. On the basis of the substitution types and sequence context of kataegis substitutions, an underlying role for APOBEC family enzymes was proposed for kataegis as well as for Signatures 2 and 136.
The 507 whole cancer genome mutation catalogs were searched for clusters of mutations. Cancers of breast (67/119), pancreas (11/15), lung (20/24), liver (15/88), medulloblastomas (2/100), CLL (15/28), B-cell lymphomas (21/24) and ALL (1/1) showed occasional (<10), small (<20 mutations) foci of _kataegis_ whilst AML (0/7) and pilocytic astrocytoma (0/101) did not. Subsets of breast (7), lung (6) and haematological cancers (3) showed numerous (>10) kataegic foci and two breast and one pancreatic cancer showed major foci of kataegis (>50 mutations) (Fig. 6 and Supplementary Figs 97 and 98).
Figure 6. Kataegis in three cancers.
Each of these “rainfall” plots represents an individual cancer sample in which each dot represents a single somatic mutation ordered on the horizontal axis according to its position in the human genome. The vertical axis denotes the genomic distance of each mutation from the previous mutation. Clusters of mutations in kataegis are arrowed.
Kataegic foci are often associated with genomic rearrangements (Supplementary Fig. 98). In yeast, introduction of a DNA double strand break greatly increases the likelihood of kataegis in its vicinity indicating a role for such breaks in initiating the process20. However, even in cancer cases with kataegis, most rearrangements do not exhibit nearby kataegis, suggesting that a double strand break is not sufficient.
In neoplasms of B-lymphocyte origin, including CLL and many lymphomas, mutation clusters recurrently occurred at immunoglobulin loci. In these cancers the mutation characteristics were different (Supplementary Fig. 98), bearing the hallmarks of somatic hypermutation associated with AID, which is operative during the generation of immunological diversity20.
DISCUSSION
The diversity and complexity of somatic mutational processes underlying carcinogenesis in human beings is now being revealed through mutational patterns buried within cancer genomes. It is likely that more mutational signatures will be extracted, together with more precise definition of their features, as the number of whole-genome sequenced cancers increases and analytic methods are further refined.
The mechanistic basis of some signatures is, at least partially, understood but for many it remains speculative or unknown. Elucidating the underlying mutational processes will depend upon two major streams of investigation. First, compilation of mutational signatures from model systems exposed to known mutagens or perturbations of the DNA maintenance machinery and comparison with those found in human cancers. Second, correlation of the contributions of mutational signatures with other biological characteristics of each cancer through diverse approaches ranging from molecular profiling to epidemiology. Collectively, these studies will advance understanding of cancer etiology with potential implications for prevention and treatment.
ONLINE METHODS
Validating Mutational Signatures
Validating a mutational signature requires ensuring that a large set of somatic mutations attributed to this signature is genuine in at least one sample. Validation is complicated as multiple mutational processes are usually operative in most cancer samples, and thus every individual somatic mutation can be probabilistically assigned to several mutational signatures. To overcome this limitation, we examined our dataset for samples that are predominantly generated by one mutational signature (i.e., more than 50% of the somatic mutations in the sample belong to an individual mutational signature) and/or for samples in which all operative mutational processes have mutually exclusive patterns of mutations (e.g., a sample with mutations only from Signature 1B, which is predominantly C>T substitutions, and Signature 18, which is predominantly C>A substitutions). We identified the optimal available sample for every mutational signature and attempted to validate the subset of somatic mutations attributed to this signature using one of three methods (Supplementary Figure 99):
- - Validation through re-sequencing with an orthogonal sequencing technology.
- - Validation through re-sequencing with the same sequencing technology (including RNA-Seq, bisulfite sequencing, etc.).
- - Validation through visual examination of somatic mutations by an experienced curator using a genomic browser and BAM files for both the tumour and its matched normal.
For some of the previously published samples, we leveraged the already reported validation data. When possible, somatic mutations were validated by either re-sequencing with orthogonal technology or re-sequencing using the same sequencing technology. We resorted to visual validation only when there was no other possibility for validating a mutational signature. 21 of the 27 originally identified mutational signatures were validated (Supplementary Table 1 and Supplementary Figure 99). 3 mutational signatures failed validation – Signatures R1 to R3 (Supplementary Figs 24 to 26). We were unable to validate 2 mutational signatures, Signatures U1 and U2 (Supplementary Figs 27 and 28), due to lack of available biological samples and access to BAM files for the samples with sufficient number of somatic mutations generated by these two mutational signatures.
Samples and curation of freely available cancer data
Informed consent was obtained from all subjects. Collection and use of patient samples were approved by the appropriate IRB of each Institution. In addition to newly generated data, we curated freely available somatic mutations from three other sources:
- - The data portal of The Cancer Genome Atlas (TCGA)
- - The data portal of the International Cancer Genome Consortium (ICGC)
- - Previously published data in peer-review journals, see additional references6,23,27-59
Filtering, estimating mutation prevalence, and generating mutational catalogs
In all examined samples, normal DNA from the same individuals had been sequenced to establish the somatic origin of variants. Extensive filtering was performed to remove any residual germline mutations and technology specific sequencing artifacts prior to analyzing the data. Germline mutations were filtered out from the lists of reported mutations using the complete list of germline mutations from dbSNP60, 1000 genomes project61, NHLBI GO Exome Sequencing Project62, and 69 Complete Genomics panel (http://www.completegenomics.com/public-data/69-Genomes/). Technology specific sequencing artifacts were filtered out by using panels of BAM files of (unmatched) normal tissues containing more than 120 normal genomes and 500 normal exomes. Any somatic mutation present in at least three well-mapping reads in at least two normal BAM files was discarded. The remaining somatic mutations were used for generating a mutational catalog for every sample. Prevalence of somatic mutations was estimated based on a haploid human genome after all filtering. Prevalence of somatic mutations in exomes was calculated based on the identified mutations in protein coding genes and assuming that an average exome has 30 megabases in protein coding genes with sufficient coverage. Prevalence of somatic mutations in whole genomes was calculated based on all identified mutations and assuming that an average whole genome has 2.8 gigabases with sufficient coverage.
The immediate 5′ and 3′ sequence context was extracted using the ENSEMBL Core APIs for human genome build GRCh37. Curated somatic mutations that originally mapped to an older version of the human genome were re-mapped using UCSC’s freely available lift genome annotations tool (any somatic mutations with ambiguous or missing mappings were discarded). Dinucleotide substitutions were identified when two substitutions were present in consecutive bases on the same chromosome (sequence context was ignored). The immediate 5′ and 3′ sequence content of all indels was examined and the ones present at mono/polynucleotide repeats or microhomologies were included in the analyzed mutational catalogs as their respective types. Strand bias catalogs were derived for each sample using only substitutions identified in the transcribed regions of well-annotated protein coding genes. Genomic regions of bidirectional transcription were excluded from the strand bias analysis.
Deciphering signatures of mutational processes
Mutational signatures were deciphered independently for each of the 30 cancer types using our previously developed computational framework5. The algorithm deciphers the minimal set of mutational signatures that optimally explains the proportion of each mutation type found in each catalogue and then estimates the contribution of each signature to each catalogue. Mutational signatures were also extracted separately for genomes and exomes. Mutational signatures extracted from exomes were normalized using the observed trinucleotide frequency in the human exome to the one of the human genome. All mutational signatures were clustered using unsupervised agglomerative hierarchical clustering and a threshold was selected to identify the set of consensus mutational signatures. Mis-clustering was avoided by manual examination (and whenever necessary re-assignment) of all signatures in all clusters. 27 consensus mutational signatures were identified across the 30 cancer types. The computational framework for deciphering mutational signatures as well as the data used in this study are freely available and can be downloaded from: http://www.mathworks.com/matlabcentral/fileexchange/38724
Factors that influence extraction of mutational signatures
Recently, using simulated and real data, we described in detail the factors that influence the extraction of mutational signatures5. These included the number of available samples, the mutation prevalence in samples, the number of mutations contributed by different mutational signatures, the similarity between the signatures of mutational processes operative in cancer samples, as well as the limitations of our computational approach. Here, we examined datasets with varying sizes from 30 different cancer types and we have taken great care to report only validated mutational signatures. However, our approach identified two similar patterns most likely representing the same biological process, viz., Signature 1A and 1B. The reasons for this is, for some cancer types, we have sufficient numbers of samples and/or mutations ( i.e., statistical power) to decipher the cleaner version ( i.e., Signature 1A) while for other cancer types we do not have sufficient data and our approach extracts a version of the signature which is more contaminated by other Signatures present in that cancer type ( i.e., Signature 1B). Nevertheless, the two Signatures are very similar, hence we call them 1A and 1B. Being almost mutually exclusive amongst cancer types ( i.e., finding either Signature 1A or 1B in each cancer type but not usually both) is supportive of the notion that they represent the same underlying process as is the fact that Signatures 1A and 1B both correlate with age and have the same overall pattern of contributions to individual cancer genomes. Indeed, in our view it is likely, that if we had sufficient data, Signature 1B would disappear and the algorithm would extract only Signature 1A.
Displaying mutational signatures
Mutational signatures are displayed using a 96 substitution classification defined by the substitution class and the sequence context immediately 3′ and 5′ to the mutated base. Mutational signatures are displayed in the main text of the report and in Supplementary Information based on the observed trinucleotide frequency of the human genome, i.e., representing the relative proportions of mutations generated in each signature based on the actual trinucleotide frequencies of the reference human genome. However, in Supplementary Information we also provide a visualization of mutational signatures based on an equal frequency of each trinucleotide (Supplementary Figs 2 to 28). The equal trinucleotide frequency representation results, in all mutational signatures, in a greater degree of prominence of C>T substitutions at NpCpG trinucleotides as major features compared to the plots based on the observed trinucleotides. This difference, may in some cases, reflect the biological reality, i.e., a propensity of the particular mutational process to be more active at Np_C_pG trinucleotides. However, please note, that it may also in some cases, be due to incomplete extraction by the algorithm of the signature in question from Signature 1A/B which is characterized by prominent features at NpCpG trinucleotides. This is likely to happen because a) Signature 1A/B is ubiquitous and b) because even a small probability of mutations at NpCpG trinucleotides will generate a prominent feature because of the severe depletion of NpCpG trinucleotides in the reference genome. In future, with larger numbers of sequences and large numbers of whole genome sequences it is anticipated that the latter effect will be reduced.
Approaches for associating cancer etiology and exposures of validated mutational signatures
Generalised linear models (GLMs) were used to fit signature exposures (i.e., number of mutations assigned to a signature) and age of cancer diagnoses. For each cancer type, all mutational signatures operative in it were evaluated using GLMs and the p-values were corrected for multiple hypothesis testing using FDR. The resulting p-values indicate that age strongly correlates with Signature 1A/B across 15 cancer types (Supplementary Table 2). Exposure to Signature 4 also correlates with age of diagnosis in kidney papillary and thyroid cancers. However, in both cancer types, we were not able to detect/extract Signature 1A/B due to low number of mutations in their samples and it is likely that Signature 1A/B is currently mixed within Signature 4. Further studies, involving whole genome sequences, will be needed to validate this hypothesis. Interestingly, in melanoma, age of diagnosis also correlates with exposure to Signature 7, which we have associated with exposure to UV-light.
Associations between all other etiologies and signature exposures were performed using two-sample Kolmogorov-Smirnov tests between two sets of samples. The first set contains the signature exposures of the samples with the “desired feature” (e.g., samples that contain a hypermutation in the Ig gene) and the second set are the signature exposures of the samples without the “desired feature” (e.g., samples that do NOT contain a hypermutation in the Ig gene). Samples with unknown features status (e.g., not knowing the status of the Ig gene) were ignored. Kolmogorov-Smirnov tests were performed for all signatures and all examined “features” in a cancer type. P-values were FDR corrected based on the performed tests in a particular cancer class.
A PCF-based algorithm for the detection of kataegis
Foci of localized hypermutation, termed kataegis, were sought in 507 whole-genome sequenced cancers. High-quality variant calls that had been previously subjected to filtering for mutational signature analysis were investigated using an algorithm developed to identify foci of kataegis.
For each sample, all mutations were ordered by chromosomal position and the intermutation distance, defined as the number of base pairs from each mutation to the next one, was calculated. Intermutation distances were then segmented using the piecewise constant fitting (PCF) method63 in order to find regions of constant intermutation distance. Parameters used for PCF were gamma=25 and kmin=2 and were trained on the set of kataegis foci that had been manually identified, curated and validated using orthogonal sequencing platforms6. Putative regions of kataegis were identified as those segments containing 6 or more consecutive mutations with an average intermutation distance of less than or equal to 1,000 bp.
Variation in number of foci of kataegis and relationship with genome-wide mutation burden
In order to examine the likelihood of kataegis occurring for different mutation burdens, the expected number of kataegis events that would be observed by chance was calculated for a range of total number of mutations per cancer, n, between 1,000 and 2,000,000. The probability that any one mutation will be followed by 5 other mutations within a distance of 5,000 bp, thereby triggering the identification of kataegis, is given by
where g is the length of the genome, in base pairs.
Supplementary Figure 97 shows the expected number of kataegis events identified in genomes with between 100,000 and 500,000 mutations. For cancers with up to 200,000 mutations, the expected number of kataegis events is extremely small (0.16 for a total mutation load of 200,000) making the detection of kataegic foci highly significant for each sample. Supplementary Table 3 presents all the samples in which kataegic foci were identified, the total mutation burden for each sample, the observed number of kataegic foci and the expected number of foci.
Post-processing: Specificity of variants in kataegis foci
Clusters of variant calls can easily occur in regions of low sequence complexity. These are not true substitution mutations but represent systematic sequencing artefacts or mis-mapping of short reads. The quality of variant calls depends on the quality of mutation-calling by individual institutions. Additional filtering was applied in order to remove likely false positive calls and then putative kataegic foci were individually curated.
1,436 kataegis foci were called by PCF, with 873 finalised as putative kataegis foci (Supplementary Table 4) involving 9,219 substitution variants. Where possible, BAM files were retrieved, inspected and substitution variants involved in kataegis foci were manually curated to remove likely false positive calls. Where BAM files were not available to us, substitution variants were strictly excluded if called in:
- - Genomic features that generate mapping errors, e.g., regions of excessively high coverage due to collapsed repeat sequences in the reference genome64.
- - Highly repetitive regions with reads consistently demonstrating low mapping qualities in 20 unrelated normal samples.
- - Locations with known germline insertions/deletions within the sequencing reads reporting the mutated base.
Reassuringly, several features were seen in the finalised putative kataegis foci, which reinforced the conviction in the validity of these calls. Although clusters of mutations identified by the PCF method were sought in an approach unbiased by mutation-type and based exclusively on intermutation distances, we find that the 873 putative foci demonstrate: first, a preponderance to C>T and C>G mutations (Supplementary Figure 97B); second, the enrichment for a TpC sequence context as previously described6(Supplementary Figure 97B); third, processivity - where consecutive mutations within a cluster were on the same strand (i.e., 6 C>T mutations in a row or 6 G>A mutations in a row) (Figure 6C); and fourth, visual curation of reads carrying these processive variants showed that the variants were usually in cis (i.e., mutations were on the same read (Supplementary Figure 97C) or on the read mate of other affected alleles within the insert size) with respect to each other indicating that they had arisen on the same allele. Finally, where data were available, we found that clusters of substitution mutations within the same kataegis foci shared approximately the same variant allele fraction indicating that they had likely arisen during a single cell cycle event.
BAM files from some samples were not accessible and therefore a proportion of substitution variants involved in kataegis foci were not visually curated. The application of the strict criteria described above and the subsequent finding of the consistency of the mutation-type, sequence context, processive nature of the mutations, with the majority in cis on individual sequencing reads, suggests that the vast majority of these foci are likely to be genuine. However, the possibility that some of the foci are not truly kataegis, particularly for the cancers which have not been validated or visually curated, remains.
Sensitivity of kataegis detection
It is acknowledged that the likelihood of detection of kataegis foci rests on the sensitivity of mutation detection. It is possible for foci to be missed because the mutations were not detected by mutation-callers of the various institutions, prior to our analysis. This is particularly relevant for subclonal mutations bearing a low variant allele fraction or for mutations that occur on a single copy of a multi-copy locus. This is because the likelihood of mutation detection is reduced when uncorrected for copy number and for aberrant cell fraction of the tumour sample. Furthermore, our stringent post-processing criteria, particularly of samples that have not been visually curated, make it more likely that kataegis is under-represented in this analysis.
Relationship between kataegis and large-scale genomic changes
Reinforcing our previous findings6, we found that some kataegic foci were very closely associated with rearrangements. For example, a breast cancer sample with 1,534 point mutations had only one focus of kataegis which contained 32 point mutations. The same breast cancer sample also had 25 large-scale genomic structural variations (SV) scattered throughout the genome. However, one tandem duplication coincided with this single locus of kataegis in this cancer. Dramatically, no other mutations or SVs were seen for 2Mb flanking this extraordinary event (Supplementary Figure 97B). Another breast cancer (Figure 6A) that contained 22,454 mutations and had 292 rearrangements altogether, had nine regions of kataegis, five of which coincided with large-scale SVs underscoring the co-localization of kataegis foci with SVs. This also highlights that not all foci of kataegis co-localized with SVs and not all SVs were associated with kataegis.
Sites of amplification represent a potential source of false variant calls. If the amplification occurred early in the evolution of a cancer, then there is an increased likelihood of substitutions accumulating randomly within the amplified genomic region. When mapped back to the reference genome, these will appear as clustered variants.
A number of features allow us to distinguish such events from ‘true’ kataegis. These mutations would not be expected to have features associated with kataegis, such as the mutation-type, predilection for a TpC sequence context and the processivity. Furthermore, if they have accumulated as random events in a multi-copy locus, then they would be less likely to occur in cis (on the same sequencing read) with respect to each other. In contrast, mutations which have occurred at the same time, during one moment of transient hypermutability in a single cell cycle event, would be expected to cluster on one copy of a multi-copy locus, to be in cis and to demonstrate approximately the same variant allele fraction. Finally, to achieve the level of hypermutation required to be called as a focus of kataegis (average intermutation distance of less than 1,000bp for six consecutive mutations equivalent to ~1,000 substitutions per Mb), the degree of copy number amplification would have to be considerable.
In order to examine this likelihood of false calls in regions of amplification, simulations were performed assuming background mutation rates of 10 per Mb, 40 per Mb and 100 per Mb for different copy number states and for different sizes of focal amplification. The expected number of kataegic foci for these different states are provided in Supplementary Table 5. For most of the samples in which kataegis was detected (all but twenty), a 10MB region of amplification would require a copy number state of 36 or above to generate 1 cluster of 6 mutations with an average intermutation distance of less than 1,000bp. For 19 of the remaining 20 samples, a 10MB region of amplification would require a copy number state of 10 or above. For the single cancer with a mutation rate exceeding 40 per Mb, a copy number state of 4 is required to generate a cluster of mutations. As mentioned previously, these clusters would have to be processive, be in cis and have roughly the same variant allele fraction in order to be called as a focus of kataegis.
Definition of kataegis
In summary, kataegis has been identified via a PCF-based method as 6 or more consecutive mutations with an average intermutation distance of less than or equal to 1,000 bp. Other salient features include a preponderance for C>T and C>G mutations, a predilection for a TpC mutation context, processivity, evidence of having arisen on the same parental allele (being in cis) on sequencing reads and additionally (but not necessarily) co-localization with large-scale genomic structural variation.
Supplementary Material
1
2
3
4
ACKNOWLEDGEMENTS
We would like to thank the Wellcome Trust for support (grant reference 098051) together with many other funding bodies and individuals (Supplementary Note 1).
REFERENCES
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. doi:10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfeifer GP. Environmental exposures and mutational patterns of cancer genomes. Genome medicine. 2010;2:54. doi: 10.1186/gm175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pena-Diaz J, et al. Noncanonical mismatch repair as a source of genomic instability in human cells. Molecular cell. 2012;47:669–680. doi: 10.1016/j.molcel.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. doi:10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell reports. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. doi:10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nik-Zainal S, et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. doi:10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson TJ, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. doi:10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 10.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. doi:10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. doi:10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 12.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. doi:10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 13.Pfeifer GP, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. doi:10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 14.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. doi:10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 15.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. e2073. doi: 10.1053/j.gastro.2009.12.064. doi:10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. doi:10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Hunter C, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer research. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomita-Mitchell A, et al. Mismatch repair deficient human cells: spontaneous and MNNG-induced mutational spectra in the HPRT gene. Mutat Res. 2000;450:125–138. doi: 10.1016/s0027-5107(00)00020-8. [DOI] [PubMed] [Google Scholar]
- 19.Taylor BJM, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. doi:10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Molecular cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 22.Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 2013;4:28. doi: 10.3389/fmicb.2013.00028. doi:10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. doi:10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. doi:10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research, N. et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. doi:10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013 doi: 10.1038/nature12213. doi:10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References (Online Methods)
- 27.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nature genetics. 2013;45:242–252. doi: 10.1038/ng.2532. doi:10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. doi:10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nature genetics. 2013;45:186–190. doi: 10.1038/ng.2508. doi:10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. doi:10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. doi:10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature genetics. 2012;44:47–52. doi: 10.1038/ng.1032. doi:10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 33.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. doi:10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nature genetics. 2013 doi: 10.1038/ng.2591. doi:10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. doi:10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. doi:10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo G, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nature genetics. 2012;44:17–19. doi: 10.1038/ng.1014. doi:10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 38.Pena-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44:751–759. doi: 10.1038/ng.2323. doi:10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. doi:10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo JS, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome research. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. doi:10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. doi:10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nature genetics. 2012;44:1321–1325. doi: 10.1038/ng.2468. doi:10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nature genetics. 2013 doi: 10.1038/ng.2611. doi:10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. doi:10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. doi:10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nature genetics. 2013;45:279–284. doi: 10.1038/ng.2529. doi:10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. doi:10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21188–21193. doi: 10.1073/pnas.1118046108. doi:10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sausen M, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nature genetics. 2013;45:12–17. doi: 10.1038/ng.2493. doi:10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. doi:10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. doi:10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44:685–689. doi: 10.1038/ng.2279. doi:10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudin CM, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nature genetics. 2012;44:1111–1116. doi: 10.1038/ng.2405. doi:10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature genetics. 2012;44:1104–1110. doi: 10.1038/ng.2396. doi:10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stark MS, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nature genetics. 2012;44:165–169. doi: 10.1038/ng.1041. doi:10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. doi:10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. doi:10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nature genetics. 2012;44:570–574. doi: 10.1038/ng.2246. doi:10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 59.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nature genetics. 2011;43:1219–1223. doi: 10.1038/ng.982. doi:10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 60.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. doi:10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. doi:10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baumbusch LO, et al. Comparison of the Agilent, ROMA/NimbleGen and Illumina platforms for classification of copy number alterations in human breast tumors. BMC Genomics. 2008;9:379. doi: 10.1186/1471-2164-9-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pickrell JK, Gaffney DJ, Gilad Y, Pritchard JK. False positive peaks in ChIP-seq and other sequencing-based functional assays caused by unannotated high copy number regions. Bioinformatics. 2011;27:2144–2146. doi: 10.1093/bioinformatics/btr354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4