Endocytosis and Autophagy: Exploitation or Cooperation? (original) (raw)
Abstract
Autophagy is a lysosome-mediated degradative system that is a highly conserved pathway present in all eukaryotes. In all cells, double-membrane autophagosomes form and engulf cytoplasmic components, delivering them to the lysosome for degradation. Autophagy is essential for cell health and can be activated to function as a recycling pathway in the absence of nutrients or as a quality-control pathway to eliminate damaged organelles or even to eliminate invading pathogens. Autophagy was first identified as a pathway in mammalian cells using morphological techniques, but the Atg (autophagy-related) genes required for autophagy were identified in yeast genetic screens. Despite tremendous advances in elucidating the function of individual Atg proteins, our knowledge of how autophagosomes form and subsequently interact with the endosomal pathway has lagged behind. Recent progress toward understanding where and how both the endocytotic and autophagic pathways overlap is reviewed here.
The endosomal pathway may play a greater role in autophagy than just providing degradative enzymes in the lysosome. Contributions from several stages of the endocytic pathway may be essential for efficient autophagy.
Autophagy is a lysosome-mediated pathway for the degradation of cytosolic proteins and organelles, which is essential for cell homeostasis, development, and for the prevention of several human diseases and infection (Choi et al. 2013). Importantly, autophagy cannot occur without an active lysosome. However, it is becoming increasingly recognized that the endosomal pathway plays a greater role than just providing the degradative enzymes found in the lysosome. Recent data suggest that in mammalian cells multiple contributions from several stages of the endocytic pathway are essential for efficient autophagy. Here we outline the autophagic pathway and then address the recent data on how different endosomal compartments contribute to autophagy, and the molecular machinery required for the interaction of the endosome and lysosome during the formation, and consumption of the autophagosome. Given the model emerging that the amino-acid-sensitive autophagic pathway originates from the endoplasmic reticulum (ER), several questions arise, including how do recognition and productive interaction occur between an ER-derived membrane and endosomes? How are these interactions mediated, and which are essential for efficient autophagy?
AUTOPHAGY AS AN INTRACELLULAR TRAFFICKING PATHWAY
The best-studied pathway responsible for the delivery of cytosolic content and organelles for lysosomal degradation in eukaryotes is macroautophagy, the focus of this review, hereafter referred to as autophagy. (Macro)autophagy is the process of “self-eating” or sequestering cytosolic material and is the canonical pathway conserved in eukaryotes. Autophagy is a unique membrane trafficking process whereby a newly formed organelle termed a phagophore, or isolation membrane, is elongated and engulfs part of the cytoplasm to form autophagosomes that are finally delivered for degradation in the lysosome (Fig. 1). Autophagy was originally considered a nonselective process; however, it is now known that it can also be a selective process and that an autophagosome can engulf specific organelles and protein aggregates, as well as bacteria, viruses, and parasites. There are other types of autophagy, for example, microautophagy (direct invagination into the lysosome) and chaperone-mediated autophagy (CMA), which is a pathway for direct import into the lysosome, as well as selective autophagy (e.g., mitophagy), but these are not discussed here (for a recent review, see Mizushima et al. 2011).
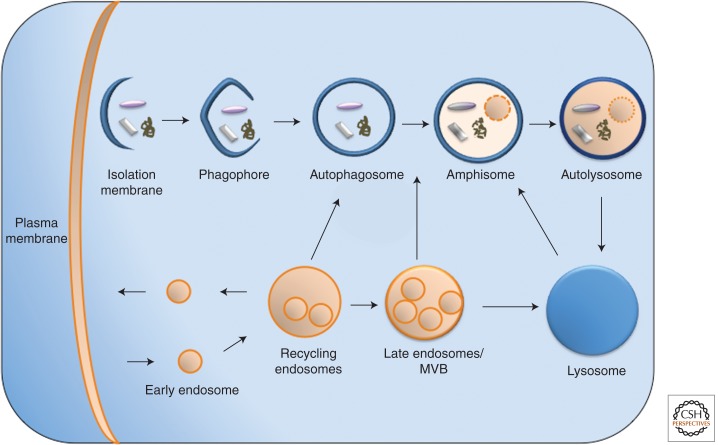
Figure 1.
Autophagy and the endocytic pathway. Autophagosomes form at the PAS (pre-autophagosome structure or phagophore assembly site) and fuse with recycling endosomes, MVBs, and lysosomes (not shown for clarity), becoming an amphisome, which upon further fusion with lysosomes becomes an autolysosome. Not shown are the membrane compartments implicated in the formation of the PAS and autophagosomes (ER, Golgi, mitochondria, and plasma membrane–derived vesicles).
The current view is that autophagosomes are formed at distinct sites termed PAS (pre-autophagosomal structure), to which many of the specific autophagic factors are recruited in an orchestrated manner (Suzuki et al. 2004). Whereas in yeast typically a single PAS is found near the vacuolar membrane, in mammals the equivalent PAS structures are likely present or formed throughout the cytoplasm (Axe et al. 2008). The PAS expands to form the phagophore membrane. Like all other intracellular membrane trafficking processes, autophagy involves multiple membrane fusion events needed for the membrane expansion and elongation, possibly closure of the phagophore membrane, and the subsequent fusion between the autophagosome and the endolysosomal system (Lamb et al. 2012; Rubinsztein et al. 2012).
The cellular machinery for such membrane fusion events usually includes members of the Rab small GTPase family, tethering proteins and complexes, and SNARE molecules needed to promote the actual membrane fusion event. Remarkably, despite conservation of the fusion machinery between yeast and mammals, none of the yeast screens performed to identify specific autophagy genes revealed any of these components. This suggests that the molecular mechanisms driving autophagy may be unique or regulated in a complex fashion. Broadly, and for the purposes of this article, the autophagy pathway downstream from nutrient deprivation in yeast and mammals is identical, and involves 18 Atg (autophagy-related) proteins (Mizushima et al. 2011). Therefore, although the molecules and, increasingly, the molecular function of the Atg proteins are becoming known, the formation of the PAS and autophagosome and the membrane trafficking events regulating these steps as well as the completion of autophagy by formation of an autolysosome remain largely unknown. Apparently, these processes are mediated by factors involved in other intracellular trafficking processes, and therefore we also expand our review to cover the molecular details available in yeast on autophagosome formation, in particular, the role of the Golgi complex.
INITIATION OF AUTOPHAGY
The best-studied signal to induce autophagy is acute amino acid starvation, which triggers the canonical, regulated pathway, resulting in restoration of the amino acid pool, energy levels, and homeostasis. Induction of autophagy by amino acid withdrawal occurs very rapidly (within 15 min) and is initiated by inactivation of mTORC1 (mammalian target of rapamycin complex 1, TORC1 in yeast) and activation of AMPK (AMP-activated protein kinase) (for a recent review, see Inoki et al. 2012).
Induction of autophagy activates two kinase complexes, the Atg1 (yeast) or ULK1/2 (mammals) complex and the class III phosphatidylinositol-3-phosphate (PI3P) Vps34 complex, including Vps30, known as Beclin1 in mammals. ULK1/2 is a serine/threonine protein kinase, which is found in a complex including Atg101, Atg13, and Atg17 (FIP200 in mammals). The Vps34–Beclin1 complex produces PI3P on the PAS and autophagosome. Both the ULK1/2 and Vps34 complexes are required for the formation of the PAS and for the recruitment of downstream Atg proteins, including WIPI1 and 2 (Atg18 in yeast), Atg9, the only membrane Atg protein, and the ubiquitin-like protein complexes Atg12–Atg5–Atg16 and Atg8-PE (see below).
UBL SYSTEMS AND AUTOPHAGOSOME FORMATION
Two autophagic-specific systems of ubiquitin-like (UBL) proteins control the early events of autophagosome formation (Shpilka et al. 2012). These UBL systems are distinctly different from the classical ubiquitination systems (see Henne et al. 2013) but do share several salient features, in particular, the formation of covalent conjugates. These UBL systems and the UBL proteins regulate elongation and sealing of the autophagic membrane. In the first system, Atg7 and Atg10, E1-like-activating, and E2-like-conjugating enzyme, respectively, conjugate the ubiquitin-like molecule Atg12 to Atg5 (Ohsumi 2001). The Atg12–Atg5 conjugate then interacts with Atg16, and the entire complex localizes on the phagophore but not to the mature autophagosomal membrane (Kuma et al. 2002; Fujioka et al. 2010). Although neither Atg12 nor Atg5 shares amino acid sequence homology with ubiquitin, their crystal structures reveal a single ubiquitin fold for Atg12 (Suzuki et al. 2005) and two adjacent ubiquitin-like folds for Atg5 (Matsushita et al. 2007). Hence, each of the Atg12–Atg5 conjugates is comprised of three ubiquitin-like domains, the function of which remains elusive. The Atg12–Atg5–Atg16 complex is mostly cytosolic, and only a small fraction is associated with the phagophore membrane (Suzuki et al. 2001; Mizushima et al. 2003). This complex is dissociated from the membrane by a yet-unknown mechanism before the completion of the double-membrane autophagosome.
The second system that acts downstream is the ubiquitin-like Atg8 protein family (Ichimura et al. 2000), including a single member in yeast and several homologs, subdivided into three subfamilies—LC3, GABARAP, and GATE-16, in mammals (Shpilka et al. 2011). Atg8s are usually synthesized as precursors that are virtually cotranslationally processed by the cysteine protease Atg4, leading to the exposure of a carboxy-terminal glycine residue (Kirisako et al. 2000). Next, Atg7, the E1 enzyme, shared by the Atg12 system, and Atg3, an E2-like conjugating enzyme, catalyze the formation of an amide bond between phosphatidylethanolamine (PE) and the Atg8 carboxyl terminus (Ichimura et al. 2000), producing the form active in autophagy. The conjugation of Atg8 to PE is transient becaues Atg4 also catalyzes its deconjugation from the membrane (Kirisako et al. 2000). In mammals, this step is subjected to regulation by reactive oxygen species (ROS) formed by the mitochondria (Scherz-Shouval et al. 2007). Unlike the Atg12–Atg5–Atg16 complex, conjugated Atg8 is found both on the outer and the inner membrane of the autophagosomes and as such serves as the only marker for both phagophore and mature autophagosome. Naturally, the Atg8 molecules that localize to the inner membrane of the vacuole are degraded in the vacuole/lysosome as part of the autophagic process. Lipidated Atg8 plays a dual role in the overall autophagic process, acting both in autophagosome biogenesis and in selective recruitment of molecules into autophagosome (Weidberg et al. 2011b). Furthermore, Atg8 molecules were implicated in selective autophagy processes such as mitophagy, xenophagy, pexophagy, and ribophagy (for recent reviews, see Johansen and Lamark 2011; Shpilka et al. 2012).
The exact function of the Atg12–Atg5–Atg16 complex is still not fully understood. Evidence is provided that this Atg12–Atg5–Atg16 complex acts as a unique E3 ubiquitin-like ligase to facilitate and dictate the subcellular localization of the Atg8 ubiquitin-like system (Fujita et al. 2008). Accordingly, deletion of Atg5 in yeast or mammals completely blocks Atg8 conjugation to PE. Studies in vitro have indicated a direct interaction between the Atg12–Atg5 complex and Atg3 (Noda et al. 2013). Moreover, using biochemical and structural information, the mechanism for its interaction with and activation of Atg3 has been revealed (Noda et al. 2013; Sakoh-Nakatogawa et al. 2013), supporting the notion that this complex, indeed, acts as a novel E3 ligase.
Atg8s have also been suggested to mediate the elongation of the expanding autophagosome. By reconstituting the entire conjugation machinery in a test tube, it has been reported that lipidated Atg8 promotes membrane tethering and hemifusion of liposomes (Nakatogawa et al. 2007). In mammalian cells, the different Atg8 subfamilies were found to promote different steps of autophagosome biogenesis (Weidberg et al. 2010, 2011a), and complete inhibition of Atg8 lipidation in mammals prevents autophagosome formation (Fujita et al. 2008). Atg8 fusion activity has been recently questioned based on the fact that it requires relatively high PE concentrations (Nair et al. 2011); however, the exact lipid and protein composition of the phagophore membrane is still unknown. It has been recently suggested that Atg8 acts together with Shp1 and Cdc48 in autophagosome biogenesis in a way that resembles the activity of these factors in the Golgi (Dargemont and Ossareh-Nazari 2012). Clearly, additional work is needed to clarify the role of Atg8 in autophagosome formation.
OVERVIEW OF ENDOCYTIC PATHWAY
Endocytosis is how cells sense their environment, obtain nutrients, and fine-tune their growth status. Decoding the signals generated by binding and internalization of external nutrients and growth factors starts at the plasma membrane and terminates on late endocytic compartments, in particular, the multivesicular body (MVB). Complete termination of the signal or utilization of the internalized nutrients occurs in the lysosome, an acidic proteolytic organelle. To maintain the endocytic pathway, broadly consisting of early endosomes, late endosomes (LEs), and lysosomes, the cell uses its biosynthetic pathway to synthesize both membrane and luminal endocytic components that are targeted to the endocytic compartments, and the cell actively recycles endocytic membranes for reformation of the compartments, thus ensuring that organelle identity is maintained.
The complexity of the endocytic pathway, detailed elsewhere in this collection, is simplified for the purposes of this review. We also simplify the molecular details, largely restricting our definitions to trafficking molecules including the Rabs and other small GTPases (Mizuno-Yamasaki et al. 2012), SNAREs (Jahn and Scheller 2006; Sudhof and Rothman 2009), and tethers (Brocker et al. 2010) in addition to a few well-studied membrane proteins. Importantly, the identity of these compartments is also created by production or conversion of membrane lipids, in particular, the phosphatidylinositols, which are controlled in the endocytic pathway by the Rab proteins (Stenmark 2009). Briefly, vesicles formed by clathrin-dependent or -independent mechanisms from the plasma membrane (see Table 1 in Doherty and McMahon 2009) fuse and deliver their membrane and content to Rab5- and EEA1-positive early endosomes. Early endosomes undergo a conversion from Rab5- to Rab7-positive endosomes (Rink et al. 2005), during which a multidestination dispersal system develops that allows (1) sorting back to the plasma membrane in Rab11-positive recycling endosomes (RE), (2) transport to the Golgi complex mediated by retromer, or (3) down-regulation by sequestration into intraluminal vesicles of the MVB using the ESCRT machinery (Hanson and Cashikar 2012). The Rab11-positive REs also form a juxtanuclear compartment called the endocytic recycling compartment, or ERC. LEs and MVBs mature into lysosomes or fuse with preexisting lysosomes that are enriched in LAMP proteins, which are highly glycosylated membrane proteins, the multisubunit vacuolar H+-ATPase driving acidification, and lysosomal hydrolases, which degrade proteins and lipids (Saftig and Klumperman 2009).
ROLE OF ENDOSOME AND LYSOSOMES IN AUTOPHAGY
The pleomorphic properties of the endosomal pathway present a challenge toward gaining a complete understanding of how endocytosis interacts with autophagy. Whereas many schemes depict the autophagosome fusing with the lysosome, the endocytic and autophagic pathways intersect when autophagosomes fuse with MVBs, forming an amphisome (see Fig. 1). The amphisome then fuses with lysosomes to generate an autolysosome. The convergence of the autophagosome and endolysomal pathways was recognized in early morphological studies (for review, see Eskelinen et al. 2011). The published models depicting direct fusion of the autophagosome with the lysosome are primarily based on the yeast system, but in mammalian cells it is less clear which is the preferred partner—amphisomes or autophagosomes. In contrast, the evidence that the endocytic pathway contributes to earlier autophagosome formation stages is just emerging and remains to be fully understood (Lamb et al. 2012).
Autophagosome Formation and Endosomes
The early work on formation of autophagosomes performed using immunocytochemical analysis clearly showed early autophagosomes labeled with antibodies against rough ER proteins, but no plasma membrane, _trans_-Golgi, or endosomal proteins (Dunn 1990a). However, recent data suggest that the endocytic pathway can contribute to phagophore formation and expansion. Coated vesicles, containing the clathrin adaptor protein AP2, formed from the plasma membrane using a dynamin-driven scission process contained Atg16L1, but not EEA1 or LAMP1 (Ravikumar et al. 2010). This suggests that an endocytic vesicle can contribute to phagophore formation. It is proposed that these vesicles undergo homotypic fusion mediated by a VAMP7-containing SNARE complex (Moreau et al. 2011), forming autophagosome precursor membranes that became LC3 positive. This is surprising in that early endocytic vesicles have not been previously shown to undergo homotypic fusion and are not expected to contain the late endosome SNARE VAMP7. The retention of VAMP7 in this autophagosome precursor as it becomes LC3 positive would, however, provide the nascent autophagosome with at least one of the four SNAREs required for fusion with later endocytic compartments. Another study showed that VAMP7 was not required for autophagosome formation, but implicated Vti1b on autophagosomes and VAMP8 on the lysosome (Furuta et al. 2010). Interestingly, VAMP7 has also been implicated in the formation of LAMP-positive, MPR-negative TGN-derived vesicles called LAMP carriers, which transport LAMPs to the LE; loss of VAMP7 causes accumulation of these vesicles (Pols et al. 2013). It will be important to distinguish and delineate the population of VAMP7 vesicles involved in autophagy and endocytosis for further progress.
Additional contributions from endosomal membranes to forming autophagosomes are via vesicles derived from Rab11-positive RE. Both ULK1 and Atg9 are present on RE, but on different subdomains (Longatti et al. 2012). Upon amino acid starvation, the ULK complex together with transferrin and the transferrin receptor are delivered in Rab11 vesicles to forming autophagosomes, which colocalize with LC3. This trafficking step is inhibited by overexpression of a Rab11 effector, TBC1D14 (Longatti et al. 2012). The RE-endosome-derived membranes are delivered to nascent autophagosomes, because transferrin-HRP was detected in between the outer membrane of the newly formed autophagosomes (Longatti et al. 2012). Delivery of vesicles from an early endocytic compartment was also observed in previous EM studies whereby within 10 min of endocytosis endocytic tracers were detected in vesicular structures fusing with nascent autophagosomes (Punnonen et al. 1993; Liou et al. 1997). Aside from the requirement for Rab11 in this vesicular transport step, it is not clear what other machinery is required. A distinction should be made between endosomes (e.g., RE) involved in formation as those that do not carry either LAMPs or lysosomal hydrolases, from those endosomes (MVB or LEs) that contain either or both. Rab11 is also required for fusion of the autophagosomes with MVBs (Fader et al. 2008), raising the possibility that Rab11 effectors on the nascent autophagosome mediate this fusion.
Autophagosome Maturation and Endosomes
Shortly after autophagosomes form, they acquire proteins and enzymes that are normally found in MVB, LEs, and lysosomes (Dunn 1990b; Tooze et al. 1990; Punnonen et al. 1993), which supports the model in which LAMPs and the V-ATPase are delivered by vesicle fusion to the outer autophagosome membrane before the lysosomal hydrolases. It has also been proposed that acidification occurs before delivery of enzymes, and presumably this enables rapid activation of the hydrolases and degradation of the inner autophagosome membrane, which facilitates its transition to an amphisome. To become an autolysosome, the amphisome can either mature or fuse with a preexisiting lysosome, but the current data do not distinguish between these possibilities. However, the most compelling data to support the formation of autolysosomes by sequential fusion are the accumulation of autophagosomes and amphisomes caused by loss of trafficking complexes that control endosomes: COPI, which disrupts early endosomal trafficking (Razi et al. 2009), or mutants in ESCRT components, which disrupt the MVB (Rusten and Stenmark 2009), and fusion of the amphisome with lysosomes (Filimonenko et al. 2007). In addition, loss of the vesicular cation release channel called MCOLN1/TRPML1 inhibits fusion of amphisomes with lysosomes causing an accumulation of amphisomes (Wong et al. 2012).
Rab7, a late endosomal Rab, is found on nascent autophagosomes, but proposed only to be required for fusion of autophagosomes with lysosomes (Gutierrez et al. 2004; Jager et al. 2004). Rab7 also mediates transport of autophagosomes along microtubules by binding FYCO, a Rab7–PI3P–LC3B-binding protein, proposed to mediate plus-end movement of autophagosomes (Pankiv et al. 2010). In addition, only Rab7, but not Vps16 (part of the HOPS-CORVET core complex; see below), is required for autophagosome maturation (Ganley et al. 2011). This study also showed that thapsigargin treatment inhibits fusion of autophagosomes with lysosomes, most probably by modulating a pool of Ca2+ that has not been identified.
Reformation of Autolysosomes and TOR
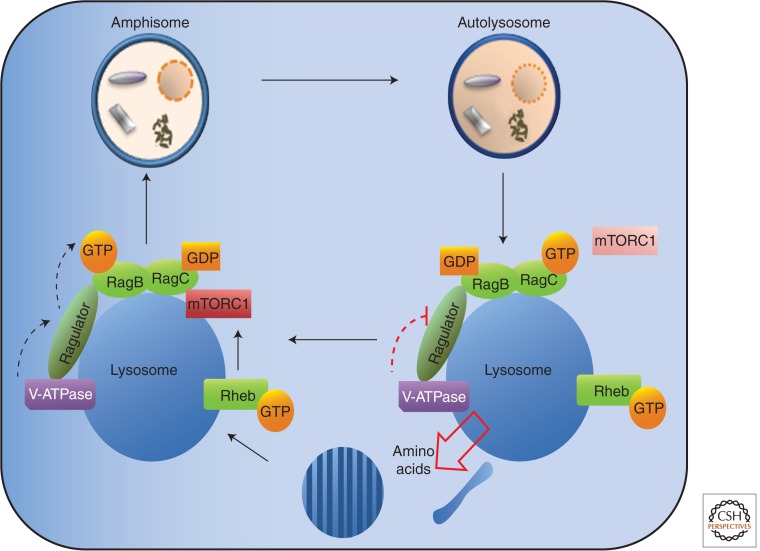
The autolysosome is the terminal stage of autophagy and is dependent on a stable cohort of lysosomes. The lysosome is adapted to uniquely provide a powerful degradative environment while maintaining active transporters in its limiting membrane. The biogenesis of the lysosome and the pool of lysosomes is coordinated by TFEB transcription factor, which regulates the CLEAR network and, in addition, coordinates the lysosomal pathway with the autophagic pathway (Settembre et al. 2011). After complete fusion with LEs or autophagosomes, lysosomes are maintained by reformation from the hybrid organelle or the autolysosome, respectively (Luzio et al. 2007; Yu et al. 2010). Interestingly, under normal conditions, lysosome reformation was detected after 30 min (Bright et al. 2005), whereas under nutrient deprivation, reformation occurred only after 4 h (Yu et al. 2010), and this latter reformation, called autophagosome lysosome reformation (ALR), required reactivation of mTORC1 (Fig. 2). Reformation in both cases would entail removal of endosomal or autophagosomal membrane components, including the SNAREs used for the fusion events and Rab 7 in particular after prolonged starvation (Yu et al. 2010).
Figure 2.
Lysosome function and reformation after autophagy. The activity of mTORC1 on the lysosome is coupled with amino acid production and reformation of the lysosome after amino acid starvation (based on data from Lamb et al. 2012).
mTORC1 integrates external signals, including amino acid and growth factor availability to control cell growth. It is not clear how amino acids control mTORC1, but recent data indicate that it involves mTORC1 recruitment to the lysosome surface (Sancak et al. 2008). In particular, the signal from amino acids within the lysosomal lumen is sensed by the V-ATPase and the Ragulator complex, leading to GTP loading and activation of the Rag GTPases, which subsequently recruit mTORC1 (Sancak et al. 2010; Zoncu et al. 2011). Although the exact mechanisms underlying this pathway remain elusive, very rapid progress is being made. During prolonged starvation, mTORC1 can be reactivated at the lysosomes by amino acid release as a result of autophagic degradation. This mechanism of ALR and autophagy inhibition provides a self-regulatory feedback mechanism whereby, once autophagy has improved the nutritional status of the cell, it is terminated to protect the cell and the autolysosome membranes are recycled to restore lysosome number.
TRAFFICKING MACHINERY REQUIRED FOR AUTOPHAGY
Tethering Factors
A fundamental question in intracellular membrane trafficking is how directionality and specificity are achieved and maintained? At least two types of protein complexes are involved: tethering factors that are regulated by the Rab-family GTPases, and SNARE proteins that provide specificity and promote membrane fusion.
Tethering factors assist in delivering vesicles to their target compartment and play an important role in conferring specificity in membrane trafficking and fusion events (for review, see Yu and Hughson 2010). They interact with membrane-associated Rab proteins. In fact, most tethering factors are Rab effectors; thus, the regulation of tethering activity occurs through GTP binding and hydrolysis. In addition, evidence has been provided that these complexes act to regulate specificity in the assembly of the SNARE complexes.
Tethering factors are found either as a distinct coiled-coil proteins such as Uso1/p115, Bug1, Golgins, and early endosomal antigen (EEA1), or large multisubunit complexes such as TRAPP, COG, HOPS, CORVET, Dsl1, Exocyst, and GRAP. Studies in yeast and mammals suggested the involvement of tethering factors both in early stages of autophagosome formation as well as in regulating the fusion between mature autophagosomes and the vacuole/lysosome (Fig. 3).
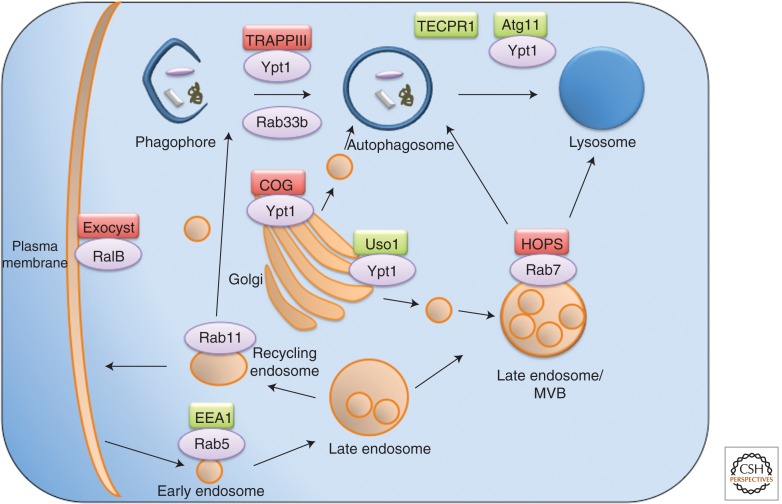
Figure 3.
Tethering complexes and small GTPases implicated in endocytosis and autophagy. See text for details.
The COG is an evolutionally conserved Golgi-associated protein complex that has been characterized both in yeast and mammalian cells. It functions as a tethering factor for vesicles that recycle within the Golgi apparatus and vesicles that recycle to the Golgi from the endosomal compartments (VanRheenen et al. 1999; Suvorova et al. 2002). The complex is composed of eight subunits (Cog1–8), which can be divided into two structurally and functionally distinct subcomplexes, lobe A (Cog1–4), which is essential in yeast (VanRheenen et al. 1999), and lobe B (Cog5–8) (Ram et al. 2002; Ungar et al. 2005). By studying the trafficking of the yeast Atg9, the only multitransmembrane autophagy protein, the involvement of lobe A in the cytosol-to-vacuole targeting (CVT) pathway, a selective yeast-specific autophagy pathway, as well as in starvation-induced autophagy was revealed (Yen et al. 2010). Other assays including EM analysis showing the accumulation of open membrane phagophore structures suggested that the COG complex was involved in autophagosome biogenesis, in particular, in PAS organization. Thus far, however, no evidence is provided for the participation of mammalian COG complexes in autophagy. Moreover, it is important to determine whether a specific form of a COG complex composed of a different subunit composition is essential for autophagy.
Another tethering complex involved in autophagosome biogenesis is the octameric TRAPPIII (Meiling-Wesse et al. 2005; Lynch-Day et al. 2010). TRAPP tethering complexes are a group of three distinct guanine-nucleotide exchange factors (GEFs) first identified in yeast that activate the Rab GTPase Ypt1 (Barrowman et al. 2010). TRAPPI and TRAPPII participate in Golgi-related trafficking events. Trs85 was recently identified as a unique subunit in the TRAPPIII complex found to regulate autophagy in yeast (Lynch-Day et al. 2010) and is known to bind Ypt31/32, the yeast ortholog of Rab11 (Jones et al. 2000). The Trs85 subunit specifically directs the complex and Ypt1 to the PAS, enabling the activity of the Rab GTPase Ypt1 in both the CVT pathway and starvation-induced autophagy. Consistently, Ypt1 and the Trs85 TRAPPIII subunit localize to Atg9 peripheral structures in yeast, suggesting their involvement in membrane recruitment to the elongating autophagosomes (Kakuta et al. 2012; Lipatova et al. 2012).
In mammalian cells, the exocyst complex was found to mediate the initial stages of autophagosome biogenesis (Bodemann et al. 2011). Similar to the COG and TRAPPIII complexes, it contains eight different subunits (Munson and Novick 2006; He and Guo 2009) and is involved in post-Golgi vesicle tethering to the plasma membrane. Exo84, an exocyst subunit, binds the small Rab GTPase RalB following nutrient starvation and interacts with early factors of autophagy induction such as Beclin1, Vps34, and Ulk1 (Bodemann et al. 2011). These interactions are inhibited under normal growth conditions by the interaction of the small Rab GTPase RalA to the Sec5 subunit, which binds the aforementioned autophagy proteins as well as mTORC1. Furthermore, additional subunits of the exocyst complex interacted with core autophagy proteins in a yeast two-hybrid screen. Accordingly, it has been hypothesized that the exocyst complex serves as a scaffold for autophagy initiation and autophagosome biogenesis at the isolation membrane.
The HOPS tethering complex has recently been reported to mediate autophagosome maturation in Drosophila and mammalian cells (Lindmo et al. 2006; Liang et al. 2008). This complex, first identified in yeast, is a Rab7 effector involved in vacuolar-related fusion events (Brett et al. 2008; Nickerson et al. 2009). Deletion of HOPS subunits leads to inhibition of LC3 accumulation in lysosomes (Liang et al. 2008). HOPS complex subunits and Rab7 were targeted to autophagosomes following overexpression of the tumor suppressor UVRAG. A more recent study has indicated that Rab7 and the HOPS subunit Vps16 are both essential for endosomal fusion with the lysosomes, whereas only Rab7 (and not Vps16) is essential for autophagosome–lysosome fusion (Ganley et al. 2011).
Increasing evidence in both yeast and mammalian cells indicates the involvement of tethering factors in different stages of the autophagic process; however, the mechanism by which these complexes act in this pathway remains largely unknown. Because these factors are essential for other intracellular trafficking pathways, it is important to exclude indirect effects. Moreover, it is possible that the tethering complexes that are specifically designated to act in autophagy are modified (e.g., by phosphorylation) or include as-yet-uncharacterized factors. How these factors are regulated is yet another issue that requires future studies.
SNAREs
Membrane fusion events are generally mediated by specific sets of SNARE complexes found on opposing sides of the fusing membranes (Jahn and Scheller 2006; Sudhof and Rothman 2009). Typically, SNARE molecules are found in a tertiary complex of a Qa-, Qb-, Qc-, and R-SNARE in one membrane that needs to be separated (primed) before membrane fusion. SNAP and NSF in mammals and Sec17 and Sec18 in yeast mediate this reaction, which requires the hydrolysis of ATP (Wickner and Schekman 2008).
SNAREs, NSF, and SNAP were implicated in the different steps of the autophagic process (Fig. 3) (Moreau et al. 2013). Early studies in yeast revealed that formation of CVT vesicles, but not autophagosomes, requires Tlg2 and Vps45 (Abeliovich et al. 1999). More recent studies in yeast revealed that the multispanning-membrane protein Atg9 cycles between the PAS and different organelles (Reggiori et al. 2005). It has been suggested that Atg9 is localized within tubulovesicular structures trafficked from the Golgi to the PAS as part of its role in autophagosome formation (Fig. 4) (Mari et al. 2010). A similar function and compartmentalization have been discovered in mammalian cells (Orsi et al. 2012). By following the GFP-Atg9-labeled structures, Klionsky and coworkers identified the yeast Q-SNAREs, Sso1, Sso2, Sec9, and Tlg2 and the R-SNARE Sec22 as being involved both in the formation of the Atg9-containing tubulovesicular structures and the overall autophagy process (Nair et al. 2011). The involvement of Sec17 and Sec18 in autophagosome formation was originally studied by Ohsumi and coworkers, who provided evidence that these proteins are needed only for autophagosome fusion with the vacuole (Ishihara et al. 2001). A more recent study suggests that Sec18 and Sec17 are also required for the formation of autophagosomes (Nair et al. 2011). These results imply a direct role for Golgi-derived membranes and SNAREs in autophagosome formation. At present, however, there is no evidence for such a role for the mammalian orthologs of these SNAREs in autophagy.
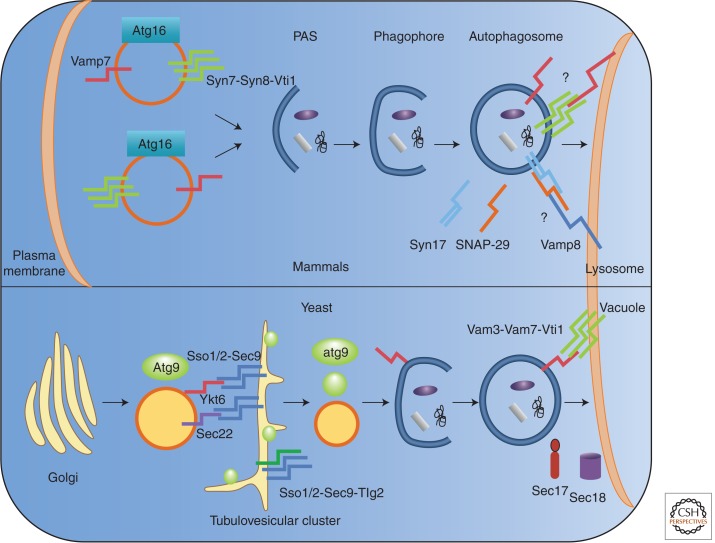
Figure 4.
SNAREs implicated in autophagosome formation and maturation. Mammalian (top) and yeast (bottom) SNAREs involved in autophagy. All SNARES have a single transmembrane spanning domain except syntaxin 17, which has two transmembrane domains (Itakura et al. 2012).
The main evidence for participation of mammalian SNAREs in autophagosome formation originated from Rubinsztein’s laboratory (Moreau et al. 2011). Accordingly, VAMP7, syntaxin 7, syntaxin 8, and Vti1B were found to mediate homotypic membrane fusion of plasma-membrane-derived vesicles to form phagophores (Moreau et al. 2011). This process was sensitive to NEM, suggesting the requirement for NSF in early stages of autophagosome formation. Here, too, a more direct approach is needed to study this process in detail. Indeed, a partial reconstitution of these membrane fusion events in the test tube was reported, and future studies should allow a resolution of this and other mechanistic aspects of autophagosome biogenesis.
As mentioned above, autophagosomes and amphisomes fuse with the lysosomal membrane. The exact machinery of this process is not yet fully understood. Studies in yeast indicated that Vam3, Vam7, Ykt6, and Vti1 participate in autophagosome–vacuole membrane fusion (Darsow et al. 1997; Fischer von Mollard and Stevens 1999; Dilcher et al. 2001; Fader et al. 2009; Ohashi and Munro 2010); however, the exact orientation of these molecules with respect to the two organelles remained elusive. A recent study by Mizushima and coworkers identified for the first time a SNARE molecule, syntaxin 17, on the outer membrane of mature autophagosomes (Itakura et al. 2012). This study provides biochemical and morphological evidence that syntaxin 17, a unique SNARE with a carboxy-terminal hairpin transmembrane domain, is localized on the autophagosomal but not phagophore membrane, raising the possibility that SNAREs needed for autophagosome–lysosome membrane fusion are targeted to the autophagosome membrane only upon its completion and closure. Although the exact mechanism for targeting of syntaxin 17 remains unknown, it provides a way to prevent premature fusion between phagophore and lysosome. It has been previously suggested that the spatial separation between phagophore and lysosome is achieved by microtubules, which transport only mature autophagosomes but not phagophores (Fass et al. 2006; Köchl et al. 2006). Incorporation of the fusion machinery after formation of the mature autophagosome will also ensure that the SNARE molecules needed for fusion with the lysosome will be only localized in the outer membrane and thus avoid lysosomal degradation following membrane fusion.
To complete its fusion activity, syntaxin 17 interacts with SNAP-29 and VAMP8 localized on the lysosomal membrane (Itakura et al. 2012). The contribution of syntaxin 17 to autophagosome biogenesis was also examined by Yoshimori and coworkers, who found that syntaxin 17 acts early in autophagosome formation, recruiting Atg14, a subunit of the autophagy-specific Vps34 PI3 kinase, to the ER–mitochondria contact sites (Hamasaki et al. 2013). Consistent with Itakura et al. (2012), however, Hamasaki et al. (2013) report that in the absence of syntaxin 17, LC3 puncta accumulate, confirming syntaxin 17’s role in autophagosome maturation. Clearly, elucidating the role of syntaxin 17 in autophagy is an important future research avenue.
CONCLUSIONS, REMARKS, AND FUTURE PERSPECTIVES
The formation and maturation of autophagosomes is a complex process, entailing vesicular trafficking and fusion events, and substantial reorganization of existing compartments, or subdomains of compartments, in particular, the ER but including the plasma membrane, Golgi, and mitochondria. In canonical autophagy, which originates in the ER, remodeling of the ER-derived autophagosome must occur to allow for their fusion with the endosome and lysosome. A particular unresolved issue is the balance between bulk conversion by compartment fusion (e.g., fusion of the nascent autophagosome with an MVB) versus conversion of an autophagosome to a “endosome”-like compartment mediated by vesicle trafficking and fusion. The former would be an efficient and rapid single-fusion event, whereas the latter would be presumably a slower process, but would allow a stepwise remodeling of the maturing autophagosome. In both cases, an unresolved issue is how recognition between fusion partners is established. Although much evidence supports a stepwise conversion of the nascent autophagosome into a hybrid autophagosome–endosome and some of the molecular components have been identified to support this process, there are still many unanswered questions that await an understanding of the full molecular mechanism.
ACKNOWLEDGMENTS
S.A.T. is funded by Cancer Research UK. Z.E. is the incumbent of the Harold Korda Chair of Biology and is funded by the Israeli Science Foundation (ISF), the German-Israeli Foundation (GIF), and the German Minerva Foundation.
Footnotes
Editors: Sandra L. Schmid, Alexander Sorkin, and Marino Zerial
REFERENCES
*Reference is also in this collection.
- Abeliovich H, Darsow T, Emr SD 1999. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J 18: 6005–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S 2010. TRAPP complexes in membrane traffic: Convergence through a common Rab. Nat Rev Mol Cell Biol 11: 759–763 [DOI] [PubMed] [Google Scholar]
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, et al. 2011. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 144: 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett CL, Plemel RL, Lobingier BT, Vignali M, Fields S, Merz AJ 2008. Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J Cell Biol 182: 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright NA, Gratian MJ, Luzio JP 2005. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol 15: 360–365 [DOI] [PubMed] [Google Scholar]
- Brocker C, Engelbrecht-Vandre S, Ungermann C 2010. Multisubunit tethering complexes and their role in membrane fusion. Curr Biol 20: R943–R952 [DOI] [PubMed] [Google Scholar]
- Choi AMK, Ryter SW, Levine B 2013. Autophagy in human health and disease. N Engl J Med 368: 651–662 [DOI] [PubMed] [Google Scholar]
- Dargemont C, Ossareh-Nazari B 2012. Cdc48/p97, a key actor in the interplay between autophagy and ubiquitin/proteasome catabolic pathways. Biochim Biophys Acta 1823: 138–144 [DOI] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD 1997. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol 138: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilcher M, Kohler B, von Mollard GF 2001. Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J Biol Chem 276: 34537–34544 [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT 2009. Mechanisms of endocytosis. Annu Rev Biochem 78: 857–902 [DOI] [PubMed] [Google Scholar]
- Dunn WA Jr 1990a. Studies on the mechanisms of autophagy: Formation of the autophagic vacuole. J Cell Biol 110: 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA Jr 1990b. Studies on the mechanisms of autophagy: Maturation of the autophagic vacuole. J Cell Biol 110: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E-L, Reggiori F, Baba M, Kovács AL, Seglen PO 2011. Seeing is believing: The impact of electron microscopy on autophagy research. Autophagy 7: 935–956 [DOI] [PubMed] [Google Scholar]
- Fader CM, Sanchez D, Furlan M, Colombo MI 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9: 230–250 [DOI] [PubMed] [Google Scholar]
- Fader CM, Sanchez DG, Mestre MB, Colombo MI 2009. TI-VAMP/VAMP7 and VAMP3/cellubrevin: Two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta 1793: 1901–1916 [DOI] [PubMed] [Google Scholar]
- Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z 2006. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 281: 36303–36316 [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 179: 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stevens TH 1999. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell 10: 1719–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F 2010. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem 285: 1508–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T 2008. The Atg16 L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta N, Fujita N, Noda T, Yoshimori T, Amano A 2010. Combinational soluble _N_-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell 21: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Wong P-M, Gammoh N, Jiang X 2011. Distinct autophagosomal–lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 42: 731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 117: 2687–2697 [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. 2013. Autophagosomes form at ER–mitochondria contact sites. Nature 495: 389–393 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Cashikar A 2012. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28: 337–362 [DOI] [PubMed] [Google Scholar]
- He B, Guo W 2009. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol 21: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Henne WM, Stenmark H, Emr SD 2013. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol 5: a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Inoki K, Kim J, Guan K-L 2012. AMPK and mTOR in cellular energy homeostasis and drug targets. Ann Rev Pharm Toxic 52: 381–400 [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell 12: 3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151: 1256–1269 [DOI] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen E-L 2004. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 117: 4837–4848 [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH 2006. SNAREs—Engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Newman C, Liu F, Segev N 2000. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell 11: 4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S, Yamamoto H, Negishi L, Kondo-Kakuta C, Hayashi N, Ohsumi Y 2012. Atg9 vesicles recruit vesicle-tethering proteins, Trs85 and Ypt1, to the autophagosome formation site. J Biol Chem 287: 44261–44269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köchl R, Hu X, Chan E, Tooze SA 2006. Microtubules facilitate autophagosomal formation and fusion of autophagosomes with endosomes. Traffic 7: 129–145 [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y 2002. Formation of the approximately 350-kDa Apg12–Apg5. Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 277: 18619–18625 [DOI] [PubMed] [Google Scholar]
- Lamb CA, Dooley HC, Tooze SA 2012. Endocytosis and autophagy: Shared machinery for degradation. BioEssays 35: 34–45 [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 10: 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K, Simonsen A, Brech A, Finley K, Rusten TE, Stenmark H 2006. A dual function for Deep orange in programmed autophagy in the Drosophila melanogaster fat body. Exp Cell Res 312: 2018–2027 [DOI] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Geelen MJH, Slot JW 1997. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol 136: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N 2012. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci 109: 6981–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S-I, Barr FA, Tooze SA 2012. TBC1D14 regulates autophagosome formation via Rab11 and recycling endosomes. J Cell Biol 197: 659–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA 2007. Lysosomes: Fusion and function. Nat Rev Mol Cell Biol 8: 622–632 [DOI] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci 107: 7811–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F 2010. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 190: 1005–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M, Suzuki NN, Obara K, Fujioka Y, Ohsumi Y, Inagaki F 2007. Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem 282: 6763–6772 [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen EL, Thumm M 2005. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem 280: 33669–33678 [DOI] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Rivera-Molina F, Novick P 2012. GTPase networks in membrane traffic. Annu Rev Biochem 81: 637–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T 2003. Mouse Apg16 L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12–Apg5 conjugate. J Cell Sci 116: 1679–1688 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y 2011. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biology 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC 2011. Autophagosome precursor maturation requires homotypic fusion. Cell 146: 303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Renna M, Rubinsztein DC 2013. Connections between SNAREs and autophagy. Trends Biochem Sci 38: 57–63 [DOI] [PubMed] [Google Scholar]
- Munson M, Novick P 2006. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol 13: 577–581 [DOI] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, et al. 2011. SNARE proteins are required for macroautophagy. Cell 146: 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178 [DOI] [PubMed] [Google Scholar]
- Nickerson DP, Brett CL, Merz AJ 2009. Vps-C complexes: Gatekeepers of endolysosomal traffic. Curr Opin Cell Biol 21: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F 2013. Structure of the Atg12–Atg5 conjugate reveals a platform for stimulating Atg8–PE conjugation. EMBO Rep 14: 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Munro S 2010. Membrane delivery to the yeast autophagosome from the Golgi–endosomal system. Mol Biol Cell 21: 3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y 2001. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley H, Robinson D, Weston A, Collinson L, Tooze S 2012. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, is required for autophagy. Mol Biol Cell 23: 1860–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun J-A, Lamark T, Overvatn A, Bjorkoy G, Johansen T 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 188: 253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols MS, van Meel E, Oorschot V, ten Brink C, Fukuda M, Swetha MG, Mayor S, Klumperman J 2013. hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nat Commun 4: 1361. [DOI] [PubMed] [Google Scholar]
- Punnonen EL, Autio S, Kaija H, Reunanen H 1993. Autophagic vacuoles fuse with the prelysosomal compartment in cultured rat fibroblasts. Eur J Cell Biol 61: 54–66 [PubMed] [Google Scholar]
- Ram RJ, Li B, Kaiser CA 2002. Identification of Sec36p, Sec37p, and Sec38p: Components of yeast complex that contains Sec34p and Sec35p. Mol Biol Cell 13: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 12: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M, Chan EY, Tooze SA 2009. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol 185: 305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Shintani T, Nair U, Klionsky DJ 2005. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 1: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell 122: 735–749 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z 2012. Mechanisms of autophagosome biogenesis. Curr Biol 22: R29–R34 [DOI] [PubMed] [Google Scholar]
- Rusten TE, Stenmark H 2009. How do ESCRT proteins control autophagy? J Cell Sci 122: 2179–2183 [DOI] [PubMed] [Google Scholar]
- Saftig P, Klumperman J 2009. Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nat Rev Mol Cell Biol 10: 623–635 [DOI] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M, Matoba K, Asai E, Kirisako H, Ishii J, Noda NN, Inagaki F, Nakatogawa H, Ohsumi Y 2013. Atg12–Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol 20: 433–439 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM 2008. The Rag GTPases bind Raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM 2010. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z 2007. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Weidberg H, Pietrokovski S, Elazar Z 2011. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol 12: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Mizushima N, Elazar Z 2012. Ubiquitin-like proteins and autophagy at a glance. J Cell Sci 125: 2343–2348 [DOI] [PubMed] [Google Scholar]
- Stenmark H 2009. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE 2009. Membrane fusion: Grappling with SNARE and SM proteins. Science 323: 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova ES, Duden R, Lupashin VV 2002. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol 157: 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Noda T, Ohsumi Y 2004. Interrelationships among Atg proteins during autophagy in Saccharomyces cerevisiae. Yeast 21: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F 2005. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy 1: 119–126 [DOI] [PubMed] [Google Scholar]
- Tooze J, Hollinshead M, Ludwig T, Howell K, Hoflack B, Kern H 1990. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J Cell Biol 111: 329–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Oka T, Vasile E, Krieger M, Hughson FM 2005. Subunit architecture of the conserved oligomeric Golgi complex. J Biol Chem 280: 32729–32735 [DOI] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Sapperstein SK, Chiang EC, Lupashin VV, Barlowe C, Waters MG 1999. Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J Cell Biol 147: 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z 2010. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 29: 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z 2011a. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell 20: 444–454 [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z 2011b. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80: 125–156 [DOI] [PubMed] [Google Scholar]
- Wickner W, Schekman R 2008. Membrane fusion. Nat Struct Mol Biol 15: 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C-O, Li R, Montell C, Venkatachalam K 2012. Drosophila TRPML is required for TORC1 activation. Curr Biol 22: 1616–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ 2010. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol 188: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IM, Hughson FM 2010. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol 26: 137–156 [DOI] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]