Improved vectors and genome-wide libraries for CRISPR screening (original) (raw)
. Author manuscript; available in PMC: 2015 Jul 1.
Published in final edited form as: Nat Methods. 2014 Aug;11(8):783–784. doi: 10.1038/nmeth.3047
Genome-wide, targeted loss-of-function pooled screens using the CRISPR (clustered regularly interspaced short palindrome repeats)–associated nuclease Cas9 in human and mouse cells provide an alternative screening system to RNA interference (RNAi) and have been used to reveal new mechanisms in diverse biological models1-4. Previously, we used a Genome-scale CRISPR Knock-Out (GeCKO) library to identify loss-of-function mutations conferring vemurafenib resistance in a melanoma model1. However, initial lentiviral delivery systems for CRISPR screening had low viral titer or required a cell line already expressing Cas9, limiting the range of biological systems amenable to screening.
Here, we sought to improve both the lentiviral packaging and choice of guide sequences in our original GeCKO library1, where a pooled library of synthesized oligonucleotides was cloned into a lentiviral backbone containing both the Streptococcus pyogenes Cas9 nuclease and the single guide RNA (sgRNA) scaffold. To create a new vector capable of producing higher-titer virus (lentiCRISPRv2), we made several modifications, including removal of one of the nuclear localization signals (NLS), human codon-optimization of the remaining NLS and P2A bicistronic linker sequences, and repositioning of the U6-driven sgRNA cassette (Fig. 1a). These changes resulted in a ~10-fold increase in functional viral titer over lentiCRISPRv11 (Fig. 1b).
Figure 1.
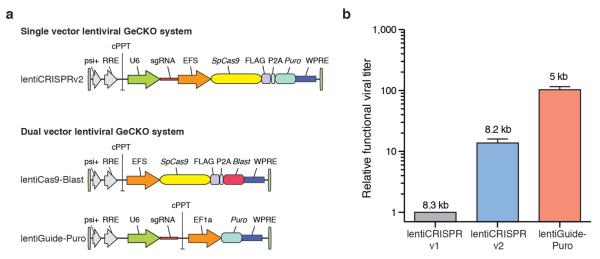
New lentiviral CRISPR designs produce viruses with higher functional titer.
(a) Lentiviral expression vector for Streptococcus pyogenes Cas9 and sgRNA in the improved one vector system (lentiCRISPR v2) and the two vector system (lentiCas9-Blast, lentiGuide-Puro). Psi packaging signal (psi+), rev response element (RRE), central polypurine tract (cPPT), elongation factor-1α short promoter (EFS), FLAG octapeptide tag (FLAG), 2A self-cleaving peptide (P2A), puromycin selection marker (puro), posttranscriptional regulatory element (WPRE), blasticidin selection marker (blast), and elongation factor-1α promoter (EF1a). (b) Relative functional viral titer of viruses made from lentiCRISPR v11, lentiCRISPR v2, and lentiGuide-Puro vector with a EGFP-targeting sgRNA (n = 3 independently-transfected virus batches with 3 replicate transductions per construct). HEK293FT cells were transduced with serial dilutions of virus and, after 24 hours, selected using puromycin (1 ug/ml). Puromycin-resistant cells were measured after 4 days from the start of selection using the CellTiter-Glo (Promega) luciferase assay. Relative titers were calculated using viral volumes that yielded less than 20% puromycin-resistant cells in order to minimize the number of cells with multiple infections. Numbers above each bar indicate the size of the packaged virus for each construct.
To further increase viral titer, we also cloned a two-vector system, in which Cas9 (lentiCas9-Blast) and sgRNA (lentiGuide-Puro) are delivered using separate viral vectors with distinct antibiotic selection markers (Fig. 1a). LentiGuide-Puro has a ~100-fold increase in functional viral titer over the original lentiCRISPRv1 (Fig. 1b). Both single and dual-vector systems mediate efficient knock-out of a genomically-integrated copy of EGFP in human cells (Supplementary Fig. 1). Whereas the dual vector system enables generation of Cas9-expressing cell lines which can be subsequently used for screens using lentiGuide-Puro, the single vector lentiCRISPRv2 may be better suited for in vivo or primary cell screening applications.
In addition to the vector improvements, we designed and synthesized new human and mouse GeCKOv2 sgRNA libraries (Supplementary Methods) with several improvements (Table 1): First, for both human and mouse libraries, to target all genes with a uniform number of sgRNAs, we selected 6 sgRNAs per gene distributed over 3-4 constitutively expressed exons. Second, to further minimize off-target genome modification, we improved the calculation of off-target scores based on specificity analysis5. Third, to inactivate microRNAs (miRNAs) which play a key role in transcriptional regulation, we added sgRNAs to direct mutations to the pre-miRNA hairpin structure6. Finally, we targeted ~1000 additional genes not included in the original GeCKO library.
Table 1.
Comparison of new GeCKO v2 human and mouse sgRNA libraries with existing CRISPR libraries.
| Wang et al. (2014)library | Shalem et al. (2014)GeCKO v1 library | Yusa et al. (2014)library | GeCKO v2human library | GeCKO v2mouse library | |
|---|---|---|---|---|---|
| Species | human | human | mouse | human | mouse |
| Number of genes targeted | 7,114 | 18,080 | 19,150 | 19,050 | 20,611 |
| Targeting constructs per gene | 10 per gene | variable (typically 3 to 4per gene) | variable (typically 4 to5 per gene) | 6 per gene(3 in Library A,3 in Library B) | 6 per gene(3 in Library A,3 in Library B) |
| Number of miRNA targeted | none | none | none | 1,864 | 1,175 |
| Targeting constructs per miRNA | n/a | n/a | n/a | 4 per miRNA | 4 per miRNA |
| Control (non- targeting) sgRNAs | 100 | none | none | 1,000 | 1,000 |
| Total sgRNA constructs | 73,151 | 64,751 | 87,897 | 123,411(65,383 in Library A,58,028 in Library B) | 130,209(67,405 in Library A,62,804 in Library B) |
| Viral plasmid vector | Dual vector:sgRNA only | Single vector:Cas9 and sgRNA(lentiCRISPR v1) | Dual vector:sgRNA only | Single and dual vector:lentiCRISPR v2 andlentiGuide-Puro | Single and dual vector:lentiCRISPR v2 andlentiGuide-Puro |
Both libraries, mouse and human, are divided into 2 sub-libraries — containing 3 sgRNAs targeting each gene in the genome, as well as 1000 non-targeting control sgRNAs. Screens can be performed by combining both sub-libraries, yielding 6 sgRNAs per gene, for higher coverage. Alternatively, individual sub-libraries can be used in situations where cell numbers are limiting (eg. primary cells, in vivo screens). The human and mouse libraries have been cloned into lentiCRISPRv2 and into lentiGuide-Puro and deep sequenced to ensure uniform representation (Supplementary Fig. 2, 3). These new lentiCRISPR vectors and human and mouse libraries further improve the GeCKO reagents for diverse screening applications. Reagents are available to the academic community through Addgene and associated protocols, support forums, and computational tools are available via the Zhang lab website (www.genome-engineering.org).
Supplementary Material
Supplementary methods and figures
Acknowledgements
We also would like to thank the entire Zhang Lab for support and helpful advice. N.S. is supported by a postdoctoral fellowship from the Simons Center for the Social Brain at MIT. O.S. is a Klarman Fellow of the Broad-Israel Partnership. F.Z. is supported by the NIH and NIMH through a Director’s Pioneer Award (5DP1-MH100706), a Transformative R01 grant (5R01-DK097768), the Keck, Merkin, Vallee, Damon Runyon, Searle Scholars, Klingenstein, and Simons Foundations, Bob Metcalfe, and Jane Pauley.
Footnotes
Author Contributions
N.S., O.S., and F.Z. conceived and designed the experiments. N.S. and O.S. performed the experiments and analyzed the data. N.S., O.S., and F.Z. wrote the manuscript.
Competing Financial Interests
A patent application has been filed relating to this work, and the authors plan on making the reagents widely available to the academic community through Addgene and to provide software tools via the Zhang lab website (http://www.genome-engineering.org/). F.Z. is a scientific advisor for Editas Medicine.
References
- 1.Shalem O, et al. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang T, Wei JJ, Sabatini DM, Lander ES. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Nat Biotechnol. 2013;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, et al. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 5.Hsu PD, et al. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, et al. Sci Rep. 2014;4:3943. doi: 10.1038/srep03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods and figures