Data use under the NIH GWAS Data Sharing Policy and future directions (original) (raw)
Dina Paltoo, Laura Lyman Rodriguez, Michael Feolo and colleagues present their analysis of the usefulness and impact of the first seven years of data sharing via the dbGaP repository and announce the extension of data-sharing provisions to other types of research funded by the NIH.
Supplementary information
The online version of this article (doi:10.1038/ng.3062) contains supplementary material, which is available to authorized users.
Subject terms: Computational biology and bioinformatics, Genome-wide association studies, Policy
Abstract
In 2007, the US National Institutes of Health (NIH) introduced the Genome-Wide Association Studies (GWAS) Policy and the database of Genotypes and Phenotypes (dbGaP) to facilitate 'controlled' access to GWAS data based on participants' informed consent. dbGaP has provided 2,221 investigators access to 304 studies, resulting in 924 publications and significant scientific advances. Following on this success, the 2014 Genomic Data Sharing Policy will extend the GWAS Policy to additional data types.
Supplementary information
The online version of this article (doi:10.1038/ng.3062) contains supplementary material, which is available to authorized users.
Main
Following on early successes with GWAS data, the NIH launched two initiatives in 2007 to leverage genomic data while respecting the privacy and autonomy of study participants. The first, the NIH Policy for Sharing of Data Obtained in NIH Supported or Conducted Genome-Wide Association Studies (GWAS Policy), created a framework for sharing data from GWAS while promoting participant protections through the establishment of a two-tiered system of unrestricted or controlled access to GWAS data. The second, dbGaP, established a central repository to store and distribute GWAS data for use by other researchers in their studies1. These initiatives aimed to maximize scientific advances and potential public benefit in a manner consistent with the informed consent of research participants and with consideration of the privacy risks associated with the sharing of genomic data. On its sixth birthday, we describe the GWAS Policy, provide an overview of dbGaP data usage, evaluate the impact of sharing dbGaP data, reflect on challenges and describe future directions of genomic data sharing.
Overview of the GWAS Policy and dbGaP
The GWAS Policy established procedures to ensure the protection of research participants. Individual-level data sets in dbGaP are deidentified by investigators before submission and are organized by 'consent group'. Consent groups represent data with the same limitations on future research (or data use limitations; see also Box 1) based on participants' consent (for example, research is limited to a specific disease). The NIH controls access to these data to ensure that data use is consistent with data use limitations. The GWAS Policy charges institutions with reviewing consent documents and collaborating with investigators to develop data use limitations before submitting data to dbGaP. NIH Data Access Committees (DACs) use data use limitations to oversee secondary research (that is, new studies) of data in dbGaP.
Each data set submitted to dbGaP is assigned an accession number consisting of a phs number and a version number. Each dbGaP study page includes the accession number, descriptive information about the study, the Data Use Certification (DUC; hereafter, certification) agreement, data use limitations, the date after which secondary research may be disseminated (up to 12 months after data release in dbGaP), related publications, attribution for contributing investigator(s) and funding source, and aggregate phenotypic data (for example, see the National Heart, Lung, and Blood Institute's Framingham Cohort study dbGaP page). Although dbGaP was originally developed to archive GWAS data, to meet the needs of the research community, it now accepts genome sequence, array-derived expression, RNA sequencing (RNA-seq) and epigenomic data.
To obtain controlled-access data from dbGaP, investigators submit a project request describing how they will use the data via the access protocol described on the Genomic Data Sharing (GDS) website. Investigators and their institution(s) agree to the terms and conditions described in the certification for each data set. dbGaP creates a data access request (hereafter, DAR or request) for each requested consent group. A project request may include multiple requests and multiple consent groups.
Each project request is reviewed by DACs with responsibility for the requested data sets. DACs are established by NIH Institutes and Centers and comprise federal employees with relevant scientific, bioethics or human subjects research expertise. DACs review requests for consistency with any data use limitations and approve, disapprove or return requests for revision. Since the GWAS Policy began, more NIH Institutes and Centers have funded GWAS, increasing the number of DACs from 8 to 16, representing 18 Institutes and Centers.
The governance structure of dbGaP and the GWAS Policy involves three NIH governance committees: the Participant Protection and Data Management Steering Committee (PPDM), the Technical Standards and Data Submission Steering Committee (TSDS) and the Senior Oversight Committee (SOC), which reports to the NIH Director. These committees assist in developing consistent 'best practices' across the NIH for the oversight of genomic data sharing, such as the submission of data and review of requests, and provide guidance for investigators and for NIH-wide policy decisions when new issues arise and as NIH oversight of genomic data evolves.
To ensure the maximum usefulness of dbGaP and related resources, the NIH provides education and outreach to investigators and their institutions' ethics committees (IRBs) and signing officials about their responsibilities under the GWAS Policy.
Box 1: Glossary of terms and abbreviations.
Consent group: Grouping of study participants whose data have the same data use limitations, as determined by the submitting institution on the basis of its review of the original informed consent documents. Data are distributed by consent group to approved users. Each data set consists of a single consent group.
Controlled-access data: Individual-level or aggregate data that may include sensitive information and have data use limitations determined by the submitting institution, requiring NIH authorization for access.
Data access committee (DAC): An NIH committee that reviews and approves or disapproves requests from researchers for proposed secondary research uses of the data sets overseen by that DAC. The DAC also reviews reports on data use submitted annually by approved users.
Data access request (DAR or request): A request submitted to a DAC for a specific consent group and specifying the data to which access is sought, the planned research use and the names of the collaborators and the institution's information technology director. The data access request is signed by the investigator requesting the data and by her/his institutional signing official. Collaborators and project team members on a request must be from the same institution or organization.
Data use certification (DUC or certification): An agreement that defines the specific terms and conditions for data use of a given data set.
Data set: Grouping of data within a study that falls under the same data use limitations, as determined by the submitting institution on the basis of its review of the original informed consent documents. Data are distributed to approved users by consent group.
Data use limitations: A description of the limitations for secondary research use of controlled-access data. The limitations are specified by the submitting institution and are based on review of informed consent materials and other relevant study information by the IRB of the submitting institution.
Unrestricted-access data: Data that are publicly available and can be browsed online or downloaded without previous permission or authorization and without limits on data use.
Project request: The overarching request from an investigator to access one or more controlled-access data sets in NIH-designated data repositories (for example, dbGaP) that are managed by one or more data access committees. A project request includes one or more data access requests for specific consent groups. All data sets requested must be listed in the application.
Requestor: The home institution or organization of the investigator who applies to dbGaP for access to data.
Research use statement (RUS): A technical statement included in a project request or single data access request describing the research objectives, study design and analysis plan. The research use statement includes an explanation of how the proposed research is consistent with the data use limitations specified by each relevant consent group.
Submission and use of dbGaP data
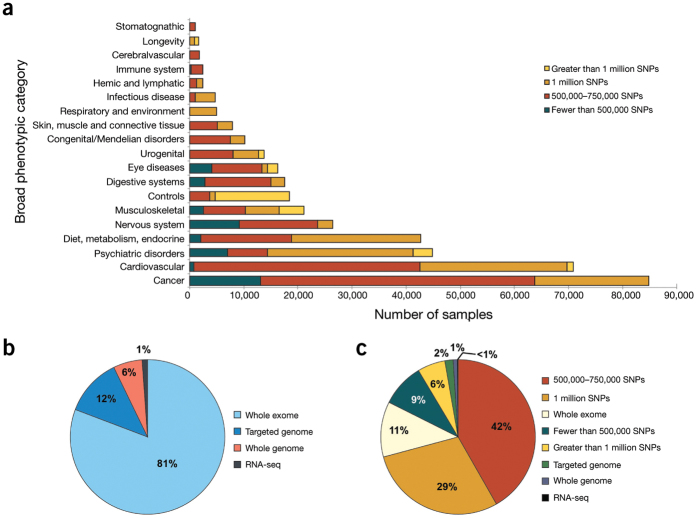
To evaluate data submission and access under the GWAS Policy, we analyzed 304 dbGaP studies deposited through 1 December 2013. Data from studies submitted to dbGaP vary widely in the number of samples from research participants, the breadth of phenotypic data and the genomic data type (Fig. 1). Data were submitted primarily by academic institutions (70%), non-academic research or non-profit organizations (22%), and government research agencies and health departments (8%). Eighty-seven percent (265/304) of the studies were funded by the NIH. Because many scientific journals now require authors to share data as a precondition for publication, NIH Institutes and Centers have sponsored the submission and oversight of 39 non-NIH-funded studies. Acceptance of non-NIH-funded studies is determined on a case-by-case basis by NIH Institutes and Centers.
Figure 1. Number of dbGaP samples by broad phenotypic category and genomic technology.
(a) GWAS samples in dbGaP by broad phenotypic category and size of SNP array. Original submitting investigators select a broad phenotypic category for their studies from standard National Library of Medicine Medical Subject Headings (MeSH) at the time of study registration. The phenotypic categories of GWAS samples are shown by the scale of genotypic array used in the study (n = 393,729). (b) dbGaP samples by type of next-generation DNA sequencing performed. In addition to GWAS data, dbGaP maintains data collected using next-generation sequencing technologies, including sequencing of all genomic DNA (whole-genome), some genomic DNA (targeted genome), DNA expressed as RNA (whole-exome) and RNA-seq studies (n = 65,770). (c) dbGaP samples by type of genomic analysis performed (n = 459,499).
Through 1 December 2013, 17,746 requests were submitted by 2,221 investigators and approximately 6,800 collaborators from 41 different countries, mostly in North America (∼77%; Supplementary Fig. 1). For transparency, each study page lists all approved users for that data set, their institutional affiliations and their research use statements. Additional information about the types of institutions approved to access dbGaP data is provided in Table 1.
Table 1.
Institutions approved for dbGaP controlled-access data (2007–2013)
| Status | Institution type | Number of institutions | Percent | Number of investigators from institution | Percent |
|---|---|---|---|---|---|
| Not for profit | |||||
| Academic | 340 | 64 | 1,473 | 70.5 | |
| Government | 37 | 7 | 151 | 7.2 | |
| Hospital | 54 | 10.2 | 186 | 8.9 | |
| Research institute | 100 | 18.8 | 279 | 13.4 | |
| For profit | |||||
| Biotechnology | 68 | 57.1 | 86 | 51.2 | |
| Genealogy | 1 | 0.8 | 1 | 0.6 | |
| Hospital | 2 | 1.7 | 3 | 1.8 | |
| Information technology | 12 | 10.1 | 16 | 9.5 | |
| Pharma | 33 | 27.7 | 59 | 35.1 | |
| Research | 3 | 2.5 | 3 | 1.8 |
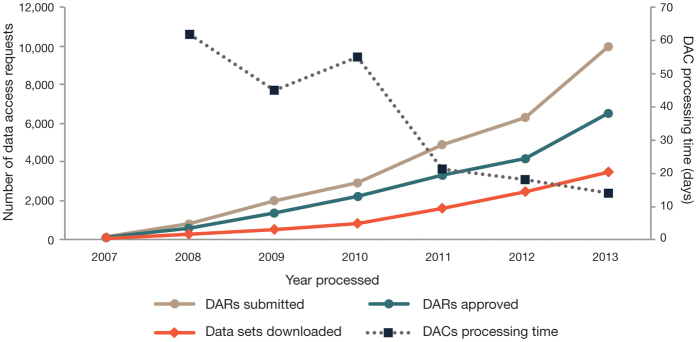
Despite an increasing rate of requests, the average time for DACs to process requests has decreased from 62 days in 2008 to 14 days in 2013 (Fig. 2). The total time to process requests includes time obtaining signatures at the requesting institution and review by the DAC (including any revisions). Often, DACs communicate with requesting investigators to seek additional information about the proposed research, increasing the time to decision. Decreased processing time reflects a greater familiarity in the scientific community and improvements to the process. DACs have also increased their efficiency as they have gained experience reviewing requests for studies under their purview (for example, see the experience of the NIH Genetic Association Information Network (GAIN) DAC)2.
Figure 2. dbGaP data access activity from April 2007 to 1 December 2013.
Shown is the number of DARs submitted to the NIH, approved DARs, downloaded data sets and average time for DACs to process DARs.
Sixty-nine percent (12,391/17,746) of requests were approved as of 1 December 2013 (Fig. 2), with the annual percentage ranging from 61% to 75% over the 6-year period. The most common reason for disapproving requests was inconsistency between the proposed research and the data use limitations of the requested data set. Interestingly, only 50% of data sets approved for access were downloaded by requestors. This percentage is expected to be below 100% because many projects involve collaborations across institutions or investigators approved to access the same data set via multiple projects, resulting in a single download for multiple requests. Another factor might be the technical challenges of downloading dbGaP data sets due to data formats, software incompatibilities or file sizes.
Investigator compliance with the certification and robust data security practices for managing dbGaP data is an essential precondition for data sharing. Nevertheless, the risk remains for intentional or unintentional violation of the terms of the certification. Although rare, several violations occurred in the six-year period. These involved errors in assigning data use limitations during data submission, investigators sharing controlled-access data with unapproved investigators and investigators using data for purposes not described in the research use statement. In every case, as soon as the NIH became aware of the problem, the relevant institution and investigators were notified and appropriate steps were taken to address the violation and prevent it from recurring. Fortunately, to our knowledge, no participants were harmed. Summary information about resolved incidents will be made available on the GDS Policy website.
Improvements have been made to dbGaP and the oversight system in response to stakeholder feedback (including investigators, IRBs and NIH staff). For example, standard data use limitations were developed to provide increased transparency, consistent implementation of the consent group categories and a simplified process for submitting data and reviewing requests. Other improvements include filters that allow investigators to search dbGaP for data sets with specific criteria (for example, data use limitations, disease area and data type). Another resource allows investigators to submit a single request for a collection of most data sets approved for general research use.
Assessing the impact of secondary use of dbGaP data
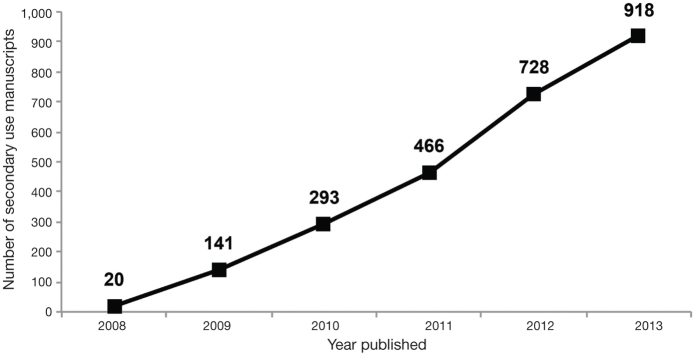
To assess the effect of the GWAS Policy on facilitating additional research, we evaluated publications with secondary use of dbGaP data cited to DACs through 1 December 2013. Secondary use of dbGaP data was reported in 924 publications with a PubMed identification number (Fig. 3). There was a large increase in such use between 2008 (n = 20) and 2009 (n = 121), followed by an annual rise in the number of publications. In 2012 (the last year with complete data for publications examined here), there were 262 publications.
Figure 3. Publications describing research involving the secondary analyses of dbGaP data.
The cumulative number of publications reported by approved users of dbGaP data was collected from annual reports from 2007 to 2013.
Publications were categorized by subject matter, and most publications focused on cancer (20%) and methods development (20%), followed by mental health disorders (15%) and cardiovascular disease (7%) (Supplementary Fig. 2). Overall, many publications appeared in top-tier journals, with approximately 25% published in journals with an impact factor of greater than 10 (238/924). PLoS One hosted the most publications, with 93 papers subsequently cited 834 times in additional publications, followed by Nature Genetics, with 69 papers subsequently cited 9,060 times (Scopus, 1 March 2014; Supplementary Table 1).
Secondary research involving dbGaP data has made significant discoveries in a wide range of fields. For example, access to dbGaP data enabled researchers to identify a previously unknown association between Parkinson's disease and the human leukocyte antigen (HLA) genetic locus3, suggesting the involvement of the immune system in Parkinson's disease, and might offer new targets for gene therapy trials and drug development. Investigators have also combined data sets to increase the statistical strength of associations. The largest independent alcohol dependence GWAS thus far combined dbGaP data with other data sets, identifying several novel loci associated with alcohol dependence4.
Research using dbGaP data has been essential in demonstrating that a small set of genes contributes to a range of psychiatric disorders, including schizophrenia, bipolar disorder and autism5,6,7. These findings contributed to an ongoing effort to transform the diagnosis of psychiatric disorders to account for their biological properties rather than relying on current diagnostic categories based solely on clinical symptoms8. Before the introduction of the GWAS Policy and dbGaP, no public resource existed capable of supporting analyses of such variation and scale in human populations.
Many reported publications involved the development of methods, often made freely available to the research community. For example, VirusSeq, an algorithm for detecting known viruses and their integration sites in the human genome using whole-exome or transcriptome data, was validated using dbGaP data9. Access to dbGaP data also contributed to the development of the SNP-set (Sequence) Kernel Association Test (SKAT), a tool for testing the association between rare variants and phenotypes in GWAS data or genome sequencing studies10.
Challenges, responses and anticipated growth
The increasing volume and complexity of genomic data creates an urgent need for alternative data management and analysis mechanisms beyond the original scope of dbGaP. To accommodate The Cancer Genome Atlas (a project to create an atlas of the genomic changes that occur in a wide variety of cancer types), the NIH developed general principles and core standards allowing external organizations to serve as a 'trusted partner' for data management through a contract. One example is the Cancer Genomics Hub (CGHub). Access to data held by trusted partners is overseen by the NIH. Another model under exploration involves the use of cloud-based resources to transfer, store and analyze genomic data. Several pilot programs are underway to explore secure, cloud-based systems for managing human-derived data. The outcome of these pilots will be considered by NIH leadership in advancing data-sharing policies and other data science initiatives, such as the Big Data to Knowledge (BD2K) initiative.
Advances in genomics and bioinformatics have occasionally led the NIH to consider its policies in light of scientific or technological developments that might alter the risks for participants. In 2008, a novel method for inferring the presence of an individual's genomic information within an aggregate data set was published11. In response, the NIH moved unrestricted aggregate genomic data sets in dbGaP into controlled access. In 2013, a study demonstrated that integrative analyses of unrestricted data sets from the 1000 Genomes Project, information from the Coriell repository and other publicly available information could be combined to deduce the individual identities of some research participants12. In response, the NIH requested that the Coriell repository move the relevant information about individuals to controlled access13. Also in 2013, after the publication of whole-genome sequence data from the HeLa cell line and a request from the family of Henrietta Lacks that the data not be available through unrestricted access, the NIH reached an agreement with the family to control access to HeLa cell whole-genome sequence data through dbGaP.
As part of the certification signed by requesting investigators and their organizations, dbGaP users agree to cite the dbGaP accession numbers for analyzed data sets and to acknowledge the contributing investigators in any public presentation or publication. Citing accession numbers facilitates validation and consistency in analyses of the same data sets. Acknowledging contributing investigators also ensures the proper attribution of data. The NIH has noted that many publications using dbGaP data do not include appropriate acknowledgment. DACs that notice missing or incomplete information may request that an investigator submit an erratum to the journal. One approach to this problem would be for journals to require citation of all relevant accession numbers.
Despite challenges, significant strides have been made in genomic data sharing since the launch of dbGaP in 2007. Improvements have streamlined the implementation of the GWAS Policy and have ensured that data users and institutions are familiar with the processes and responsibilities of data use. A dbGaP user base of more than 2,200 approved investigators and their collaborators, accessing data from more than 300 studies resulting in more than 900 publications thus far, and very few policy violations are a positive testament to the GWAS Policy and dbGaP.
Future directions
The rapid growth in the genomic data generated, advances in genomics and bioinformatics that raise the risk of the reidentification of deidentified data and the experience of six years of the GWAS Policy led the NIH to initiate significant changes to its data-sharing practices. The NIH GDS Policy extends data-sharing expectations to additional genomic data types from both humans and non-humans. An important change in the GDS Policy is the expectation for obtaining explicit informed consent for research and the sharing of genomic data, even data derived from deidentified clinical specimens and cell lines. The draft GDS Policy was released for public comment in September 2013, and comments received helped prepare the final GDS Policy, which will be implemented in 2015.
The experience thus far with the NIH GWAS Policy and dbGaP provides a foundation for the next phase of developing broader data-sharing policies for biomedical research. The ethical use of genomic data and the public's perceptions of that use will continue to be of paramount concern. The NIH remains committed to developing and improving its policies and oversight for sharing biomedical research data to maximize the public benefit of federally funded research.
Supplementary information
Acknowledgements
The authors thank the NIH GWAS governance committees and DACs for their contribution to the implementation and oversight of the GWAS Policy and assistance in developing this manuscript. This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine.
Author Contributions
D.N.P. and L.L.R. coordinated the overall analysis and writing process. Data were generated and contributed by M.F., E.G., E.M.R., J.L.R., V.O.W., M.C., E.L.H., K.L., E.L., T.P., T.R. and K.T. Analyses of the data and results were contributed by D.N.P., L.L.R., M.F., E.G., E.M.R., J.L.R., S.S., V.O.W., A.B., R.B., E.L.H., K.L., H.L., E.L., T.P., T.R., K.T., A.P. and E.D.G. All authors contributed to writing, reviewing and editing the manuscript.
Competing interests
The author declare no competing financial interests.
Footnotes
A full list of members and affiliations appears in the Supplementary Note.
Dina N Paltoo, Laura Lyman Rodriguez and Michael Feolo: These authors contributed equally to this work.
Contributor Information
the National Institutes of Health Genomic Data Sharing Governance Committees:
Dina N Paltoo, Laura Lyman Rodriguez, Michael Feolo, Elizabeth Gillanders, Erin M Ramos, Joni L Rutter, Stephen Sherry, Vivian Ota Wang, Alice Bailey, Rebecca Baker, Mark Caulder, Emily L Harris, Kristofor Langlais, Hilary Leeds, Erin Luetkemeier, Taunton Paine, Tamar Roomian, Kimberly Tryka, Amy Patterson, and Eric D Green
References
- 1.Mailman MD, et al. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramos EM, et al. Am. J. Hum. Genet. 2013;92:479–488. doi: 10.1016/j.ajhg.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamza TH, et al. Nat. Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelernter J, et al. Mol. Psychiatry. 2014;19:41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy SE, et al. Nat. Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon FJ, et al. Nat. Genet. 2010;42:128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lancet381, 1371–1379 (2013).
- 8.Adam D. Nature. 2013;496:416–418. doi: 10.1038/496416a. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, et al. Bioinformatics. 2013;29:266–267. doi: 10.1093/bioinformatics/bts665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S, et al. Am. J. Hum. Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homer N, et al. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. Science. 2013;339:321–324. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez LL, Brooks LD, Greenberg JH, Green ED. Science. 2013;339:275–276. doi: 10.1126/science.1234593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.