Human thymic epithelial primary cells produce exosomes carrying tissue-restricted antigens (original) (raw)
Abstract
Exosomes are nano-sized vesicles released by cells into the extracellular space and have been shown to be present in thymic tissue both in mice and in humans. The source of thymic exosomes is however still an enigma and hence it is not known whether thymic epithelial cells (TECs) are able to produce exosomes. In this work, we have cultured human TECs and isolated exosomes. These exosomes carry tissue-restricted antigens (TRAs), for example, myelin basic protein and desmoglein 3. The presence of TRAs indicates a possible role for thymic epithelium-derived exosomes in the selection process of thymocytes. The key contribution of these exosomes could be to disseminate self-antigens from the thymic epithelia, thus making them more accessible to the pool of maturing thymocytes. This would increase the coverage of TRAs within the thymus, and facilitate the process of positive and negative selection.
As a primary lymphoid organ the human thymus is an arena for intensive cell interactions between developing thymocytes and stromal cell populations. Before export as mature T cells into the blood and peripheral lymphoid tissues, thymocytes undergo stromal-dependent maturation, proliferation, selection and lineage commitment within the thymus.1, 2 A key stromal cell population is the medullary thymic epithelial cells (mTECs), which express the autoimmune regulator (AIRE)3 transcription factor that controls the expression and presentation of a battery of otherwise tissue-restricted antigens (TRAs).
One intriguing question is how a small population of mTECs is able to school/purge the manifold larger population of developing thymocytes, especially since not every TRA is expressed in every mTEC.4 One hypothesis among several is that exosomes could contribute as a mean of spreading antigens within the thymus.5, 6 Exosomes are nano-sized membrane enclosed vesicles of endocytic origin, and are released into the extracellular space by cells when multivesicular bodies fuse with the cell plasma membrane. Previous work have identified and characterized exosomes in murine7 as well as human thymic tissue.8, 9 Unlike other human epithelial cell types10 it is still however, not formally shown that the TEC population is able to produce these extracellular vesicles. Following the finding that B cells secrete antigen-presenting exosomes11 several studies have shown exosomes to be capable of presenting antigens for T cells by themselves12, 13 or indirectly via uptake by dendritic cells (DCs).14
In this report, we describe the isolation and characterization of exosomes produced by primary cultures of human TECs. The exosomes were found to carry antigen presentation molecules and TRAs, among them known autoantigens such as myelin basic protein (MBP), desmoglein 1 and 3 (DSG1 and DSG3).
RESULTS
Cultured cells display TEC morphology and carry TEC surface markers and mRNAs
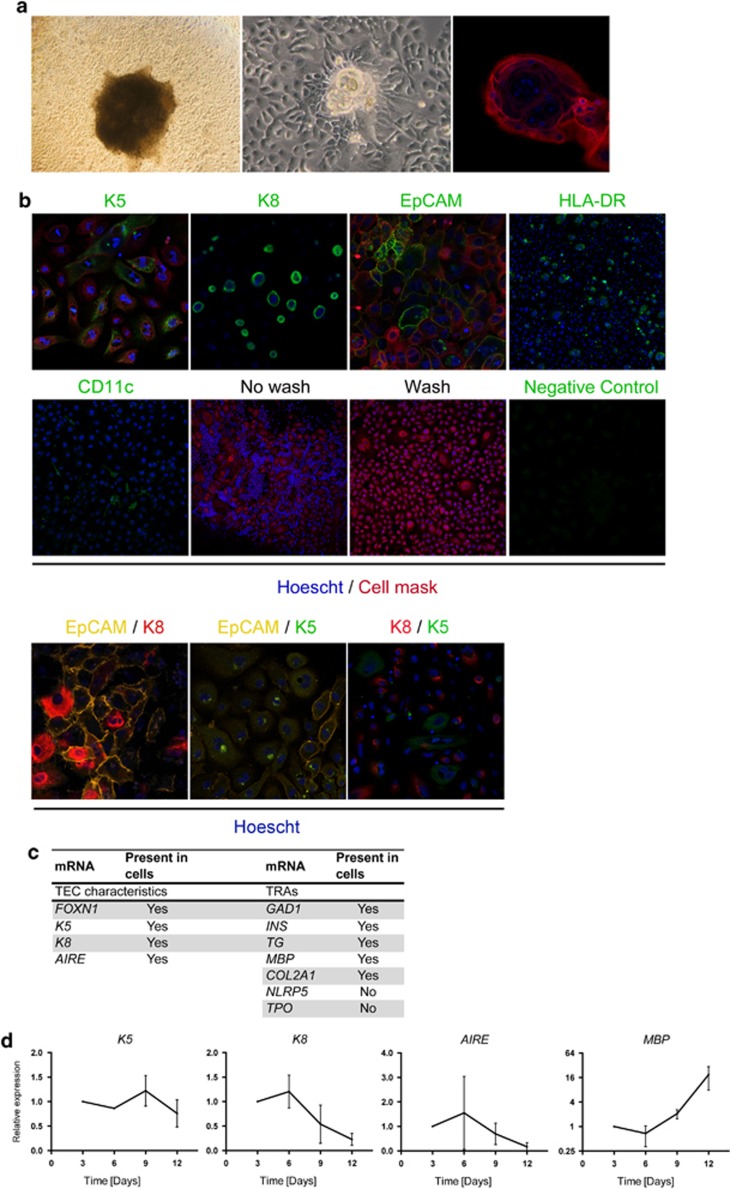
To clarify whether TECs are able to produce exosomes, cultures were set up where cells grew from pieces of thymic tissue in TEC favoring serum-free culture media (see Methods). The resulting cells showed a morphologic appearance distinct of cultured epithelial cells (Figure 1a), which resembled previously described cultured TECs as in Ropke.15 In addition, Hassall's Corpuscle (HC)-like structures spontaneously appeared after approximately 1 week of culturing (Figure 1a) and displayed HC typical swirl-like inner membrane formation. Further, cultured cells stained positive for the TEC markers EpCAM, Keratin 5 (K5) and Keratin 8 (K8) (Figure 1b). Also EpCAM/K8 and EpCAM/K5 double positive cells are detected in the cultures (Figure 1b). Most cells labeled with K5 or K8 were single positive but K8/K5 double positive cells were also present (Figure 1b). The mTEC-typical transcription factor AIRE and TRAs represent essential components for thymic function. To further investigate the cultured cells, and more specifically confirm the presence of cells with mTEC phenotype, we performed RT–PCR analysis for FOXN1, K5, K8, AIRE and TRA-mRNAs. Indeed, FOXN1, K5, K8 as well as AIRE were present in the analyzed cultures (Figure 1c). Also, TRA-mRNAs were found, exemplified with MBP and INS (Figure 1c). Cultured cells were negative for the TRAs TPO and NLRP5 (Figure 1c). Over time, the cultured TECs keep their expression of K5 and K8 (Figure 1d). While AIRE was detected throughout the 12-day culture period, although highly variable and descending, MBP expression was stably elevated during the culture period (Figure 1d).
Figure 1.
Cultured cells display the phenotype, cell-surface markers and mRNAs of TECs. (a) Light microscopy and immunofluorescent stainings of cultured human TECs. Morphology of cells growing from a piece of thymic tissue (left), and formed HC-like structures (middle and right). (b) Immunofluorescent stainings for K5 (mTECs; green, × 25), K8 (mTECs, cTECs; green, × 25), EpCAM (TECs; green, × 25), HLA-DR (TECs; green, × 10). CD11c (DCs; green, × 10) was included to estimate DC contamination. No wash and wash: illustration of non-adherent cell (thymocyte) removal by wash of cultures (× 10). For applicable pictures: Hoescht (nuclei; blue) and cell mask (cell membranes; red). Negative controls were performed by replacement of primary antibodies with isotype-matched antibodies. Double immunostainings: EpCAM/K8 (TECs; yellow, cTECs; red × 25), EpCAM/K5 (TECs; yellow, mTECs; green × 25) and K8/K5 (cTECs; red, mTECs; green × 25). Representative pictures are shown. (c) mRNA profile of cultured cells include TEC-mRNAs and TRA-mRNAs. (d) mRNA profile for K5, K8, AIRE and MBP in cultured TECs over time analyzed by RT–qPCR. GAPDH was used as an endogenous control and expression was related to expression at day 3. Data are presented as mean±s.e.m. (_n_=3).
As the cells were highly autofluorescent when analyzed with flow cytometry, limited information could be drawn from this methodological approach (Supplementary Figure S1). Still though, unwanted populations of cells such as CD45+ leukocytes, ER-TR7+ fibroblasts and CD11c+ DCs were low in presence (Supplementary Figure S1).
Characteristics of exosomes released by cultured TECs
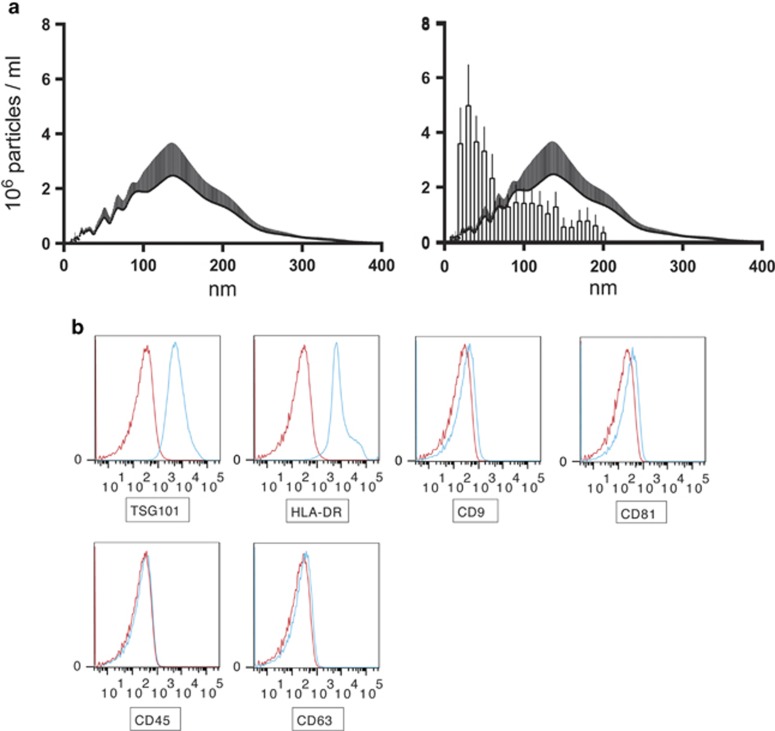
The presence of small vesicles in the culture supernatants was assessed with a dynamic light scattering approach using a Nanosight LM10. A peak size of 136 nm was observed and the size profile ranged from 50 to 200 nm in diameter with few vesicles exceeding 200 nm (Figure 2a). When compared with exosomes isolated from human thymic explant tissue,8 54% of the exosomes isolated from human thymic explant tissue are covered by the size distribution of exosomes produced by cultured TECs (Figure 2a). To investigate exosomal membrane bound markers, the vesicles were attached to latex beads and analyzed with flow cytometry. The presence of the typical exosomal markers TSG101, HLA-DR, CD9 and CD81 was confirmed (Figure 2b). The exosomes were negative for CD45 (Figure 2b).
Figure 2.
Characteristics of exosomes from cultured TECs. (a) Left panel: Size distribution of TEC derived exosomes was investigated using Nanosight LM10 instrument. Distribution peaked at 136 nm and the majority of particles were smaller than 200 nm. The number of tracks exceeded 200 in each measurement and the minimum expected particle setting was 30 nm. Data are presented as mean+s.e.m. as a result of four analyzed cultures. Right panel: An overlay comparison of exosomes isolated from human thymic tissue8 (bars+s.e.m.) and exosomes isolated from cultured TECs (bars+s.e.m.). (b) Flow-cytometry analysis with blue histograms representing samples (exosomes+beads+antibody) and red histograms negative controls (beads+antibody). The exosomes stained positive for TSG101 and HLA-DR, weakly positive for CD9 and CD81 and negative for CD45. The exosomes were close to negative for CD63. Gating strategy was: all beads with coupled exosomes were gated upon and investigated for stained markers. Representative stainings are shown.
The proteomic profiles of cultured cells and exosomes include typical TEC proteins, TEC typical proteasome subunits, TRAs and autoantigens
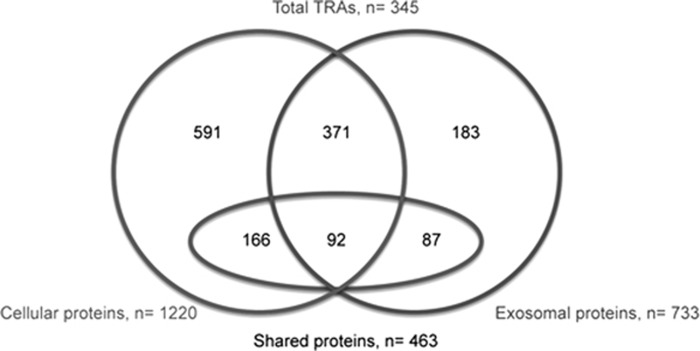
Analyzes of the protein content of cultured cells and exosomes using mass spectrometry (MS) identified 1220 cellular proteins and 733 proteins of exosomal origin (culture 1). In all, 463 proteins were shared between exosomes and the cultured cells from which the exosomes were derived (Figure 3). Complete lists of the identified proteins are found in Supplementary Table S1 for cellular proteins and Supplementary Table S2 for exosomal proteins. Several cytokeratins with a known TEC expression were identified. Of special interest is that the typical mTEC-associated cytokeratins K5 and K14 were found in both cells and exosomes, while the typical cortical thymic epithelial cell (cTEC) associated cytokeratin K8 was only identified in cells. Besides the cytokeratins, other typical TEC proteins were found in the cells, for example, involucrin, a marker for late-stage mTEC differentiation and CLAUDIN-1.16 Amidst exosome-specific proteins (_n_=270), not found in the cells, classical exosomal markers such as TSG101, CD82, CD63, MFG-E8 and FLOTILIN-1 were present. The immunoproteasome subunits PSMB8 (cells and exosomes), PSMB9 and PSMB10 (exosomes) were also found.
Figure 3.
Proteomic analysis of cells and exosomes compared in a Venn diagram (culture 1). The diagram illustrates the distribution of the identified proteins between cultured cells (left circle) and exosomes (right circle) as well as the numbers of TRAs (ellipse) in the different categories.
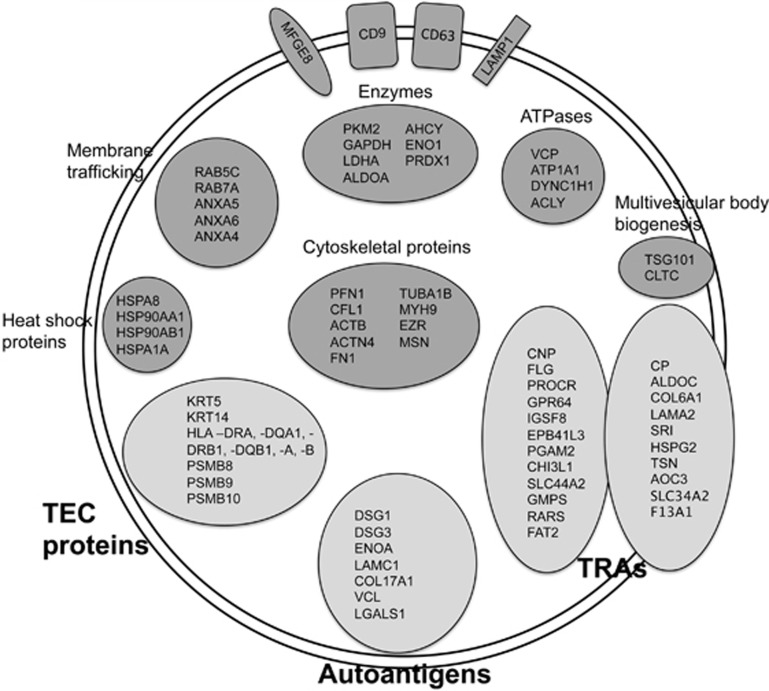
Genes with expression restricted to up to five tissues were considered as TRAs as defined by Derbinski et al.17 Proteins with a known expression in the thymus were not considered as TRAs. A large group of TRAs were identified (_n_=345 in total), both among the cellular (_n_=258) and exosomal (_n_=179) proteins. The fractions of the proteins classified as TRAs differed somewhat between the cellular proteins (21%) and exosomal proteins (24%). Among the proteins that were only found in exosomes an even higher fraction was TRAs (32%, _n_=87, compared with 22% for cells _n_=166) (Figure 3). Examples of TRAs with a previously described mTEC expression found in the cultured cells were Adenylate kinase 2, Alpha actin (ACTC1), Antithrombin III (SERPINC1), Apolipoproteins (APOB and APOM), Fatty acid-binding protein (FABP4), Glutathione S-transferases (GSTM1 and GSTM3), LDL receptor, Monocarboxylate transporter 4 (SLC16A3), Mucins (MUC1 and MUC18), Myosins (MYO1B, MYO6, MYO5A), Retinol-binding protein 1 and S100 proteins (S100A2, S100A8, S100A9 and S100A11). Also in the exosomes, previously reported mTEC-enriched TRAs such as Glutathione S-transferase M3 (GSTM3), LDL receptor, Monocarboxylate transporter 4 (SLC16A3), Mucins (MUC5B and MUC18), Myosin 1B (MYO1B), Apolipoprotein A1 and S100 proteins (S100A8, S100A9 and S100A11) were identified.3, 18 Further examples of exosomal TRAs and also TEC-typical proteins including antigen-presenting molecules are provided in Figure 4. Known disease-associated autoantigens, for example, Collagen type II (COL2A1), DSG3 and heat-shock protein 60 (HSPD1), were also present in cells and exosomes (Table 1, Figure 4).
Figure 4.
Schematic illustration of protein examples found in the TEC exosome proteome (culture 1). TEC exosomes contain TEC proteins, autoantigens and TRAs (light gray fields). TRAs shown in the figure are either exosome specific (left TRA circle) or detected in both exosomes and cultured cells (right TRA circle). Examples of several typical exosomal proteins are found within the TEC exosomes (dark gray fields). The depicted typical exosomal proteins are represented in the core exosomal protein list retrieved from the Exocarta database and presented with gene symbols.31
Table 1. Proteomic examples of autoantigens found in cultured TECs and TEC-derived exosomes (cultures 1 and 2).
| Protein | Gene | Associated disease | Present in cells | Present in exosomes | TRA | Ref. |
|---|---|---|---|---|---|---|
| Collagen type II | COL2A | Rheumatoid arthritis | Yes | No | Yes | 32 |
| Desmoglein 1 and 3 | DSG1, DSG3 | Pemphigus vulgaris | No, Yes | Yes, Yes | Yes, Yes | 33 |
| Alpha-Enolase | ENOA | Various; RA, peridontitis, hypophysitis | Yes | Yes | No | 34, 35, 36 |
| Titin | TTN | Myasthenia gravis | Yes | No | Yes | 37 |
| Laminin gamma 1 | LAMC1 | Pemphigoid | Yes | Yes | No | 38 |
| Collagen 17 | COL17A1 | Bullous pemphigoid | Yes | Yes | Yes | 39 |
| Vinculin | VCL | Systemic sclerosis | Yes | Yes | Yes | 40 |
| Galectin 1, 3 and 7 | LGALS1, LGALS3, LGALS7 | SLE | Yes, Yes, Yes | Yes, No, No | No, No, Yes | 41, 42 |
| Heat-shock protein 60 | HSPD1 | Various; Vasculitis, T1D, RA | Yes | Yes | No | 43 |
| Tissue transglutaminase 2 | TGM2 | Coeliac disease | nd | Yes | Yes | 20 |
| Myelin basic protein | MBP | Multiple sclerosis | nda | Yes | Yes | 44 |
In culture 2 experiment, exosomes were isolated from cultured cells and the protein content of the exosomes was determined without simultaneous analysis of the proteome of the cells of origin. This second analysis identified 447 exosomal proteins with similar characteristics as described for the first exosomal proteome regarding the presence of typical TEC proteins (and also involucrin), TRAs and autoantigens (Supplementary Table S3). Interestingly, MBP and tissue transglutaminase 2 that are autoantigens associated with multiple sclerosis and coeliac disease were identified (Table 1).19, 20 The presence of MBP mRNA in the cells of origin was confirmed by RT–PCR (Figure 1c).
DISCUSSION
In the present study, we report that cultured human thymic cells with TEC characteristics release exosomes that harbor TRAs, and among them known autoantigens. The study supports the hypothesis that thymic exosomes of mTEC origin may participate in the selection of developing thymocytes by spreading mTEC-derived antigens.
The cultured primary cells presented in this article display several TEC characteristics: morphology, HC-like structure formation, characteristic markers (EpCAM, K5, K8) (Figure 1), mRNA expression (K5, K8, FOXN1, AIRE, TRAs) which for K5 and K8 were kept over time (Figure 1) and proteomic footprint (K5, K14, Claudin-1, involucrin, HLA-A, HLA-DRB1, TRAs; Supplementary Table S1). Hence, it is plausible that the cultured cells are predominantly TECs.
Although it is known that cTECs express TRAs, mTECs are the cell type that expresses the highest number of these antigens.18, 21 The presence of TRA mRNAs in the cultured cells indicates that the cultured cells are predominantly mTECs. In addition, the presence of the immunoproteasome subunits PSMB8, PSMB9 and PSMB10 in the cultures confirms epithelial origin since these subunits are restrained to TECs and not found other thymic cell populations.22, 23 In the proteomic analysis, peptides are found that could point both toward a cTEC and mTEC cell origin, this may imply mixed epithelial cultures.
It is notoriously difficult to maintain the expression of Aire in murine mTEC cultures,24, 25, 26 this also in the recent approach using organotypic cocultures.27 However, none of these studies cultured human TECs, and the present study differs in approach regarding the presence of whole thymic tissue pieces in the culture during the first half of the culture period. Even so, we observe a discrepancy between a clearly detectable presence of AIRE mRNA (during the entire culture period of 12 days), by PCR and the lack of detectable AIRE protein by immunofluorescence (not shown). This can be explained by the differing sensitivity between the methods and indicates that the cultured cells do not express AIRE at the same level as mTECs in tissue sections do. However, both detection of _AIRE_-mRNA in the cultured cells and the presence of AIRE-controlled TRAs in cells and exosomes strengthen presence of AIRE activity in the cultured cells.
Thymocytes and fibroblasts ought to represent the major sources of contaminating cell types in these cultures. A majority of the thymocytes are flushed away by the PBS washing of the cultures, see Figure 1b. Fibroblast growth is counteracted both by careful selection of thymic pieces free from connective tissue and also by the use of serum-free medium. Despite these endeavors, contaminating fibroblasts occur within the cultures (Supplementary Figure S1).
The TEC-derived exosomes contain a number of typical exosomal markers (TSG101, CD82, CD63, MFG-E8, and Flotilin-1), which verifies the identity of the secreted vesicles as exosomes. The TEC origin of the exosomes can be further strengthened by the identification of TEC-typical markers in the exosomes (K5, K14). Importantly, K8, a more pronounced cTEC marker found at the cellular level, is not found in the exosomes, which may indicate that the exosomes are predominantly of mTEC, as opposed to cTEC, origin. In addition, involucrin was present in the exosomes from culture 2 (Supplementary Table S3); this indicates that terminally differentiated TECs possibly also release exosomes. The size distribution of the exosomes isolated from cultured TECs is not identical to the size distribution of exosomes isolated from human thymic explant tissue (Figure 2a). This is expected since the exosomal sources are different, and this gives a hint toward how much of the exosomes isolated from human thymic explant tissue that are derived from TECs. The 54% match between explant tymic exosomes and TEC exosomes is probably an overestimation of the contribution of TEC exosomes to the total exosome content of the thymus since, for example, circulating exosomes from the periphery are contributing to the pool of thymic exosomes in vivo.
Thymocyte selection via exosomes could potentially occur via a number of different routes: direct presentation of peptide–MHC complexes to the developing thymocytes, engulfment and processing of exosomal antigens by DCs that subsequently present the antigens to thymocytes or co-stimulation of thymocytes by DCs while exosomes present the peptides. The high number of TRAs present within the TEC–exosome proteome strengthens the hypothesis that exosomes participate in the negative selection process by spreading mTEC-derived TRAs via one or more of the above routes. The strong surface expression of MHC II on the exosomes speaks in favor of MHC bound TRA peptides being transported by the exosomes. This mechanism would enhance the efficacy of TEC antigen presentation by spreading preselected MHC binding self-peptides in the thymic medulla.
The presence of several known disease-associated autoantigens, for example, MBP, transglutaminase 2, Collagen type II and DSG3, within the exosomal proteome further strengthen the potential role for exosomes in thymic tolerance induction (Table 1). Both AIRE-dependent and AIRE-independent autoantigens were identified in the exosomes. The presence of disease-associated autoantigens in thymic exosomes may indicate that thymic exosomes have a therapeutic potential by mediating tolerance induction against autoantigens in patients with autoimmune diseases.
In summary, we report that cultured primary human TECs are able to produce exosomes carrying TRAs and known autoantigens. These findings support the hypotheses that mTECs are a potent source of thymic exosomes and that thymic exosomes participate in the thymic selection and maturation processes.
METHODS
Collection of human thymic tissue
Human thymuses were removed during cardiac surgery in children 0–6 months of age at Sahlgrenska University Hospital, Gothenburg, Sweden. The tissue was immediately put in 1:1 mixture of DMEM/F-12 Ham (Sigma-Aldrich, Saint-Louis, MO, USA) on ice. Parents gave informed consent, and the study was approved by the local ethics committee (no. 217-12, 2012-04-26).
Cell culture
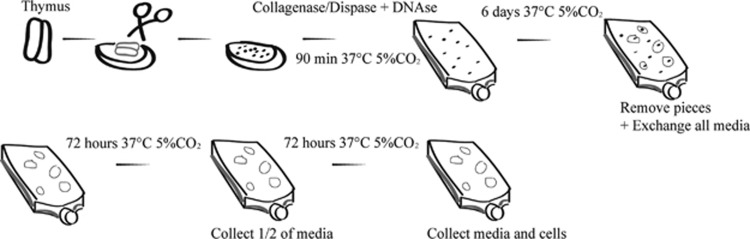
Thymic epithelial cell cultures were set up as previously described15 with minor modifications. Briefly, tissue was washed with PBS, connective tissue and fat was trimmed off and remaining tissue was cut into millimeter-seized pieces. The pieces were washed twice in 1:1 DMEM/F-12 Ham medium and placed in 37 °C 5% CO2 during 90 min in Collagenase/Dispase 0.1 U ml−1 respective 0.8 U ml−1 and DNase 0.2 mg ml−1 (Roche, Mannheim, Germany). The rugged pieces were washed twice with 37 °C 1:1 DMEM/F-12 Ham medium and transferred into Ti75 collagen (Sigma-Aldrich) coated flasks in complete medium: 1:1 DMEM/F-12 Ham with 1% L-glutamine, 1% PEST, 0.5 μg ml−1 hydrocortisone, 10 ng ml−1 cholera toxin, 20 ng ml−1 epidermal growth factor, 3 μg ml−1 insulin (Sigma-Aldrich). At day 6, thymic pieces were removed from the cultures and all media was exchanged to remove all exosomes that originates from thymic tissue pieces. From this point media was completed with addition of 0.1 μg ml−1 RANK ligand (Life Tech, Frederick, MD, USA). After another 72 h of culturing, half of the media volume was collected for isolation of exosomes. After a final 72-h culture the complete media volume was collected for exosomal isolation and the cells were trypsinized 0.5 mg ml−1 (15 min reaction stopped with PBS-5% FCS) for analysis by flow cytometry, RNA isolation or proteomic analysis. A flowchart of the culture method is shown in Figure 5.
Figure 5.
Scheme of thymic culture approach, tissue removal and exosome-supernatant collection. Thymic tissue was washed, trimmed and cut into pieces. The pieces were washed, treated with enzymes by incubation for 90 min. The pieces were washed, and grown in complete media for 6 days. All pieces of tissue were removed and all media was discarded and replaced with fresh complete media. The remaining cells were cultured for another 72 h after which half of the media was collected for exosomal isolation and replaced with fresh complete media. Finally, another 72 h of culture took place and at the end all media was collected for exosomal isolation and cells were released by trypsinization for downstream analysis.
Isolation of exosomes from thymic epithelial culture media
Exosomes were isolated as previously described by Thery et al.28 Briefly, cultures were centrifuged 10 min at 850 g. Supernatants were collected and centrifuged for 15 min at 3000 g to remove cell debris. Further, the supernatants were spun for 30 min at 10 000 g followed by filtration through 0.2-μm filter. Finally, the supernatants were ultracentrifuged for 70 min at 100 000 g to pellet the exosomes. The protein content of the exosome fraction was measured using Bradford total protein assay (Bio-Rad, Hercules, CA, USA).
Confocal microscopy
Cells were cultured as above, but poly-L-lysine coated 96-well glass bottom plates (MatTek, Ashland, MA, USA) were used with 200 μl medium per well and 1–2 pieces of thymic tissue per well. Cells were fixed using 100 μl 1:1 4% paraformaldehyde/PBS for 5 min. Unspecific binding was blocked with serum-free protein block for 15 min (Dako, Copenhagen, Denmark). Cells were stained with Abs against EpCAM (HEA125), K8 (EP1628Y, Abcam, Cambridge, UK), K5 (D5/16 B4, Dako), HLA-DR, (G46-6, BD Pharmingen, Franklin Lakes, NJ, USA) or CD11c (3.9, eBioscience, San Diego, CA, USA) for 30 min at 37 °C. The wells were gently washed twice with PBS followed by incubation with alexa fluor-conjugated anti-mouse or anti-rabbit secondary antibodies for 30 min in 37 °C and finally washed in PBS and refilled with PBS. The stained cells were analyzed using a Zeiss lsm 700 at × 10x or × 25 magnification (Zeiss, Oberkochen, Germany).
Flow cytometry
Immunostaining and flow-cytometry analysis were performed according to standard procedures. At day 7, the cultured cells were detached with Liberase TM (0.5 U ml−1) and DNAse I (0.2 mg ml−1) for 15–20 min in 37 °C and gently pipetted to achieve single-cell suspensions. The cells were washed, filtered and counted. Fc receptor blocking solution (Biolegend, San Diego, CA, USA) was used and the cells were then immunostained with the following antibodies: CD45 PerCP (2D1, BD Biosciences, Franklin Lakes, NJ, USA), HLA-DR APC (L243, BD Biosciences), ER-TR7 AF647 (sc-73355, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and CD11c BV421 (3.9, Biolegend). For the analysis of exosomes, 5 μg of exosomes was attached to 0.125 μl latex beads per staining (Invitrogen, Carlsbad, CA, USA) overnight at 4 °C with agitation. Unspecific binding to the latex beads was blocked with 0.5% BSA (Sigma-Aldrich) followed by Fc-blocking (Biolegend). The exosomes/beads were then incubated with one of the following Abs; TSG101 (5B7, Abnova, Taipei, Taiwan), CD63 (H5C6), CD81 (JS-81), CD9 (M-L13), HLA-DR, (G46-6, BD Pharmingen) and CD45 (2D-1, BD Bioscience). After a wash step, the staining was completed with a FITC-labeled anti-mouse secondary ab (Sigma-Aldrich). Flow-cytometric analyses were performed using a FACS CANTO II (exosomes) and a FACSVerse (cells) (BD Biosciences) and FlowJo software (Tree Star, Ashland, OR, USA).
RNA isolation and cDNA preparation
Trypsin-treated cells were pelleted at 350 g for 5 min, counted using tryptophan blue and lysed in RLT lysis buffer (1% betamerkapthoethanol) (Qiagen, Hilden, Germany). Samples were centrifuged at 10 000 g to pellet any debris and RNA was isolated from the supernatant using RNeasy kit (Qiagen) on a Qiacube (Qiagen) with the following settings: RNeasy Mini, animal tissue and cells, standard, 30 μl eluate volume. cDNA was prepared from 1 μg of RNA per sample with QuantiTect Reverse Transcription kit (Qiagen) according to the manufacturers instructions. For the quantitative RT–PCR expression time series, cells were cultured as above, but poly-L-lysine coated 96-well glass bottom plates (MatTek) were used with 200 μl medium per well and 1–2 pieces of thymic tissue per well.
RT–PCR and quantitative RT–PCR
Taqman assays for analysis of cultured cells: K5 (Hs00361185_m1), K8 (Hs01670053_m1), FOXN1 (HS00186096_m1) AIRE (Hs00230829_m1), INSULIN (Hs02741908_m1), THYROGLOBULIN (TG) (Hs00968042_m1), THYROID PEROXIDASE (TPO) (Hs00892519_m1), GAD1 (Hs01065893_m1), MBP (Hs00921945_m1), COL2A1 (Hs01060345_m1) or NLRP5 (Hs00411266_m1), RNAse-free water (Qiagen) and Taqman Universal Master Mix II (Life Technologies Carlsbad, CA, USA) were prepared as master mixes. Each 20 μl PCR consisted of 18 μl master mix together with 2 μl of cDNA. Plates were run on a ViiA7 (Life Technologies) with the setting: Fast 96-well Block, Presence/Absence, TaqMan Reagents, Standard. For the quantitative RT–PCR time series, cells were harvested at days 3, 6, 9 and 12. cDNA preparation and PCRs were set up as above and the following ViiA7 settings were used: Fast 96-well Block, Comparative CT, TaqMan Reagents, Standard. GAPDH (Hs99999905_m1) was used as an endogenous control and expression was related to expression at day 3. Non-template control and control wells excluding the reverse transcriptase enzyme were included in each run. All samples were run in triplicates.
Computation of exosomal size distribution
Size distribution profile was computed using measurements of particle Brownian motion in a NanoSight LM10 instrument with Nanoparticle Tracking Analysis software (NanoSight, Amesbury, UK). Samples were diluted with PBS in the optical chamber to reach a suitable concentration for the analysis. Each sample was run at least three times to yield a mean sample distribution. Continuous variable in nanoparticle tracking analysis is presented with mean±s.e.m.
Protein identification by tandem MS
In all, 50 μg of exosomal material (two separate experiments, designated culture 1 and culture 2), or 200 000 cells in cell sample (culture 1) was separated by one-dimensional SDS-PAGE (4–12% Bis-Tris Novex mini-gel, Invitrogen) and visualized by Coomassie staining (Novex, Invitrogen). The complete gel lanes were excised and divided into equal slices and subjected to in-gel protein digestion with trypsin overnight at 37 °C.29 Peptides were extracted with 50% acetonitrile in 1% formic acid and the supernatant was lyophilized in a vacuum centrifuge and reconstituted in 0.2% formic acid. Two-microliter sample injections were made with an HTC-PAL autosampler (CTC Analytics AG, Zwingen, Switzerland) connected to an Agilent 1200 binary pump (Agilent Technologies, Palo Alto, CA, USA). The peptides were trapped on a precolumn (45 × 0.075 mm i.d.) and separated on a 200 × 0.050 mm column packed with 4 μm Reprosil-Pur C18-AQ particles (Dr Maisch, Ammerbuch, Germany). The flow through of the analytical column was passively split to approximately 100 nl per minute. A 40-min gradient 5–35% acetonitrile in 0.2% formic acid was applied for peptide separation. The LTQ-Orbitrap was operated in a data-dependent mode automatically switching between MS and MS/MS mode. Full MS scans were acquired in the orbitrap (from m/z 400 to 2000) with a resolution of 60.000 at m/z 400. The top six most intense double or triple protonated ions were selected for fragmentation in the linear ion trap using CID (collision induced dissociation) fragmentation. All tandem mass spectra were searched using MASCOT (v.2.3, Matrix Science, London, UK) and matched against the SwissProt database (release 2011_04). The search parameters were set to: species Human, MS accuracy 5 p.p.m., MS/MS accuracy 0.5 Da, enzyme trypsin allowing one missed cleavage, fixed modification of propionamide on cysteine and variable modifications of oxidized methionine and acetylation protein N-terminal. The threshold for protein identification was set to <1% FDR for both peptide and protein identification based on a minimum of two unique peptides.
Identification of tissue-restricted genes was performed using gene expression data from the BioGPS database (www.biogps.org).30
Acknowledgments
We wish to thank Angela Hanson at the Queen Silvia Children's Hospital for assistance in collecting of thymic material and the Proteomics Core Facility at Sahlgrenska Academy, Gothenburg University for performing proteomic analyses. The financial support from the Swedish Research Council (contract 80409601), the Marianne and Marcus Wallenberg Foundation, Region Västra Götaland (ALFGBG-771712), AFA Försäkring (contract 100258), IngaBritt and Arne Lundbergs Research Foundation, AnnMari and Per Ahlqvists Foundation, The Gothenburg Medical Society and Wilhelm and Martina Lundgrens research foundation is greatly acknowledged. Proteomics Core Facility at Sahlgrenska academy, University of Gothenburg for performing proteomic analysis.
The authors declare no conflict of interest.
Footnotes
The Supplementary Information that accompanies this paper is available on the Immunology and Cell Biology website (http://www.nature.com/icb)
Supplementary Material
Supplementary Figure S1
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
References
- 1Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol 2014; 14: 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Anderson G, Jenkinson EJ. Lymphostromal interactions in thymic development and function. Nat Rev Immunol 2001; 1: 31–40. [DOI] [PubMed] [Google Scholar]
- 3Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 2002; 298: 1395–1401. [DOI] [PubMed] [Google Scholar]
- 4Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci USA 2008; 105: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Koble C, Kyewski B. The thymic medulla: a unique microenvironment for intercellular self-antigen transfer. J Exp Med 2009; 206: 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Collado JA, Guitart C, Ciudad MT, Alvarez I, Jaraquemada D. The Repertoires of Peptides Presented by MHC-II in the Thymus and in Peripheral Tissue: A Clue for Autoimmunity? Front Immunol 2013; 4: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Wang GJ, Liu Y, Qin A, Shah SV, Deng ZB, Xiang X et al. Thymus exosomes-like particles induce regulatory T cells. J Immunol 2008; 181: 5242–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Skogberg G, Gudmundsdottir J, van der Post S, Sandstrom K, Bruhn S, Benson M et al. Characterization of human thymic exosomes. PLoS ONE 2013; 8: e67554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Skogberg G, Lundberg V, Lindgren S, Gudmundsdottir J, Sandstrom K, Kampe O et al. Altered Expression of Autoimmune Regulator in Infant Down Syndrome Thymus, a Possible Contributor to an Autoimmune Phenotype. J Immunol 2014; 193: 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 2001; 121: 337–349. [DOI] [PubMed] [Google Scholar]
- 11Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med 1996; 183: 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Hwang I, Shen X, Sprent J. Direct stimulation of naive T cells by membrane vesicles from antigen-presenting cells: distinct roles for CD54 and B7 molecules. Proc Natl Acad Sci USA 2003; 100: 6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Admyre C, Johansson SM, Paulie S, Gabrielsson S. Direct exosome stimulation of peripheral human T cells detected by ELISPOT. Eur J Immunol 2006; 36: 1772–1781. [DOI] [PubMed] [Google Scholar]
- 14Vincent-Schneider H, Stumptner-Cuvelette P, Lankar D, Pain S, Raposo G, Benaroch P et al. Exosomes bearing HLA-DR1 molecules need dendritic cells to efficiently stimulate specific T cells. Int Immunol 2002; 14: 713–722. [DOI] [PubMed] [Google Scholar]
- 15Ropke C. Thymic epithelial cell culture. Microsc Res Tech 1997; 38: 276–286. [DOI] [PubMed] [Google Scholar]
- 16Ichimiya S, Kojima T. Cellular networks of human thymic medullary stromas coordinated by p53-related transcription factors. J Histochem Cytochem 2006; 54: 1277–1289. [DOI] [PubMed] [Google Scholar]
- 17Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med 2005; 202: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med 2004; 199: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Allegretta M, Nicklas JA, Sriram S, Albertini RJ. T cells responsive to myelin basic protein in patients with multiple sclerosis. Science 1990; 247: 718–721. [DOI] [PubMed] [Google Scholar]
- 20Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med 1997; 3: 797–801. [DOI] [PubMed] [Google Scholar]
- 21Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res 2014; 24: 1918–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Groettrup M, Kirk CJ, Basler M. Proteasomes in immune cells: more than peptide producers? Nat Rev Immunol 2010; 10: 73–78. [DOI] [PubMed] [Google Scholar]
- 23Oh KI, Seo JN. Expression pattern of immunoproteasome subunits in human thymus. Immune Netw 2009; 9: 285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Palumbo MO, Levi D, Chentoufi AA, Polychronakos C. Isolation and characterization of proinsulin-producing medullary thymic epithelial cell clones. Diabetes 2006; 55: 2595–2601. [DOI] [PubMed] [Google Scholar]
- 25Chin RK, Lo JC, Kim O, Blink SE, Christiansen PA, Peterson P et al. Lymphotoxin pathway directs thymic Aire expression. Nat Immunol 2003; 4: 1121–1127. [DOI] [PubMed] [Google Scholar]
- 26Kont V, Laan M, Kisand K, Merits A, Scott HS, Peterson P. Modulation of Aire regulates the expression of tissue-restricted antigens. Mol Immunol 2008; 45: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Pinto S, Schmidt K, Egle S, Stark HJ, Boukamp P, Kyewski B. An organotypic coculture model supporting proliferation and differentiation of medullary thymic epithelial cells and promiscuous gene expression. J Immunol 2013; 190: 1085–1093. [DOI] [PubMed] [Google Scholar]
- 28Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006; Chapter 3 Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 29Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 1996; 68: 850–858. [DOI] [PubMed] [Google Scholar]
- 30Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 2009; 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010; 73: 1907–1920. [DOI] [PubMed] [Google Scholar]
- 32Terato K, Shimozuru Y, Katayama K, Takemitsu Y, Yamashita I, Miyatsu M et al. Specificity of antibodies to type II collagen in rheumatoid arthritis. Arthritis Rheum 1990; 33: 1493–1500. [DOI] [PubMed] [Google Scholar]
- 33Amagai M, Stanley JR. Desmoglein as a target in skin disease and beyond. J Invest Dermatol 2012; 132: 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34de Pablo P, Dietrich T, Chapple IL, Milward M, Chowdhury M, Charles PJ et al. The autoantibody repertoire in periodontitis: a role in the induction of autoimmunity to citrullinated proteins in rheumatoid arthritis? Ann Rheum Dis 2014; 73: 580–586. [DOI] [PubMed] [Google Scholar]
- 35Magrys A, Anekonda T, Ren G, Adamus G. The role of anti-alpha-enolase autoantibodies in pathogenicity of autoimmune-mediated retinopathy. J Clin Immunol 2007; 27: 181–192. [DOI] [PubMed] [Google Scholar]
- 36Falorni A, Minarelli V, Bartoloni E, Alunno A, Gerli R. Diagnosis and classification of autoimmune hypophysitis. Autoimmun Rev 2014; 13: 412–416. [DOI] [PubMed] [Google Scholar]
- 37Chen XJ, Qiao J, Xiao BG, Lu CZ. The significance of titin antibodies in myasthenia gravis—correlation with thymoma and severity of myasthenia gravis. J Neurol 2004; 251: 1006–1011. [DOI] [PubMed] [Google Scholar]
- 38Dainichi T, Koga H, Tsuji T, Ishii N, Ohyama B, Ueda A et al. From anti-p200 pemphigoid to anti-laminin gamma1 pemphigoid. J Dermatol 2010; 37: 231–238. [DOI] [PubMed] [Google Scholar]
- 39Iwata Y, Komura K, Kodera M, Usuda T, Yokoyama Y, Hara T et al. Correlation of IgE autoantibody to BP180 with a severe form of bullous pemphigoid. Arch Dermatol 2008; 144: 41–48. [DOI] [PubMed] [Google Scholar]
- 40Dib H, Tamby MC, Bussone G, Regent A, Berezne A, Lafine C et al. Targets of anti-endothelial cell antibodies in pulmonary hypertension and scleroderma. Eur Respir J 2012; 39: 1405–1414. [DOI] [PubMed] [Google Scholar]
- 41Sarter K, Janko C, Andre S, Munoz LE, Schorn C, Winkler S et al. Autoantibodies against galectins are associated with antiphospholipid syndrome in patients with systemic lupus erythematosus. Glycobiology 2013; 23: 12–22. [DOI] [PubMed] [Google Scholar]
- 42Lim Y, Lee DY, Lee S, Park SY, Kim J, Cho B et al. Identification of autoantibodies associated with systemic lupus erythematosus. Biochem Biophys Res Commun 2002; 295: 119–124. [DOI] [PubMed] [Google Scholar]
- 43Mallard K, Jones DB, Richmond J, McGill M, Foulis AK. Expression of the human heat shock protein 60 in thyroid, pancreatic, hepatic and adrenal autoimmunity. J Autoimmun 1996; 9: 89–96. [DOI] [PubMed] [Google Scholar]
- 44Reindl M, Linington C, Brehm U, Egg R, Dilitz E, Deisenhammer F et al. Antibodies against the myelin oligodendrocyte glycoprotein and the myelin basic protein in multiple sclerosis and other neurological diseases: a comparative study. Brain 1999; 122: 2047–2056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3