Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 1.
Published in final edited form as: Nat Med. 2015 Jun 8;21(7):808–814. doi: 10.1038/nm.3871
Abstract
Candida albicans colonization is required for invasive disease1-3. Unlike humans, adult mice with mature intact gut microbiota are resistant to C. albicans gastrointestinal (GI) colonization2,4. But the factors that promote C. albicans colonization resistance are unknown. Here we demonstrate that commensal anaerobic bacteria – specifically Clostridial Firmicutes (Clusters IV and XIVa) and Bacteroidetes – are critical for maintaining C. albicans colonization resistance in mice. Using Bacteroides thetaiotamicron as a model organism, we find that HIF-1α, a transcription factor important for activating innate immune effectors, and the antimicrobial peptide LL37-CRAMP are key determinants of C. albicans colonization resistance. While antibiotic treatment enables C. albicans colonization, pharmacologic activation of colonic Hif1a induces CRAMP expression and results in a significant reduction of C. albicans GI colonization and a 50% decrease in mortality from invasive disease. In the setting of antibiotics, Hif1a and Cramp are required for _B. thetaiotamicron_-induced protection against CA colonization of the gut. Thus, C. albicans GI colonization modulation by activation of gut mucosal immune effectors may represent a novel therapeutic approach for preventing invasive fungal disease in humans.
Commensal fungi, mostly Candida spp., have been detected in the GI tract of various mammals5. While reportedly 40-60% of humans are colonized with Candida albicans (CA) in the GI tract, adult mice are resistant to GI colonization by CA2,4. Since colonization is a prerequisite for CA invasive disease1-3, gaining a better understanding of the factors that modulate CA colonization could lead to novel methods for preventing CA dissemination.
Commensal anaerobic bacteria in the GI tract provide an important defense mechanism against infections by inhibiting growth of potentially pathogenic bacteria6-8. One mechanism for GI colonization resistance involves stimulation of the mucosal immune system by members of “beneficial” microbiota9. Yet there have been no studies examining a commensal bacteria or host mediated immune response that modulates commensal fungal colonization. Thus, we asked whether identifying a single bacterial species that promotes CA colonization resistance could help unveil host immune effectors critical for maintaining CA colonization resistance in the mouse GI tract.
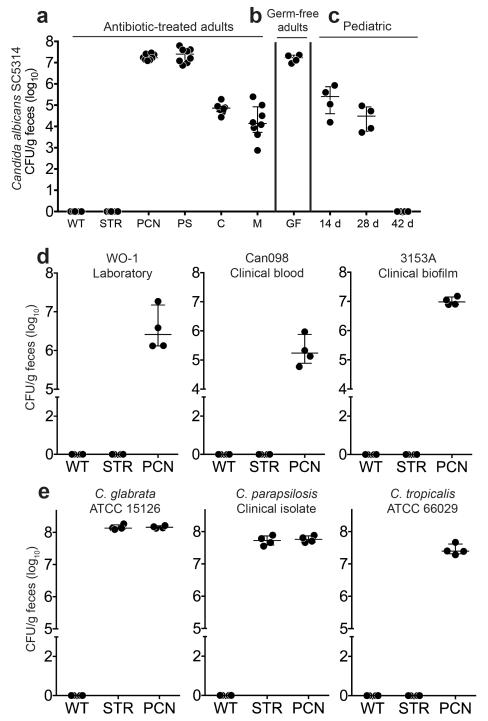
To determine the effect of specific antibiotics on CA colonization resistance, we treated mice with various antibiotics for five days, orally challenged with CA, and then assessed susceptibility to CA colonization. CA was unable to establish sustained GI colonization in adult control (no antibiotic) mice (Fig. 1a), regardless of mouse strain (Supplementary Table 1). In the treatment groups, CA colonization levels were directly proportional to the anaerobic depleting efficacy of the antibiotics used: penicillin (PCN) > clindamycin (C) > metronidazole (M) > streptomycin (STR) (Fig. 1a, Supplementary Table 2)10. In fact, CA GI colonization levels in PCN treated mice were comparable to colonization levels in germ-free mice (Fig. 1b). Even CA strains that had been serially passaged through an antibiotic-treated mouse GI tract could not persistently colonize a mouse GI tract with an intact gut microbiome (Supplementary Fig. 1a-b).
Figure 1. Candida spp. gastrointestinal colonization levels in antibiotic-treated adult mice, germ-free adult mice, and infant-adolescent mice.
CA SC5314 colonization levels in (a) adult C57/BL6 mice (Harlan) pre-treated with (streptomycin, STREP; penicillin G, PCN; penicillin and streptomycin, PS; clindamycin, C; or metronidazole, M) or without antibiotics and then oral gavaged with CA. Mice continued on antibiotic water throughout the experiment. n=8. (b) Adult germ-free mice (C57/BL6). n=4. (c) Postnatal day (P)14, P28, and P42 mice (C57/BL6, Harlan). n=4. (d, e) Colonization levels of (d) other CA strains and (e) other Candida spp. Adult C3H/HeN mice (Harlan) pre-treated with or without antibiotics in the drinking water for 5 days then oral gavaged with (d) CA or other (e) Candida spp. Mice continued on antibiotic water throughout the experiment. n=4. Candida levels were measured every 7 days (a-c) or 21 days (d-e) after oral gavage. Points represent results from individual animals. Horizontal lines with bars represent the median with interquartile range.
The gut microbiota in infant humans and mice have significantly fewer commensal anaerobes than adults11,12. Hence, CA established persistent GI colonization in postnatal day (P)14 and P28 mice but not in adolescent (P42) animals (Fig. 1c, Supplementary Table 2). Other CA strains, including 2 clinical isolates, (Fig. 1d, Supplementary Table 3) and other Candida spp that infect humans (Fig. 1e) were also unable to colonize mice with intact gut microbiota (Supplementary Table 2). Altogether, these findings indicate that a mature adult bacterial microbiota, particularly commensal anaerobes, is essential for maintaining CA colonization resistance.
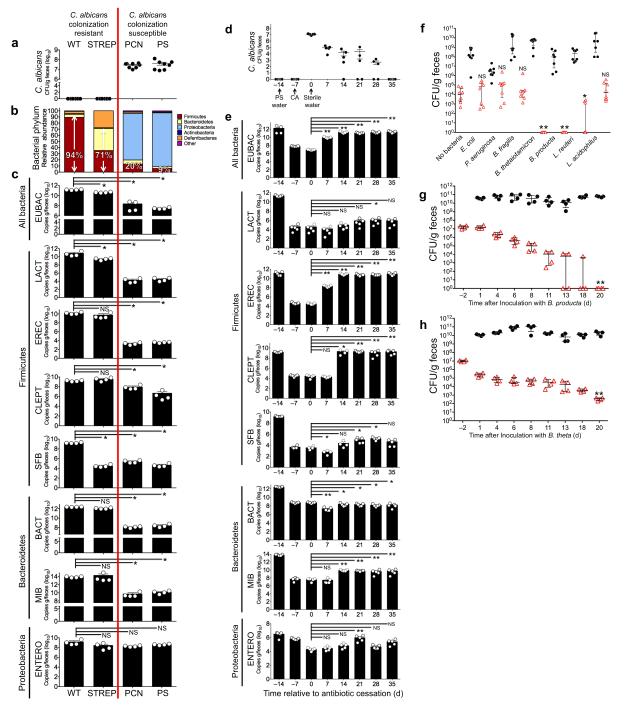
To identify specific members of the gut microbiota essential for maintaining CA colonization resistance, we profiled the gut microbiomes (using 16S rRNA sequencing and bacterial group qPCR) of CA colonization resistant (no antibiotics or STR) or CA colonization susceptible (PCN or penicillin-streptomycin, PS) mice (Fig. 2a). The bacterial phyla Firmicutes and Bacteroidetes account for >95% of the bacteria in the distal guts of healthy adult mice and humans12. The Firmicutes-Bacteroidetes abundance in CA colonized mice markedly decreased compared to colonization resistant mice after 5 days of antibiotic treatment (Fig. 2b). PCN treated mice exhibited the most significant decrease (3-4 log fold) in total gut bacteria (Eubacteria, EUBAC) by qPCR compared to the more modest 0.5 log-fold reduction seen in the STR group (Fig. 2c). Streptomycin is an antibiotic effective against gram-negative bacteria13 and completely ineffective against obligate anaerobic bacteria14. PCN mice had significant decreases in all bacterial groups, with the exception of ENTERO (Phylum Proteobacteria, Enterobacteriaceae). Penicillin-G has activity against gram-positive bacteria and anaerobes but is ineffective against gram-negative bacteria15 (Fig. 2c). Interestingly, endogenous mouse gut fungi (which include some Candida spp. but not CA) increased significantly in mice treated with antibiotics (e.g. PCN) active against Bacteroidetes (mouse intestinal Bacteroides, MIB) and Clostridial-Firmicutes (Clostridial Cluster XIVa, EREC; Clostridial Cluster IV, CLEPT ) (Supplementary Fig. 2). Thus, only antibiotic treatment that sufficiently depletes anaerobic bacteria (e.g. PCN) is sufficient to overcome CA colonization resistance in the mouse GI tract.
Figure 2. Clostridial Firmicutes and Bacteroidetes Promote Candida albicans GI Colonization Resistance.
(a) CA colonization levels of mice pre-treated with antibiotics and oral gavaged with CA. CA levels measured 7 days after oral gavage. n=8. (b) Relative abundance of bacterial Phyla as determined by 16S rRNA sequencing of fecal specimens collected from C3H/HeN mice treated with sterile water (WT), STREP, PCN, or PS. n=3. (c) Bacterial group qPCR (copies/g feces) performed on fecal gDNA collected from mice treated with sterile water or oral antibiotics. n=4. (d) CA colonization levels and (e) bacterial group qPCR in antibiotic-treated CA-colonized mice after cessation of oral antibiotics. Antibiotic water was discontinued on Day 0. CA levels and bacterial group qPCR measured every 7 days. n=4-5. (f) Bacterial (black circles) and CA (red triangle) levels in antibiotic-treated mice colonized with CA and gavaged with one bacterial commensal species. Colonization levels measured 14 days after bacterial gavage. n=8. Bacterial and CA levels in germ-free mice co-colonized with (g) Blautia producta and CA or (h) Bacteroides thetaiotamicron and CA. n=4. For all experiments, points represent results from individual animals. Horizontal lines represent the median with interquartertile range (a, d, f, g, h). Bars represent the mean with SEM (c, e). * p<0.05, ** p<0.01, *** p<0.001 Statistical analysis by Mann-Whitney test.
We postulated that cessation of antibiotics in CA colonized mice would allow suppressed bacterial groups to regrow and restore CA colonization resistance. Indeed, CA colonization levels steadily decreased and were undetectable 35 days after antibiotic cessation (Fig. 2d). Overall bacterial levels (EUBAC) returned to pre-antibiotic treated levels 14 days after stopping antibiotics (Fig. 2e). Notably, Bacteroidetes and Clostridial Firmicutes increased significantly (though still remained lower than baseline), while LACT (Firmicute, Lactobacillus), SFB (Firmicute, segmented filamentous bacteria) and ENTERO remained suppressed throughout the duration of the experiment (Fig. 2e). In total, these findings suggest that Bacteroidetes and Clostridial Firmicutes may be the bacterial groups most effective in promoting CA colonization resistance.
To test this hypothesis, we performed oral bacterial add-back experiments in antibiotic-treated CA colonized mice. Interestingly, among Bacteroidetes, the addition of Bacteroides fragilis did not significantly change CA colonization levels compared to a no bacteria control group, whereas the presence of Bacteroides thetaiotamicron resulted in undetectable CA levels in all mice fourteen days after bacterial inoculation (Fig. 2f, Supplementary Table 3). Similarly, among Firmicutes, Blautia producta promoted complete elimination of CA, while the Lactobacillus spp (L. acidophilus and L. reuteri) did not consistently eliminate CA colonization. Both members of the phylum Proteobacteria (E. coli, P. aeruginosa), however, had no significant effect on CA. Collectively, these experiments show that individual Firmicute and Bacteroidetes species are sufficient to promote CA colonization resistance in the gut.
Since B. thetaiotamicron and B. producta had the most significant effect in our model, we replicated these experiments in germ-free mice (that can be colonized with CA without antibiotic pretreatment). While B. producta and B. theta GI levels remained consistent throughout the duration of the experiment, C. albicans levels steadily decreased and were undetectable by Day 20 in the B. producta group (Fig. 2g) and significantly decreased (nearly 5-log fold) but remained detectable in the B. theta group (Fig. 2h).
Overall, we found that only antibiotics that deplete anaerobic bacteria were sufficient to overcome CA colonization resistance. Among those bacteria we tested, Clostridial Firmicutes and Bacteroidetes most effectively fostered colonization resistance, although individual bacterial species varied greatly in this ability. Ultimately, we identified two genetically distinct bacterial species, B. theta and B. producta that individually promoted CA colonization resistance.
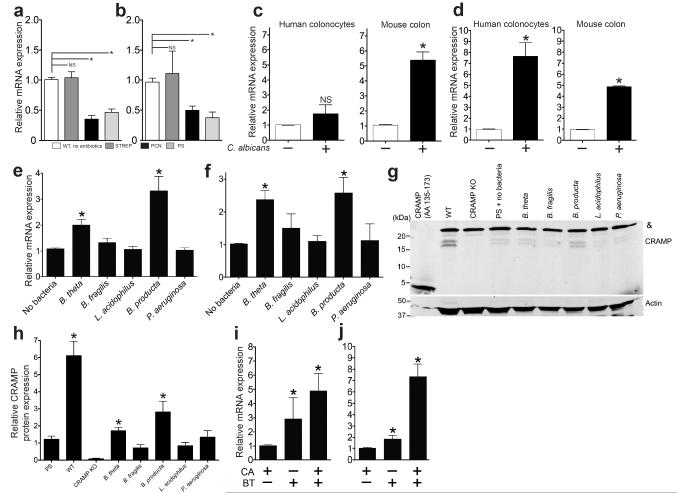
We postulated that B. theta induces host immune effectors critical for maintaining colonization resistance and focused on the transcription factor, hypoxia-inducible factor (HIF)-1α, and the antimicrobial peptide LL-37-CRAMP. HIF-1α is an essential regulator of mammalian innate defense16 and increases expression of antimicrobial cathelicidin peptides17 in myeloid cells. Cathelicidin-related antimicrobial peptides are a family of polypeptides that serve a critical role in mammalian innate immune defense against bacterial infection16,17. The human cathelicidin LL-37 has been shown to have anti-Candida activity18 and inhibits CA adhesion to epithelial surfaces19. Interestingly, Hif1a and Cramp (cathelicidin-related antimicrobial peptide, LL-37 ortholog) expression significantly increased in colonization resistant mice compared to CA colonized mice (Fig. 3a-b). Both HIF1a-Hif1a and _LL-37_-Cramp expression significantly increased in both human colonocytes (HT-29) exposed to CA and in the colon of CA-colonized, antibiotic-treated mice (Fig. 3c-d). The colon had the highest concentration of fungi (Supplementary Fig. 3), thus all of our experiments utilize colons. Of note, other Candida spp. also induced mouse colonic expression of Hif1a and Cramp (Supplementary Fig. 4). Interestingly, B. theta and B. producta induced a significantly greater degree of mouse colonic Hif1a (mRNA) and _Cramp_-CRAMP (mRNA and protein) expression (Fig. 3e-h) compared to other commensal bacteria that we tested in the bacterial add-back experiments. In germ-free mice, co-colonization with B. theta and CA induced greater colonic Hif1a and Cramp expression (Fig. 3i-j) compared to mono-colonization with either B. theta or CA. We concluded that this additive mucosal immune stimulatory effect might explain how B theta facilitates CA colonization reduction in germ-free mice. Our findings suggest that Hif1a and Cramp may be critical immune effectors for maintaining CA colonization resistance.
Figure 3. Bacteroidetes thetaiotamicron induces Hif1a and Cramp in mouse colons.
(a) Hif1a and (b) Cramp mRNA expression in colons resected from CA colonization resistant mice (WT or STREP) and CA susceptible mice (PCN or PS). n=4. (c) _HIF1a_-Hif1a and (d) _LL-37_-Cramp mRNA expression measured in cultured human colonocytes exposed to ± CA and colons of antibiotic-treated mice ± CA colonization. n=4. (e) Hif1a and (f) Cramp mRNA expression measured in the colons of antibiotic treated mice ± oral gavage with commensal bacteria. n=4 (g) A representative western blot using an anti-CRAMP antibody (amino acids 135-173) against protein extracts from the distal colon of wild-type, Cramp KO, and antibiotic-treated mice ± oral gavage with commensal bacteria. Synthetic CRAMP peptide (5 ng, amino acids 135-173, 4.419 kDa) was used as a positive control. Cramp KO protein extracts were used as a negative control. Actin used as a loading control. Mouse CRAMP (amino acids 28-173) has a molecular mass of 16.422 kDa. A non-specific band (&) ~25kDa was detected in all lanes loaded with mouse colon protein extract. (f) Quantitative western blot analysis. Values obtained for CRAMP immunoblots were normalized to the optical density of corresponding immunoblots for actin. n=4. (i) Hif1a and (j) Cramp mRNA expression measured in the distal colon of germ-free mice colonized with CA, B. theta, or B. theta and CA. For all experiments, n=4. All data shown are means ± SEM. Statistical analysis by Mann-Whitney test. * p< 0.05; ** p<0.01; ns, not significant.
In addition to indirect host effects, B. theta may have a direct inhibitory effect on CA. Both B. producta and B. theta produce small-chain fatty acids (SCFAs) that have numerous immunomodulatory properties20-22. In fact, bacterial-produced SCFAs at physiologically-relevant doses23 inhibited CA growth in vitro and diminished CA colonization in mice but not to the degree seen with B. theta, suggesting that the host response is important to completely suppress CA colonization (Supplementary Figure 5)
To further test whether HIF-1α or LL-37 is required to prevent CA colonization, we first used a well-established HIF-1α agonist, L-mimosine24, in an in vitro fungicidal assay using cultured human colonocytes. mRNA and protein expression of both HIF1a and LL-37 increased in colonocytes exposed to mimosine (Supplementary Figure 6a-c). L-mimosine stabilizes HIF-1α through inhibition of prolyl hydroxylases and inhibits HIF-1α degradation16; this likely explains the modest increase in HIF1a gene expression compared to the more pronounced increase in protein expression. Candidacidal activity of LL-37, confirmed by using a spot assay25, is enhanced at lower pH, which has been previously reported with LL-37 and other anti-fungal agents18 (Supplementary Figure 6d). Of note, pH is acidic in the more distal segments of the intestine, ranging from 6.4-7.5 in the small intestine to 6.4 to 7.0 in the colon26. Furthermore, the distal intestine has the lowest oxygen tension in the GI tract,27 and, in principle, this could directly impact HIF-1α activation and LL-37 expression.
HT-29 colonocytes pretreated with L-mimosine significantly reduced fungal levels in a dose-dependent manner (Supplementary Figure 6e), but this effect was diminished when knocking down HIF1a mRNA expression (Supplementary Figure 6f-h). Also, L-mimosine did not inhibit the growth of CA in the absence of co-cultured colonocytes (Supplementary Figure 6i). Thus, pharmacological activation of HIF-1α in colonocytes is required for L-mimosine-dependent killing of CA.
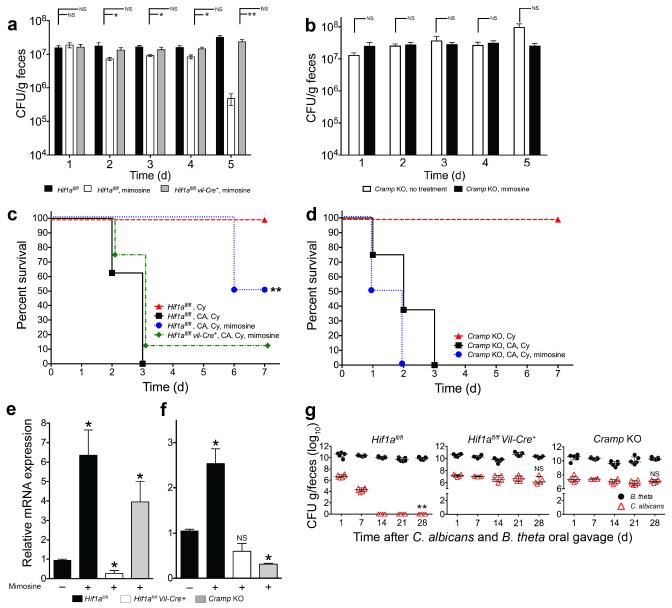
We next tested the importance of HIF-1α and CRAMP in vivo by using genetically engineered mice. We demonstrated that CA GI colonization levels significantly decreased in mice treated with mimosine, but this effect was nullified in mimosine-treated mice that had Hif1a specifically deleted from their intestinal epithelium (Hif1afl/fl Vil-Cre+ mice, Fig. 4a). In prior observations, mortality from invasive disease significantly decreased in mice with CA GI colonization levels < 107 cfu/g feces (data not shown). Thus, we administered cyclophosphamide to induce disseminated disease4. Strikingly, mice treated with mimosine had a 50% reduction in overall mortality (p=0.038 by Fisher’s exact test) and significantly increased length of survival (p<0.001 by log-rank test) compared to untreated counterparts (Fig. 4c). Further, mimosine treatment had no measurable effect on survival in Hif1afl/fl Vil-Cre+ mice (Fig 4c). We confirmed a reduction in colonization levels and increased length of survival (p=0.0082 by log-rank test, Supplementary Figure 7) when using a second CA strain, CAF2-1.
Figure 4. L-mimosine activation of HIf1a and Cramp in vivo decreases Candida albicans GI colonization and dissemination.
Candida albicans GI colonization levels in (a) Hif1afl/fl, Hif1afl/fl Vil-Cre+, and (b) Cramp KO mice treated with antibiotics, colonized with CA and then treated ± L-mimosine. n=8. Bars represent the mean ± SEM. Statistical analysis performed by Mann-Whitney test. * p<0.05; ** p<0.001; ns, not significant. Survival curves of (c) Hif1afl/fl, Hif1afl/fl Vil-Cre+, and (d) Cramp KO mice treated with antibiotic water, colonized with CA, treated ± L-mimosine for five days and then given cyclophosphamide. L-mimosine treatment continued for an additional 7 days after the first cyclophosphamide dose. n=8. Statistical analysis performed by log-rank test. * p< 0.05; ** p<0.01; ns, not significant. (e) Hif1a and Cramp mRNA levels in colons of Hif1afl/fl, Hif1afl/fl Vil-Cre+, and Cramp KO mice treated ± L-mimosine. n=4. All data shown are means ± SEM. Assays were performed in triplicate. Statistical analysis performed by Mann-Whitney test. * p< 0.05; ** p<0.01; ns, not significant. (g) C. albicans (red triangles) and B. theta (black circles) GI colonization levels in antibiotic-treated Hif1afl/fl, Hif1afl/fl Vil-Cre+, and Cramp KO mice. n= 6 for Hif1afl/fl and Cramp KO mice. n=5 for Hif1afl/fl Vil-Cre+. Black circles (B. theta) and red triangles (CA) represent results from individual animals. Horizontal lines represent the median with interquartertile range. * p<0.05, ** p<0.01. Statistical analysis by Mann-Whitney test.
We next examined whether CRAMP was necessary for the mimosine-induced antifungal effects that we observed. First, we verified that colonic Hif1a and Cramp mRNA expression significantly increased in mimosine-treated mice (Fig. 4e). Second, CRAMP knockout mice treated with mimosine showed neither a significant decrease in C. albicans GI colonization levels (Fig. 4b) nor a decrease in mortality after administration of cyclophosphamide (Fig. 4d) compared to untreated controls. As expected, colonic Hif1a mRNA expression significantly increased, and Cramp expression was negligible in mimosine-treated CRAMP knockout mice (Fig. 4e-f).
In mice pre-treated with antibiotics (PS) and then co-colonized with CA and B. theta, HIF-1α and CRAMP were required for _B. theta_-induced protection against CA colonization of the gut (Fig. 4g). In the absence of antibiotics, HIF-1α or CRAMP was not necessary to maintain CA colonization resistance in mice which we attribute to the presence of other redundant immune pathways that may aid in maintaining CA colonization resistance when a mature and intact gut microbiota is present (Supplementary Figure 8).
Humans are considered the main reservoir of CA28,29. Reportedly 40-80% of humans living in Westernized societies are colonized with CA30-32, but more recent studies of humans living in remote and traditional societies exhibit a CA GI carriage rate of less than 10%33-35. An ongoing study of human subjects suggests that the CA carriage rate in humans may be lower than previously reported as well (data not shown). Thus, CA might not be a “normal” commensal of the human gut, but a more recently acquired commensal resulting from medical advances (e.g. antibiotics) and adoption of Western diets36,37.
Commensal anaerobes, which account for > 99% of all gut bacteria38-40, are needed to maintain CA colonization resistance in mice**.** Notably, we show that two anaerobic species belonging to the same Genus, Bacteroides fragilis and Bacteroides thetaiotamicron, had markedly different effects on CA colonization: B. theta reduced CA colonization while B. fragilis did not. This discrepancy is likely explained by differing commensal bacteria-host immune response effects (i.e. HIF-1α, CRAMP; Fig 3e-h). Furthermore, B. theta has been shown to stimulate production of other antimicrobial peptides (RegIIIγ41, Ang 442) that may result in fungal killing. In contrast, B. fragilis has been shown to control activation of T-cell-dependent immune responses43, which we have previously shown to have no effect on CA GI colonization in mice4. Similarly, L. acidophilus reduces Candida colonization in the stomach of mice44, but we did not see this effect of L. acidophilus in the distal gut. In contrast, L. reuteri, significantly decreased CA colonization and merits further study. Identifying distinct bacterial species may allow us to uncover other host immune effectors that promote CA colonization resistance.
In a stressed system (exposure to antibiotics and resultant gut dysbiosis), the magnitude of the microbiota stimulus that activates the gut’s redundant systems for maintaining CA colonization resistance is markedly diminished and we find the elimination of select immune effectors (HIF-1α, CRAMP) is enough to nullify the CA-protective effect mediated by single bacterial species such as B. theta. These findings underline the importance of an intact gut microbiota in maintaining effective gut mucosal defenses against microbial pathogens, and may have significant clinical ramifications for those patients (i.e. cancer and transplant patients) who receive broad-spectrum antibiotics and are at high risk for developing invasive fungal infections_._
Probiotic therapy would seem to be an obvious approach for modulating CA colonization and has had some efficacy in both mice44 and humans45,46. In cancer patients, the introduction of probiotics raises the concern that the probiotic itself may cause infection. In fact, when using B. theta or B. producta as probiotic therapy in antibiotic-treated mice, we were able to prevent CA dissemination only to find occasional evidence of B. theta or B. producta dissemination (data not shown).
Pharmacologic modulation of HIF-1α levels has been explored extensively in the context of cancer therapy and angiogenesis47,48. HIF-1α agonists boost the bactericidal capacity of phagocytes and we show they may augment the fungicidal capacity of GI epithelial cells. As noted above, another potential approach could use commensal anaerobe metabolites, such as SCFAs. Finally, since SCFAs are naturally occurring metabolites of the gut, adverse effects, typically associated with pharmacologic agents, should not be a concern.
In conclusion, the emergence of invasive fungal disease in humans correlates with advances in medical therapy, particularly antibiotics and invasive surgical procedures. Where augmenting innate cellular function or mucosal integrity is difficult, if not impossible, in immunocompromised patients, boosting GI mucosal immune effectors to reduce fungal burden may be the key to tipping the balance back towards homeostasis and preventing invasive fungal disease.
Online Methods
Fungal and Bacterial strains
The Candida spp. strains and bacteria strains used are listed in Supplementary Table 3. Unless otherwise noted, C. albicans refers to strain SC5314.
Mouse Studies
Mouse strains used are listed in Supplementary Table 1. All animal experiments were done in accordance with NIH guidelines, the Animal Welfare Act and US federal law. The University of Texas Southwestern Medical Center’s Institutional Animal Care and Use Committee approved the experimental protocol “2009-0243” that was used for this study. All animals were housed in a centralized and AAALAC-accredited research animal facility that is fully staffed with trained husbandry, technical and veterinary personnel. Unless otherwise noted, mice used for experiments were C3H/HeN (Harlan), sex-matched, 6-8 weeks of age. Mice within an experiment were littermates that remained co-housed in the same cage to ensure a shared microbiota. Germ-free C57BL/6 mice were maintained in isolators as described.49
For germ-free and neonatal/infant mouse experiments, a sample size of 4 per group was chosen as the best compromise between providing an adequate sample size for assessing differences in colonization balanced with the limited availability of germ-free and neonatal mice. With the antibiotic-treated mice, we used a sample size of 5-8. Inclusion/exclusion criteria were not established. No animals were excluded from analysis. Randomization was not used. For the murine studies, age and sex-matched mice were arbitrarily assigned to a treatment group (i.e. antibiotics, bacteria, or mimosine). No blinding was done for mouse studies.
Conditional knockout of Hif1a in the murine intestine
Hif1a_f_l/fl (B6.129-Hif1atm3Rsjo/J, C57/BL background, Jackson Laboratories, floxed at exon 2) were bred with a transgenic strain expressing Cre recombinase under the control of the murine villin promoter (B6.SJL-Tg(Vil-cre)997Gum/J, C57/BL background, Jackson Laboratories). HIF-1α deletion in intestinal tissues (versus non-intestinal tissues) was confirmed by PCR on mouse genomic DNA (Supplementary Figure 9). Hif1a mRNA levels were markedly decreased in the ileums and colons, but not in the spleens, of Hif1afl/fl vil-Cre+ mice compared with their WT littermates (Supplementary Figure 9).
Antibiotic Experiments
Mice were fed sterile water or antibiotic water and colonized with C. albicans as previously described4. As for oral antibiotic treatment, mice were fed sterile water with 1) 2 mg streptomycin /ml (STREP), 2) 1500 U penicillin G /ml (PCN), 3) PCN/STREP, 4) 0.5 mg clindamycin/ml (C), or 5) 1 mg metronidazole/ml (M) for 5 days prior to C. albicans administration (2 × 108 cfu via oral gavage) and throughout the duration of the experiment. C. albicans GI colonization levels were initially checked 7 days after oral gavage (and at other indicated times) and were enumerated by culturing fecal contents on YVG agar (yeast-peptone-dextrose agar with 0.010 mg/ml vancomycin and 0.100 mg/ml gentamicin to suppress bacterial growth)4. For experiments utilizing bacteria, obligate anaerobic strains (B. fragilis, B. theta, B. producta, C. leptum) were cultured in GMM50, TYG51 or BHI/Blood media under anaerobic conditions (Coy anaerobic chamber) at 37°C; E. coli was grown in LB and MacConkey media under aerobic conditions at 37°C; P. aeruginosa was grown in LB and Cetrimide media under aerobic conditions at 37°C; and Lactobacilli spp. were grown in Lactobacilli MRS media (Difco) under aerobic or anaerobic conditions at 37°C. Bacterial colonization levels were enumerated by growth on the appropriate selective media and identity confirmed by gram-stain, enzymatic analysis (Rapid One for Enterobacteriaceae and RapID ANA II for anaerobes, Remel) and/or 16S RNA sequencing. For dissemination experiments, cyclophosphamide (Cy) was administered as previously described2. Mice were monitored for mortality for 7 days. Moribund mice were euthanized. Livers were resected, homogenized and plated on YVG, MacConkey, TSA, and BHI/Blood agars. The presence of a homogeneous population of creamy-white colonies on YVG and a complete absence of bacterial growth on the MacConkey (aerobic), TSA (aerobic), and BHI/Blood (anaerobic) plates were used for confirmation of C. albicans dissemination.
Antibiotic-Cessation Experiments
C3H/HeN mice (female, 6-8 weeks, Harlan, n=5) were treated with PS water (starting on Day -14). C. albicans (2 × 108 cfu ) was given by oral gavage on Day -7. PS water was replaced with sterile water on Day 0. C. albicans GI colonization levels were quantified every 7 days thereafter.
Bacterial Add-back in Antibiotic-treated Mice
PS-treated adult mice (C3H/HeN, female, 6-8 weeks, n=8 per group) were colonized with C. albicans. Mice were then gavaged with a single bacterial strain (2 × 108 cfu). A no bacteria control was included. All mice were then transitioned to sterile water. Bacterial and C. albicans GI colonization levels were measured after 14 days of co-colonization.
Bacterial Add-back in Germ Free Mice
Germ-free mice (no antibiotic treatment) were first colonized with C. albicans by oral gavage (2 ×108 cfu). B. thetaiotamicron or B. producta was then administered by oral gavage (2 × 108 cfu). Bacterial and Candida colonization levels were checked every 2-3 days up to 20 days after bacterial oral gavage.
SCFA Experiments
PS-treated adult mice (C3H/HeN, female, 6-8 weeks, n=4 per group) were colonized with C. albicans by oral gavage. Mice were then treated with PS water ± 50 mM SCFAs (butyric, acetic, or propionic acid). The no treatment control group received pH adjusted (with HCl) PS water. Colonization levels were checked after 14 days of SCFA water treatment.
L-Mimosine Experiments
Hif1afl/fl and Hif1afl/fl Vil-Cre+ were bred, genotyped, and functionally confirmed as noted above. Congenic Cramp knockout mice (B6.129X1-Camptm1Rlg/J, Jackson Laboratories) were obtained. Decreased Cramp mRNA expression in intestinal tissues was confirmed (Supplementary Figure 9c). Mice were treated with PS and then colonized with C. albicans strains SC5314 or CAF2-1. Mice were treated with PS water or PS water/500 uM L-mimosine ad libitum. PS and PS/mimosine water was changed every 2-3 days for the duration of the experiment. C. albicans GI colonization levels were checked daily for the first five days of mimosine treatment. Cyclophosphamide was administered to all groups on days 1, 3, and 5 of mimosine treatment to induce disseminated disease
Creation of murine-adapted C. albicans strains
C. albicans strains SC5314 and CAF2-1 were orally gavaged to adult mice (C3H/HeN, female, 6-8 weeks, Harlan) pre-treated with penicillin/streptomycin, recovered from fecal cultures, and archived (18% glycerol frozen stock). Archived C. albicans strains were grown and administered to a new set of antibiotic-treated mice, recovered, and archived again. This process was repeated for a third time. Final archived cultures were used for murine experiments.
Isolation of bacterial and fungal genomic DNA
Fecal specimens or intestinal tract segments were collected, flash frozen with liquid nitrogen, weighed, and immediately suspended in extraction buffer (200 mM NaCl, 200mM Tris, 20 mM EDTA, 6% SDS) and 0.5 ml of phenol/chloroform/isoamyl alcohol, pH 7.9 (Ambion).50 Cells were lysed by bead-beating (0.1 mm zirconia/silica beads for bacterial gDNA (BioSpec), 0.6 mm acid-washed glass beads for fungal gDNA (Sigma)) and subjected to additional phenol/chloroform extractions. Crude DNA extracts were treated with RNAseA (Qiagen) and column-purified (PCR Purification Kit, Qiagen). DNA concentrations were quantified by fluorescence-based assay (Quant-iT PicoGreen dsDNA, Life Technologies).
Isolation of ileal and colonic intestinal epithelial cells (IECs)
To confirm deletion of Hif1a (Exon 2) in intestinal epithelial cells of Hif1afl/fl Vil-Cre+ mice, colons and ileums were isolated, opened longitudinally and rinsed with PBS. The epithelial integrity was disrupted by treatment with 1 mM dithiothreitol (DTT) for 30 min at 37°C on a shaker, followed by vortexing for 1 min. The liberated IECs were collected, resuspended in 5 ml of 20% Percoll and overlaid on 2.5 ml of 40% Percoll in a 15-ml Falcon tube. Percoll gradient separation was performed by centrifugation at 780 g for 20 min at 25°C. The interface cells were collected and used as IECs20. gDNA was immediately isolated from IECs with the protocol detailed above.
Quantitative PCR (qPCR) for microbiota analysis
Bacterial and fungal loads were quantified by qPCR analysis (SsoAdvanced SYBR Green Supermix, Bio-Rad) of microbial gDNA using universal 16S rRNA gene or fungal internal transcribed spacer (ITS1-2) primers52 (Supplementary Table 4). The abundance of specific bacterial groups was determined by qPCR using group-specific 16S rRNA gene primers53. The abundance of specific Candida spp. was determined by qPCR using species-specific ITS1-2 gene primers and species-specific probes (Supplementary Table 4). To confirm specificity, Candida spp. specific qPCR was performed using gDNA from all the Candida species used in this study, and no false positives were noted (Supplementary Figure 10). Bacterial and fungal abundance was determined using standard curves constructed with reference to cloned DNA corresponding to a short segment of the 16s rRNA or ITS1-2 genes, respectively, that was amplified using conserved specific primers (Supplementary Table 4). Note that qPCR measures gene copies/g tissue, not actual bacterial/fungal numbers or colony forming units.
Preparation of murine colon protein extracts and Western blot analysis
Protein extracts for Western blot analysis were generated from mouse distal colon. A 2 cm piece of freshly isolated intestinal tissue was flushed, flash frozen and pulverized under liquid N2. The pulverized tissue was resuspended in 1 ml of ice-cold extraction buffer (8M urea, 1% SDS, 0.15M Tris–HCl, pH 7.5) with protease inhibitors (Complete Protease Inhibitor Cocktail, Roche) and lysed by homogenization, sonication, and passing the suspension through an 18 gauge needle 3–5 times, followed by 3–5 passages through a 21 gauge needle. Total protein was precipitated with tricholoroacetic acid and resuspended in electrophoresis sample buffer (50mM Tris-HCl pH 8.8, 2% SDS, 10% glycerol, 2mM EDTA, 100 mM DTT). Equal amounts of protein (25 ug), as quantitated using the Micro BCA Protein Assay (Pierce), were loaded onto 10-20% Tris-Tricine gel (Biorad) subjected to electrophoresis and transferred to a 0.45 μM PVDF (Biorad) membrane. Membranes were blocked in Odyssey Blocking Buffer (Li-Cor) for 1 hour at room temperature and incubated overnight at 4°C with a polyclonal rabbit anti-CRAMP antibody (amino acids 135-173, PA-CRCL-100, Innovagen). Incubation with a fluorescently-labeled secondary antibody (IRDye 800CW goat anti-rabbit IgG, 925-32211, Li-Cor) was performed for 1 hour at room temperature. Immunoreactivity was detected using the Odyssey CLx Imaging System (Li-Cor). Synthetic CRAMP peptide (amino acids 135-173) was used as a positive control (SP-CRPL, Innovagen). Actin (pan-Actin rabbit monoclonal antibody, D18C11, Cell Signaling) was used as an internal control to confirm equal protein loading. To calculate relative protein levels, the density of specific bands was quantified using a densitometer imaging system ImageJ software version 1.43s (NIH, Bethesda, MD); http://imagej.nih.gov. Values obtained for CRAMP immunoblots were normalized to the optical density of corresponding immunoblots for actin.
16S rRNA gene PCR amplification and sequencing
16S rRNA genes (variable region 4, V4) were amplified from each sample using a composite forward primer and a reverse primer containing a unique 10-base barcode that was used to tag PCR products from respective samples54. We used the forward primer 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG_-NNNNNNNNNN-AYTGGGYDTAAAGNG-3′: the italicized sequence is 454 Life Sciences® primer A; NNNNNNNNNN designates the unique 10-base barcode used to tag each PCR product; and the bold sequence is the broad-range bacterial primer 563F. The reverse primer used was 5′_CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCCGTCAATTYYTTTRAGTTT-3′: the italicized sequence is 454 Life Sciences’ primer B, and the bold sequence is the broad-range bacterial primer 926BSR. PCR reactions consisted of 2.5U FastStart High Fidelity Taq and 1× buffer (Roche), 400 nM of each primer, 50 ng template, and reaction conditions were 3 min at 95°C, followed by 15 cycles of 30s at 95°C, 45s at 65-50°C (decreasing by 1°C/cycle) and 60s at 72°C; then 20 cycles of 30s at 95°C, 45s at 57°C and 60s at 72°C on an Eppendorf Mastercycler. Four independent PCRs were performed for each sample, combined and purified with Ampure magnetic purification beads (Agencourt), and products visualized by gel electrophoresis. No-template extraction controls were analyzed for absence of visible PCR products. Products were quantified using Quant-iT PicoGreen dsDNA (Life Technologies). A master DNA pool was generated from the purified products in equimolar ratios to a final concentration of 10 ng ml-1. The pooled products were sequenced using the Roche 454 Titanium platform (University of Texas at Austin Genome Sequencing and Analyis Facility) using Roche/454 Titanium chemistry.
16S rRNA gene sequence analysis
Sequences generated from pyrosequencing barcoded 16S rRNA gene PCR amplicons were quality filtered. Sequences shorter than 200 nucleotides or longer than 1000 nucleotides were removed. Sequences containing ambiguous bases, primer mismatches, homopolymer runs in excess of 6 bases and uncorrectable barcodes were also removed. Sequences that passed the quality filtration were denoised and analyzed using the open source software package Quantitative Insights Into Microbial Ecology (QIIME)55. 16S rRNA gene sequences were assigned to operational taxonomic units (OTUs) using UCLUST (www.drive5.com/usearch/), with a threshold of 97% pair-wise identity, then classified taxonomically using Greengenes (greengenes.lbl.gov). 16S rRNA data (analyzed data and trimmed fasta sequences for each sample) was deposited at Figshare (http://figshare.com/articles/Fan_et_al_NMED_A69544_16S_rRNA_Data/950989).
qPCR for quantifying gene expression
Total RNA was isolated from the distal small intestine or colon using the Qiagen RNEasy RNA isolation kit and from HT-29 human colonocytes with Trizol reagent (Life Technologies). Total RNA was used to synthesize cDNA (iScript, BioRad). qPCR analysis was performed using the SsoAdvanced SYBR Green Supermix (Bio-Rad) and specific primers (Supplementary Table 4). Signals were normalized to 18S rRNA levels within each sample and normalized data were used to calculate relative levels of gene expression using ΔΔCt analysis.
HT-29 human colonocyte assays
HIF1a and LL-37 expression in response to C. albicans
HT-29 cells were propagated in RPMI-1640 with 10% FCS. HT-29 cells were exposed to C. albicans strain SC5314 at an MOI of 1 for 5 hours. Cells were then washed with PBS × 3, and total RNA was extracted with Trizol reagent (Life Technologies). HIF1a and LL-37 gene expression was evaluated by qPCR as noted above.
_HIF1a_-HIF-1α and LL-37 - LL-37 mRNA and protein expression in response to L-mimosine
HT-29 cells were propagated in RPMI-1640 with 10% FCS. HT-29 cells were exposed to L-mimosine (0-500 μmol/L) for 5 hours at 37°C. Cells were then washed with PBS. For gene expression experiments, total RNA was extracted with Trizol reagent (Life Technologies) and qPCR for selected genes was carried out as noted above. For protein analysis, protein was extracted with RIPA buffer, and concentrations determined using BCA assay (Pierce). Proteins were separated by 4-12% Tris-tricine (Invitrogen) gel electrophoresis. Western blot analysis was performed using anti HIF-1α (NB100-449, Novus) and anti LL-37 (NBP1-76864, Novus) rabbit polyconal antibodies. The secondary Ab was a horseradish (HRP) peroxidase-conjugated goat anti-rabbit (7074, Cell Signaling). Actin (pan-Actin rabbit monoclonal antibody, D18C11, Cell Signaling) was used as an internal control to confirm equal protein loading. Immunoreactive proteins were detected using the ECL-chemiluminescent system (EMD Millipore Immobilon).
Effect of L-mimosine on C. albicans growth
Candida albicans strain SC5314 cells were grown overnight, washed, and resuspended in PBS. Cells were added to RPMI-1640 with 10% FCS at a concentration of 1 × 106 cfu/ml. L-mimosine (0-500 uM) was added, and samples were incubated at 37°C for 8 hours. 1 ml of culture was collected at 0, 2, 4, 6 and 8 hours after addition of L-mimosine. Quantification of C. albicans was calculated by serial dilution and plating on YPD.
HT-29 Extracellular Killing assays
C. albicans strain SC5314 grown to mid-log phase (OD600 ~ 1.0-1.5) in YPD media at 30°C. Cells were harvested, washed with and resuspended in PBS, and enumerated with a hemocytometer. 1 × 105 cfu of C. albicans was placed in each well (24-well plate) containing confluent HT-29 colonocytes (MOI = 1.0). 24-well plates were quickly centrifuged and then incubated at 37°C for 1, 3, or 5 hours. Supernatants were collected, and cells were washed with PBS × 3 (and washes pooled with the original supernatant). C. albicans level (remaining cfu) was calculated by serial dilution and plating on YPD agar. For experiments with L-mimosine, HT-29 cells were treated with L-mimosine for 3 hours prior to the addition of C. albicans.
HIF-1α siRNA Knockdown
HT-29 cells (1 × 105 cells) were freshly seeded into each well of a 24-well plate. After 24 hours of incubation at 37°C, HT-29 cells were transfected with 5nM of HIF1a siRNA (Qiagen) or scrambled control using HiPerFect transfection reagent (Qiagen). Fungicidal assays were performed 48 hours later.
Candida albicans spot assay
Candida albicans strain SC5314 cells were grown overnight, washed, and resuspended. Cells were incubated with 30 ug recombinant LL-37/ml (Anaspec) in sodium phosphate buffer at 37°C for 1 hr, serially diluted and spotted onto YPD-agar plates25. Plates were incubated for 2-3 days at 30°C.
Candida albicans SCFA in vitro Growth Experiments
C. albicans strain SC5314 was grown overnight in YPD at 30°C under aerobic conditions. Overnight cultures were then harvested, washed in PBS and then diluted to a starting OD600 ~0.2. SCFAs (butyric acid, acetic acid, or propionic acid) were added to the culture at the desired concentration resulting in pH ranging from 3-6. pH adjusted controls were created by titrating concentrated HCl into YPD. Cultures were grown at 30°C, 250 rpm under aerobic conditions for 12 hours. Final OD600 readings were then measured.
Candida albicans/B. theta co-cultured in vitro Growth Experiments
Candida albicans strain SC5314 and B. theta were first grown individually in TYG media at 37°C under anaerobic conditions. Overnight cultures were then diluted to an OD600 = 0.2 for the initial time point (0 hours). CA and B. theta co-cultures were created by combining equivalent volumes (5 ml) of CA and B. theta individual diluted cultures. Cultures were samples every 24 hours (for a total of 96 hours), and CA and B. theta (CFU) were enumerated by serial dilution and plating on YVG agar (aerobically) and BHI/Blood agar (anaerobically), respectively
Statistical analyses
A comparison of GI colonization levels was analyzed by Mann-Whitney tests, and when multiple comparisons or more than two groups were analyzed, Bonferroni’s correction to the significance level α was invoked. Bacterial 16S rRNA gene copy numbers and qPCR gene expression analyses were compared using a Mann-Whitney test. Survival data were analyzed by both Fisher’s exact test (overall survival) and log-rank test (length of survival, Kaplan-Meier curves). Statistical analyses were carried out using the GraphPad Prism Software (San Diego, CA).
Supplementary Material
1
Acknowledgements
We would like to K. Nickerson (University of Nebraska, Lincoln, Nebraska), J. Patton-Vogt (Duquesne University, Pittsburgh, Pennsylvania), R. Wheeler (University of Maine, Orono, Maine) and M. Lorenz (University of Texas Health Science Center, Houston, Texas) for the provision of the C. albicans strains SN152 (KN), BWP17 (JPV), Can098 (RW), WO-1 (ML), 3153A (ML). We would like to thank C. Doern (Children’s Medical Center Dallas, Dallas, Texas) for the C. glabrata, C. parapsilosis, and C. tropicalis strains. We would like to thank Y. Iwakura (Institute of Medical Science, Tokyo University, Tokyo, Japan) and J. Kolls (Children’s Hospital of Pittsburgh, Pittsburgh, Pennsylvania) for providing the IL-17A knockout mice. We would like to thank S. Skapek, J. Amatruda, R. DeBerardinis, and D. Greenberg for providing helpful comments on the manuscript. This study was supported by the Roberta I. and Norman L. Pollock Fund (A.Y.K), the Global Probiotics Council, Young Investigator Grant for Probiotics (A.Y.K), NIH R01 DK060855 (L.V.H), the Howard Hughes Medical Institute (L.V.H.), and P30CA142543 (Y.X.)
Footnotes
Author contributions. AYK and LVH conceived and designed the experiments. LVH provided gnotobiotic mouse and mucosal immunology instruction and support. AYK, DF, LAC, TRSW, and MMN performed the experiments. AYK, JK, MK, XZ, and YX conducted microbial profiling and statistical analysis. AYK and LVH analyzed the data. AYK wrote the paper.
Competing interests. The authors have declared that no competing interests exist.
References
- 1.Miranda LN, et al. Candida colonisation as a source for candidaemia. J Hosp Infect. 2009;72:9–16. doi: 10.1016/j.jhin.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Nucci M, Anaissie E. Revisiting the source of candidemia: skin or gut? Clin Infect Dis. 2001;33:1959–1967. doi: 10.1086/323759. [DOI] [PubMed] [Google Scholar]
- 3.Pizzo PA, Poplack D, editors. Principles and Practice of Pediatric Oncology. 6th Edition Lippincott Williams & Wilkins; Philadelphia: 2011. p. 1531. [Google Scholar]
- 4.Koh AY, Kohler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS pathogens. 2008;4:e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samonis G, et al. Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrob Agents Chemother. 1993;37:51–53. doi: 10.1128/aac.37.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Waaij D. Colonization resistance of the digestive tract: clinical consequences and implications. The Journal of antimicrobial chemotherapy. 1982;10:263–270. doi: 10.1093/jac/10.4.263. [DOI] [PubMed] [Google Scholar]
- 8.Dubos R. The microbiota of the gastrointestinal tract. Gastroenterology. 1966;51:868–874. [PubMed] [Google Scholar]
- 9.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg RD. Promotion of the translocation of enteric bacteria from the gastrointestinal tracts of mice by oral treatment with penicillin, clindamycin, or metronidazole. Infect Immun. 1981;33:854–861. doi: 10.1128/iai.33.3.854-861.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother. 1999;43:727–737. doi: 10.1128/aac.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ringel-Kulka T, et al. Intestinal microbiota in healthy U.S. young children and adults--a high throughput microarray analysis. PLoS One. 2013;8:e64315. doi: 10.1371/journal.pone.0064315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrod LP. The relative antibacterial activity of four penicillins. British medical journal. 1960;2:1695–1696. doi: 10.1136/bmj.2.5214.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol. 2009;9:609–617. doi: 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peyssonnaux C, et al. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. The Journal of clinical investigation. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Garcia B, Lee PH, Yamasaki K, Gallo RL. Anti-fungal activity of cathelicidins and their potential role in Candida albicans skin infection. The Journal of investigative dermatology. 2005;125:108–115. doi: 10.1111/j.0022-202X.2005.23713.x. [DOI] [PubMed] [Google Scholar]
- 19.Tsai PW, Yang CY, Chang HT, Lan CY. Human antimicrobial peptide LL-37 inhibits adhesion of Candida albicans by interacting with yeast cell-wall carbohydrates. PLoS One. 2011;6:e17755. doi: 10.1371/journal.pone.0017755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 22.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinkernagel AS, Peyssonnaux C, Johnson RS, Nizet V. Pharmacologic augmentation of hypoxia-inducible factor-1alpha with mimosine boosts the bactericidal capacity of phagocytes. J Infect Dis. 2008;197:214–217. doi: 10.1086/524843. [DOI] [PubMed] [Google Scholar]
- 25.Argimon S, Fanning S, Blankenship JR, Mitchell AP. Interaction between the Candida albicans high-osmolarity glycerol (HOG) pathway and the response to human beta-defensins 2 and 3. Eukaryot Cell. 2011;10:272–275. doi: 10.1128/EC.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans DF, et al. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He G, et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odds FC. Molecular phylogenetics and epidemiology of Candida albicans. Future Microbiol. 2010;5:67–79. doi: 10.2217/fmb.09.113. [DOI] [PubMed] [Google Scholar]
- 29.Lott TJ, Fundyga RE, Kuykendall RJ, Arnold J. The human commensal yeast, Candida albicans, has an ancient origin. Fungal Genet Biol. 2005;42:444–451. doi: 10.1016/j.fgb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Bougnoux ME, et al. Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. Journal of clinical microbiology. 2006;44:1810–1820. doi: 10.1128/JCM.44.5.1810-1820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kam AP, Xu J. Diversity of commensal yeasts within and among healthy hosts. Diagn Microbiol Infect Dis. 2002;43:19–28. doi: 10.1016/s0732-8893(02)00364-4. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, et al. Species and genotypic diversities and similarities of pathogenic yeasts colonizing women. Journal of clinical microbiology. 1999;37:3835–3843. doi: 10.1128/jcm.37.12.3835-3843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angebault C, et al. Candida albicans is not always the preferential yeast colonizing humans: a study in Wayampi Amerindians. The Journal of infectious diseases. 2013;208:1705–1716. doi: 10.1093/infdis/jit389. [DOI] [PubMed] [Google Scholar]
- 34.Reichart PA, et al. Oral Candida species and betel quid-associated oral lesions in Padaung women of Northern Thailand. Mycoses. 2005;48:132–136. doi: 10.1111/j.1439-0507.2004.01071.x. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Mitchell TG. Geographical differences in human oral yeast flora. Clin Infect Dis. 2003;36:221–224. doi: 10.1086/345672. [DOI] [PubMed] [Google Scholar]
- 36.Rosenbach A, Dignard D, Pierce JV, Whiteway M, Kumamoto CA. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryotic cell. 9:1075–1086. doi: 10.1128/EC.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White SJ, et al. Self-regulation of Candida albicans population size during GI colonization. PLoS pathogens. 2007;3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 39.Beaugerie L, Petit JC. Microbial-gut interactions in health and disease. Antibiotic-associated diarrhoea. Best practice & research. Clinical gastroenterology. 2004;18:337–352. doi: 10.1016/j.bpg.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Vedantam G, Hecht DW. Antibiotics and anaerobes of gut origin. Curr Opin Microbiol. 2003;6:457–461. doi: 10.1016/j.mib.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 43.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Zwolinska-Wcislo M, et al. Are probiotics effective in the treatment of fungal colonization of the gastrointestinal tract? Experimental and clinical studies. J Physiol Pharmacol. 2006;57(Suppl 9):35–49. [PubMed] [Google Scholar]
- 45.Manzoni P, et al. Oral supplementation with Lactobacillus casei subspecies rhamnosus prevents enteric colonization by Candida species in preterm neonates: a randomized study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2006;42:1735–1742. doi: 10.1086/504324. [DOI] [PubMed] [Google Scholar]
- 46.Hatakka K, et al. Probiotics reduce the prevalence of oral candida in the elderly--a randomized controlled trial. J Dent Res. 2007;86:125–130. doi: 10.1177/154405910708600204. [DOI] [PubMed] [Google Scholar]
- 47.Semenza GL. Development of novel therapeutic strategies that target HIF-1. Expert opinion on therapeutic targets. 2006;10:267–280. doi: 10.1517/14728222.10.2.267. [DOI] [PubMed] [Google Scholar]
- 48.Zarember KA, Malech HL. HIF-1alpha: a master regulator of innate host defenses? The Journal of clinical investigation. 2005;115:1702–1704. doi: 10.1172/JCI25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 49.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodman AL, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS pathogens. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barman M, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Claesson MJ, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic acids research. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1