Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity (original) (raw)
Significance
Infections with any of the four dengue virus serotypes (DENV 1–4) are the most prevalent and rapidly spreading mosquito-borne viral infections in humans. There is no treatment or vaccine currently available. We found that the virus-specific cells display a highly polarized cytotoxic phenotype that correlated with expression of a protective HLA DR allele. Although the occurrence of cytotoxic CD4+ T cells in humans has been described in the context of some chronic viral infections, to our knowledge, this is the first report of ex vivo cytotoxic CD4+ activity after exposure with an acute virus. These results will help shed light on the specific role of CD4+ T cells in DENV infection and may help in finding a correlate of protection.
Keywords: dengue virus, T cell memory, HLA DR, CD4, cytotoxic
Abstract
Dengue virus (DENV) is a rapidly spreading pathogen with unusual pathogenesis, and correlates of protection from severe dengue disease and vaccine efficacy have not yet been established. Although DENV-specific CD8+ T-cell responses have been extensively studied, the breadth and specificity of CD4+ T-cell responses remains to be defined. Here we define HLA-restricted CD4+ T-cell epitopes resulting from natural infection with dengue virus in a hyperepidemic setting. Ex vivo flow-cytometric analysis of DENV-specific CD4+ T cells revealed that the virus-specific cells were highly polarized, with a strong bias toward a CX3CR1+ Eomesodermin+ perforin+ granzyme B+ CD45RA+ CD4 CTL phenotype. Importantly, these cells correlated with a protective HLA DR allele, and we demonstrate that these cells have direct ex vivo DENV-specific cytolytic activity. We speculate that cytotoxic dengue-specific CD4+ T cells may play a role in the control of dengue infection in vivo, and this immune correlate may be a key target for dengue virus vaccine development.
All four of the dengue virus serotypes (DENV 1–4) have spread rapidly within countries and across regions in the past few decades, resulting in an increased frequency of epidemics and severe dengue disease. Multiple serotypes circulate simultaneously in many tropical countries, and recent outbreaks have been reported in Europe and the continental United States (1, 2). These circumstances make dengue the most prevalent and rapidly spreading mosquito-borne viral disease in humans (3). Recent reports estimate that 390 million infections occur each year, with ∼25% of cases resulting in symptomatic disease (2).
All four dengue serotypes can cause a spectrum of disease, ranging from self-limiting dengue fever to potentially lethal severe dengue disease, such as states of dengue hemorrhagic fever and Dengue shock syndrome, which are associated with the plasma leakage syndromes leading to visceral organ injury (4). It is not yet fully understood why only a subset of people infected with DENV progresses to severe disease. One risk factor for severe disease is the acquisition of DENV-reactive Abs before secondary infection with a different serotype (heterologous infection). These Abs can either be acquired from a previous infection with a different serotype or, in the case of infants, acquired from an immune mother (5, 6). It has been shown that subneutralizing levels of DENV-specific Abs exacerbate disease in a phenomenon termed Ab-dependent enhancement of infection (7, 8). In brief, dengue-specific cross-reactive Abs produced after an initial DENV infection combine with those produced after a second viral infection to form immune complexes that perpetuate infection by increasing the number of infected cells and, therefore, viral output per cell (6).
The observation that only a minority of patients develops severe disease suggests that host genetic factors may play an important role in disease severity. Relatedly, a role for T cells in control of disease has been suggested by several studies that correlate the expression of certain HLA molecules with susceptibility to or protection from DENV disease (9–15). HLA molecules are one of the most polymorphic host factors in humans, with several thousand variants thus far known (16, 17). Each HLA variant is present with variable frequency, depending on ethnic lineage and geographic locality. For HLA class I MHC restricted responses, it has been recently shown that different allelic variants are associated with differential magnitude of anti-DENV responses and that HLA alleles known to be associated with increased risk of severe DENV disease are associated with weaker CD8+ T-cell responses (18). Similarly, certain MHC class II alleles have been described as associated with increased protection from severe disease (9, 15, 19–21). Thus, T-cell responses are likely to contribute to protection from dengue disease.
In this study, we defined HLA-restricted CD4+ T-cell responses resulting from natural infection with DENV. We identified cytotoxic subsets of DENV-specific effector memory CD4+ T cells that are specifically expanded as a function of history of DENV infection and in donors carrying an HLA allele associated with protection from severe DENV disease.
Results
Definition of a Screening Strategy for HLA Class II DENV-Specific CD4+ T-Cell Responses.
DENV is hyperendemic in Sri Lanka, with ∼80% of the general population seropositive by the age of 18, with the majority of people showing neutralizing Ab patterns characteristic of multiple infections (18). To analyze DENV-specific CD4 responses, we selected a set of DENV peptides predicted to bind the DRB1*0701 allele, which is highly prevalent in Sri Lanka (33% phenotypic frequency). These peptides were tested by using HLA-matched peripheral blood mononuclear cell (PBMC) donations from blood donors from the Colombo blood bank.
In contrast to the case of previously described CD8+ T-cell responses (18), we did not readily detect CD4+ responses directly ex vivo using a standard IFN-γ enzyme-linked immunospot (ELISPOT) assay. This result was expected, because it has been shown for several human viral infections that the frequency of CD4+ T cells is generally lower than CD8+ T cells, and therefore in vitro expansion is often required. Accordingly, PBMCs from HLA DRB1*0701 individuals—either seronegative or associated with primary or secondary infections—were stimulated in vitro with DRB1*0701-restricted peptide pools. To minimize the potential for detecting responses arising from primary in vitro stimulation, naïve CD4+ T cells were removed before stimulation based on expression of CD45RA.
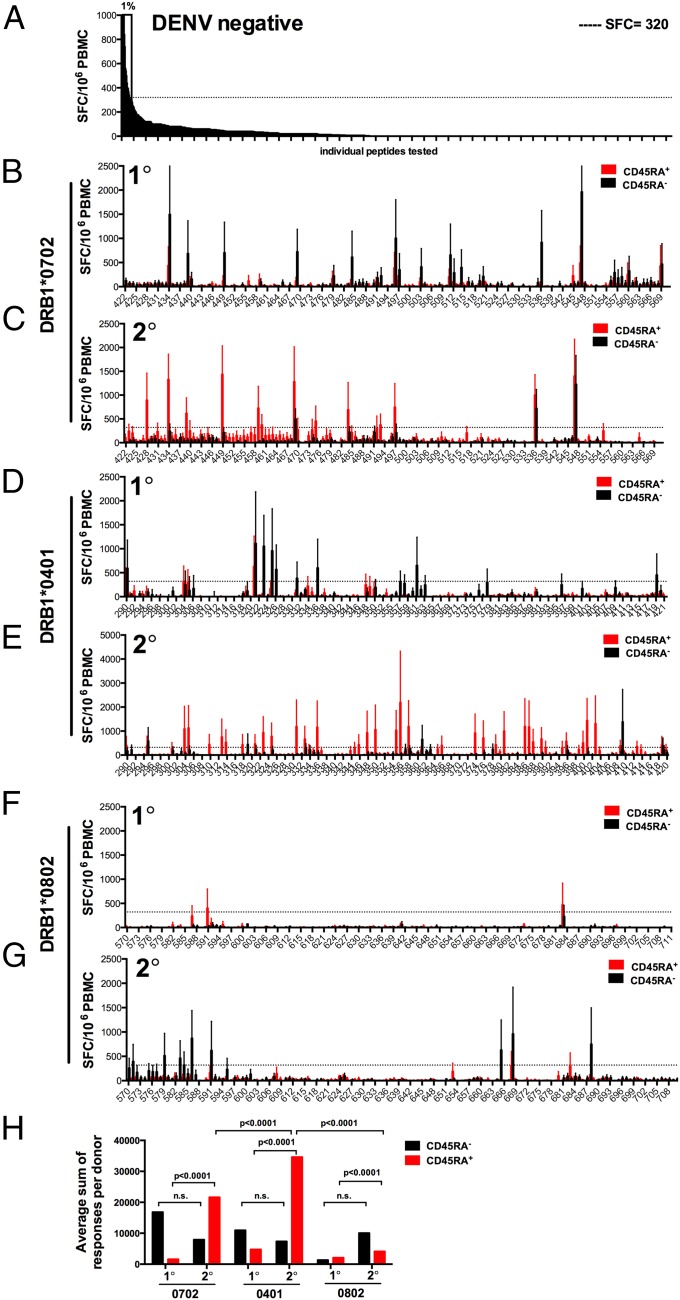
Using reactivity of seronegative individuals, we established a threshold for positivity of 320 spot-forming cells (SFCs). This threshold was >99% of individual responses; thus, responses above this threshold are expected to occur with a false positive rate of 0.01 (Fig. 1_A_). As expected, we could detect multiple positive memory responses in the CD45RA− subset in donors associated with primary DENV infection (Fig. 1_B_). However, to our surprise, the majority of CD4+ T-cell responses after secondary infection were detected in the CD45RA+ CD4+ T-cell subset (Fig. 1_C_).
Fig. 1.
Subset-specific CD4 T-cell responses after in vitro restimulation were determined by HLA restriction and DENV infection history. PBMC were thawed, and CD4+ T cells were isolated by magnetic bead negative selection. CD45RA+ and CD45RA− cells were subsequently isolated by magnetic bead selection and cocultured with autologous APCs and with DENV-specific pools (averaging 20 peptides per pool). On day 14, cells were harvested and screened for reactivity against individual DENV-specific peptides in an IFN-γ ELISPOT. (A) Eight DENV-negative donors were stimulated with HLA-matched peptide pools and tested for reactivity against individual peptides. The reactivity of seronegative individuals that was >99% of all individual responses (SFC = 320; dashed line) was used to define a threshold value for positivity corresponding to a P value of 0.01 for false positives. (B_–_G) CD45RA+ (red bars) and CD45RA− (black bars) CD4+ T cells from donors expressing the DRB1*0701 (B, n = 4; and C, n = 7), DRB1*0401 (D, n = 5; and E, n = 6), or DRB1*0801 (F, n = 2; and G, n = 6) allele and have experienced either primary (1°) or secondary (2°) infection with DENV were stimulated with HLA-matched peptide pools and tested for reactivity against individual peptides. Error bars represent mean ± SEM. (H) The sum of responses adjusted by the number of donors tested for each HLA is shown as a function of infection history. Significance of CD45RA+ (red bars) and CD45RA− (black bars) responses was compared in a two-tailed Mann–Whitney test.
In the same series of experiments, we also tested peptides predicted to bind DRB1*0401 and *0802 alleles. These two alleles were selected because they are associated with increased resistance (DRB1*0401) or susceptibility (DRB1*0802) to severe DENV clinical outcomes (9, 15, 22) In the case of the DRB1*04-protective allele and secondary infections, responses were most vigorous and associated with the CD45RA+ subset, thus resembling the pattern observed in the DRB1*07 responses (Fig. 1 D and E). By contrast, DRB1*08 restricted responses were weak in the primary infection and mostly associated with the CD45RA− subset in the case of secondary infections (Fig. 1 F and G). Statistical analyses are shown in Fig. 1_H_.
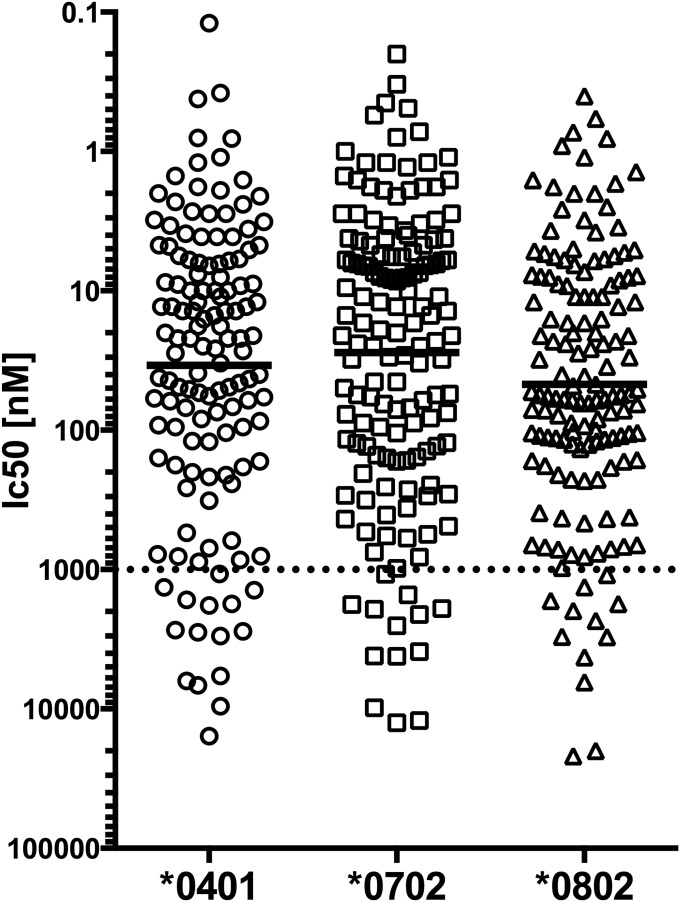
To exclude that differences in MHC binding account for the allele-specific differences, we measured the binding affinity of all predicted peptides to the respective HLA allele. For all HLA alleles, >89% of the predicted peptides bound with 1,000 nM or less, which corresponds to the biological threshold for efficient binding to a MHC class II molecules (23) (Fig. S1). No major differences in binding affinity between HLA alleles were observed, with the average IC50 being 28 for DRB1*0701, 34 for DRB1*0401, and 47 for DRB1*0802.
Fig. S1.
MHC binding of predicted peptides. Binding affinity of all predicted peptides to the respective HLA allele was measured by using a high-affinity radiolabeled peptide in a competition assay as described in Materials and Methods. The concentration of peptides yielding 50% inhibition of the binding of the radiolabeled peptide was calculated (IC50). The dashed line indicates the threshold of 1,000 nM, which corresponds to the biological threshold for efficient binding to a MHC class II molecules (24). Black bold lines indicate the geometric mean for all peptides tested for each allele (n = 132, 148, and 142 for DRB1*0401, *0702, and *0802, respectively).
Because secondary infection is associated with more consistent immunity from both homologous and heterologous infection, the response magnitude and the CD45RA+ phenotype seemed to correlate with protection from severe DENV disease.
Expansion of Memory T-Cell Subsets in DENV Secondary Infection.
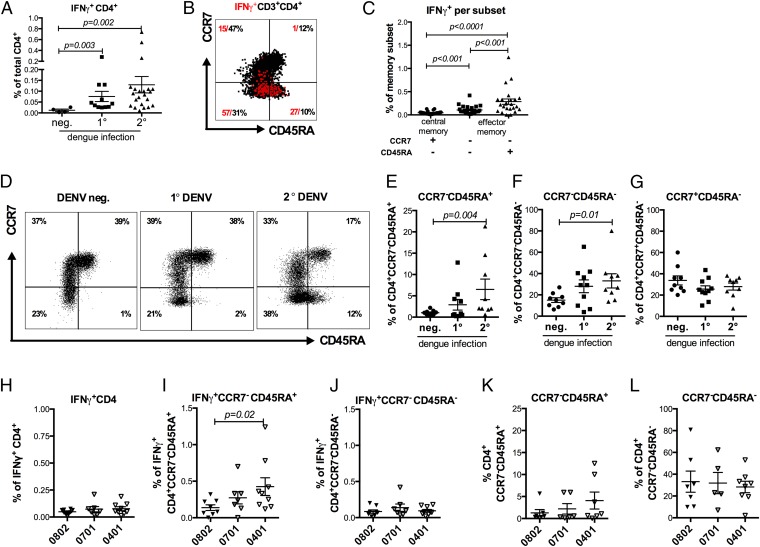
The design of HLA-specific epitope pools to enhance the frequency of responding T cells (as opposed to generic peptide pools) allowed us to readily and consistently detect ex vivo reactivity using intracellular cytokine staining (ICS). First, we examined the magnitude of response as a function of the donor infection history in a total of 37 different donors (Fig. 2_A_). No responses (<0.02%) were observed in DENV-seronegative donors, whereas on average 0.07% of total CD4+ T cells from primary infection cases produced IFN-γ upon epitope stimulation. As expected, the most vigorous responses were observed in the donors associated with secondary infections (0.13% on average). We then used the surface markers CCR7 and CD45RA to establish to which memory subset the responding T cells belonged (24). Fig. 2_B_ shows representative data for one donor, showing the expression patters of CCR7 and CD45RA in total CD4+ T cells (black dots) and antigen-specific cells after stimulation with a pool of DR-restricted epitopes (IFN-γ; red dots). Effector memory T-cell subsets, defined by the loss of CCR7, were associated with 57% (CCR7− CD45RA−) and 27% (CCR7−CD45RA+) of the response, respectively, whereas negligible amounts of the DENV-specific responses were attributed to naïve (CCR7+ CD45RA+) and central memory (CCR7+CD45RA−) T-cell subsets. Interestingly, in this donor 10% of the total CD4+ T cells were associated with the CCR7−CD45RA+ effector memory subset. Previous studies reported this subset to be present at 2.3 ± 1.1% (CD4+CD45RA+CCR7–) in a group of randomly selected healthy donors, such that the expansion of this subset in DENV-infected donors was somewhat unexpected (25). When gated on the individual memory subset, the CCR7−CD45RA+ subset produced significantly more IFN-γ compared with the other two memory populations. (Fig. 2_C_; P < 0.001 in a Mann–Whitney test).
Fig. 2.
DENV-specific responses and memory T-cell subsets change as a function of infection history and restricting HLA alleles. (A) PBMCs from the Sri Lanka cohort (n = 37) were stimulated with HLA-matched peptides for 6 h, and the IFN-γ responses were measured by ICS. Responses are shown as a function of the donors’ exposure to the dengue virus [DENV-negative (n = 4) and primary (1°; n = 11) and secondary (2°, n = 22) DENV infection]. (B) Representative staining of the memory CD4+ T cells subsets and IFN-γ–producing cells of a secondary donor is shown. Memory subsets were defined as naïve T cells (CCR7+CD45RA+), central memory T cells (CCR7+CD45RA+), and the two effector memory subsets (CCR7−CD45RA− and CCR7−CD45RA+), according to their expression of these markers. (C) The IFN-γ response of each memory T-cell subset was measured by ICS after stimulation with HLA-matched peptides (n = 23). (D) Representative FACS plots of CD4+ T-cell subsets in donors with a DENV-negative, (1°) primary or (2°) secondary DENV infection history are shown. (E_–_G) Distribution of CD4+ memory T-cell subsets in negative, primary, and secondary donors are shown (E, CCR7−CD45RA+; F, CCR7−CD45RA−; G, CCR7+CD45RA+, n = 28). (H) Percentage of total CD4+ T cells that produce IFN-γ upon stimulation with HLA-matched peptides in donors carrying the DRB1*0802, DRB1*0701, or DRB1*0401 allele. (I and J) The ability of CCR7−CD45RA+ (I) and CCR7−CD45RA− (J) subsets to produce IFN-γ upon peptide stimulation was measured for donors expressing DRB1*0802, DRB1*0701, and DRB1*0401 alleles (n = 24). (K and L) The size of the total CD4+ CCR7−CD45RA+ (K) and CCR7−CD45RA− (L) subsets were also measured for each allele. Error bars represent mean ± SEM. Statistical significance was determined by using the two-tailed Mann–Whitney test.
We extended these observations by measuring the percentage of total CD4+ T cells associated with each memory subset in seronegative individuals or in persons determined to have previous primary or secondary DENV infections. Multiple DENV infections (2° DENV) were marked by a significant and progressive increase of the CCR7−CD45RA+ subset (Fig. 2_D_). The fraction of CD4+ T cells associated with this phenotype was negligible in DENV-seronegative individuals, increased in individuals associated with primary infection, and reached an average of 6.5% (0.6–21.3% range) in donors associated with secondary infection (Fig. 2_E_). A similar trend was also noted in the case of the other effector memory subset (CCR7−CD45RA−; Fig. 2_F_). By contrast, the fraction of CD4+ T cells associated with a central memory phenotype (CCR7+CD45RA−) did not vary appreciably as a function of DENV infection history (Fig. 2_G_).
We then compared the percentage of total CD4+ T cells that produce IFN-γ upon stimulation with HLA matched DENV peptides in donors carrying either the DRB1*0802, DRB1*0701, or DRB1*0401 allele. No significant difference between the HLA alleles could be observed (Fig. 2_H_). However, when we compared the magnitude of IFN-γ DENV-specific responses for memory subsets for each HLA DR allelic variant, we found that in the case of the DRB1*04-protective allele, CCR7−CD45RA+ effector memory cells were expanded compared with the DRB1*08-susceptible allele (P = 0.02; Fig. 2_I_), whereas no HLA-specific differences have been detected in IFN-γ+CCR7−CD45RA− memory cells (Fig. 2_J_). This trend was specifically associated with DENV-specific IFN-γ+ cells, because no significant association between protective and susceptible alleles was noted in the case of total CCR7−CD45RA+ and CCR7−CD45RA− subsets (Fig. 2 K and L). In conclusion, an expansion of the CCR7−CD45RA+ effector memory subsets is associated with measured IFN-γ responses as a function of DENV infection history and associated with expression of protective HLA alleles.
Expanded CD4 T-Cell Populations in DENV-Exposed Individuals Are Associated with a Cytotoxic Phenotype.
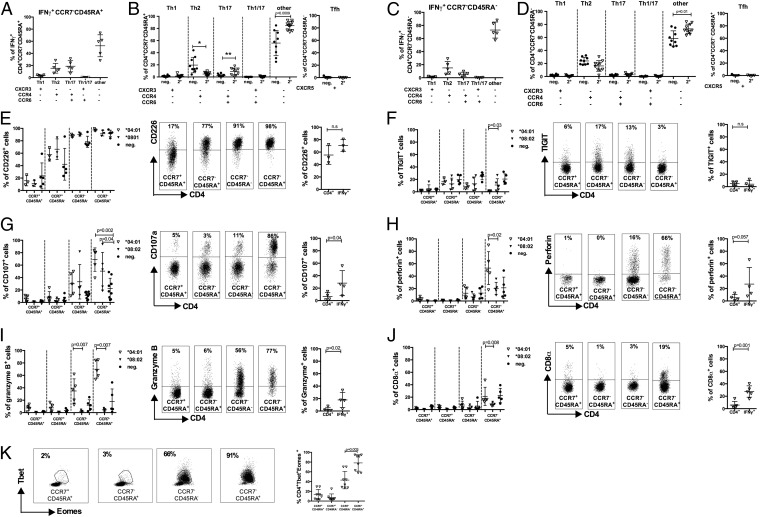
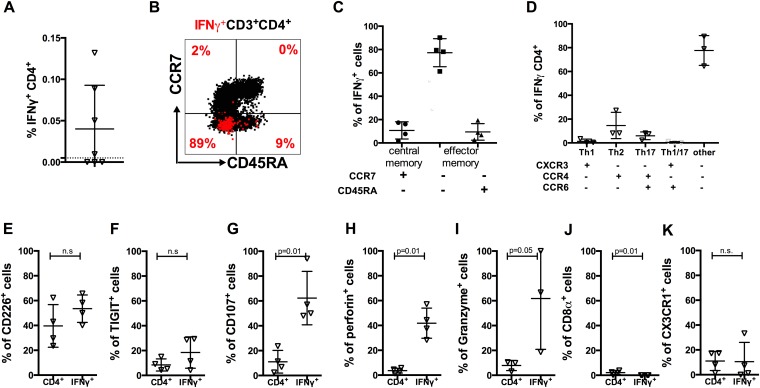
Given the marked increase in CCR7−CD45RA+ CD4 T-cell frequencies in dengue 2° subjects, and particularly the association of DENV-specific IFN-γ+CCR7−CD45RA+ CD4 T cells with protective alleles, we sought to further characterize the biology and function of these cells. The function of effector and memory T cells is dependent on the capacity to migrate to sites of antigen encounter. Chemokine receptors have been particularly useful for dissecting T-cell subsets with distinct migratory capacity and effector function (26). Accordingly, in the next series of experiments, we characterized DENV-specific T cells for the expression of chemokine receptors that are commonly used to define functional subsets of T helper (Th) cells, defined as CXCR3+CCR6− (Th1), CCR4+CCR6− (Th2), CXCR3+CCR6+ (Th1/17), CCR4+CCR6+ (Th17), and CXCR5+ (Tfh) cells, respectively (26, 27). Surprisingly, the majority of IFN-γ–producing cell were not found in any of the conventional Th subsets. In the case of CCR7−CD45RA+cells, >50% of the DENV-specific IFN-γ T cells were negative for CCR6, CCR4, and CXCR3 (Fig. 3_A_). When we examined the expression of CCR6, CCR4, and CXCR3 of total CD4+ T cells from DENV-seronegative and in samples from individuals with secondary DENV infection, a highly significant expansion of CCR6−, CCR4−, and CXCR3− cells was detected (P = 0.0009; Fig. 3_B_). These results were essentially mirrored in the case of CCR7−CD45RA− cells (Fig. 3 C and D).
Fig. 3.
Expanded CD4 T-cell populations in DENV-exposed individuals are associated with a cytotoxic phenotype. (A_–_D) PBMC were stimulated with HLA-matched peptides for 6 h, and the IFN-γ responses were measured by ICS. CD4 effector memory cells (CCR7−CD45RA+ and CCR7−CD45RA−) were defined as Th1 (CXCR3+CCR4−CCR6−), Th2 (CXCR3−CCR4+CCR6−), Th17 (CXCR3−CCR4+CCR6+), Th1/17 (CXCR3+CCR4−CCR6+), and Tfh (CXCR5+) subsets, according to the expression of these surface markers. The chemokine receptor expression for IFN-γ–producing CCR7−CD45RA+ (A) and CCR7−CD45RA− (C) effector memory cells is shown (n = 5). The distribution of CD4+ Th subsets in DENV-negative (filled circles; n = 9) and donors experiencing secondary infection with DENV (2°; open triangles; n = 10) for the effector memory subsets CCR7−CD45RA+ (B) and CCR7−CD45RA− (D) is shown (n = 10). (E_–_J) PBMCs from donors who were found to be dengue-negative (neg.; filled circles) or having experienced a secondary infection (2°; DENV infection; triangles) were stained for memory markers, IFN-γ production, and markers for effector function. Shown are expression of the specific marker in CD4 T-cell subsets (Left), representative FACS plots (Center), and the expression in DENV-specific IFN-γ–producing CD4 T cells (Right). (E) CD226 expression was measured and compared among DRB1*04:01 secondary donors (n = 5), DRB1*08:02 secondary donors (n = 3), and DENV-negative donors (n = 5). Expression was compared between naïve cells and memory subsets, as well as between bulk CD4+ and IFN-γ–producing CD4+ T cells. Similar analysis was carried out for TIGIT (F), CD107a (G), perforin (H), granzyme B (I), and CD8α (J). (K) Transcription factors Tbet and Eomes were stained, and their joint expression was compared among CD4+ subsets, with representative plots shown. Error bars represent mean ± SEM. Statistical significance was determined by using the two-tailed Mann–Whitney test.
Because we could not associate the majority of DENV-specific CD4+ T cells with common Th subsets, we hypothesized that they might be associated with cytotoxic function, as has been described in a murine system (28, 29). Accordingly, we further characterized the expression of various phenotypic markers associated with cytotoxic T-cell function. Each marker was evaluated by flow cytometry, comparing donors with secondary DENV infection expressing either DRB1*0401 or DRB1*0801 alleles. DENV-negative donors were used as a control (Fig. 3 E_–_K). Each panel shows expression of the specific marker in CD4 T-cell subsets as well as in DENV-specific IFN-γ–producing CD4 T cells.
T cells with cytotoxic function have been previously characterized to express a number of cell-surface and intracellular markers including high CD107, CD226, Perforin, and low TIGIT. As shown in Fig. 3_E_, CD226 was highly expressed in the CCR7− effector memory subsets. Conversely, TIGIT expression was correspondingly low, being most pronounced in the case of secondary infection and the protective DRB1*0401 allele (Fig. 3_F_; P = 0.03). The degranulation marker CD107 was also significantly up-regulated in donors that had experienced secondary infection with DENV (P = 0.002 for *0401 and P = 0.04 for *0802, respectively; Fig. 4_G_) and specifically up-regulated in DENV-specific IFN-γ–producing cells compared with total CD4+ T cells (P = 0.04).
Fig. 4.
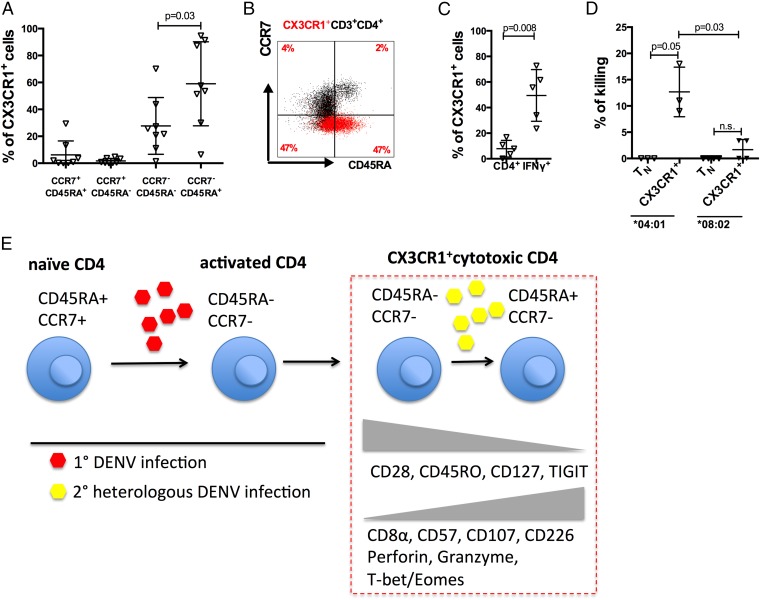
DENV-specific CD4+ T cells express CX3CR1 and mediate direct cytotoxic activity. (A) The relative expression of CX3CR1 was measured in the four T cells subsets in DRB1*0401 donors with a history of secondary DENV infection donors (n = 8). (B) A representative dot plot of total CD4+ (black dots) is overlaid with CX3CR1 expression (red dots). (C) CX3CR1 expression in total CD4+ T cells and DENV-specific IFN-γ–producing cells is shown (n = 5). (D) CX3CR1+ and naïve CD4+ T cells were sorted and cultured with peptide pulsed APCs overnight at 37 °C. Cells were then harvested, stained for HLA-DR, and analyzed by flow cytometry. The difference in absolute numbers of HLA-DR expressing APCs cocultured with either naïve CD4+ T cells or CXCR3+ cells was then expressed as percentage of killing (n = 3). Error bars represent mean ± SEM. Statistical significance was measured by using a two-tailed Mann–Whitney test. (E) Proposed model of cytotoxic CX3CR1+ T-cell differentiation. Naïve CD4+ T cells (CD45RA+CCR7+) get activated during primary infection with DENV (red hexagons) and differentiated into effector T cells (CD45RA−CCR7−). Upon continuous reexposure to heterologous DENV infections (yellow hexagons), effector memory T cells down-regulate CD28, CD45RO, CD127, and TIGIT. CD8α, CD57, CD107, and CD226, as well as perforin, granzyme B and the transcription factors T-bet and Eomes are up-regulated in highly differentiated effector memory T cells (CD45RA+CCR7−).
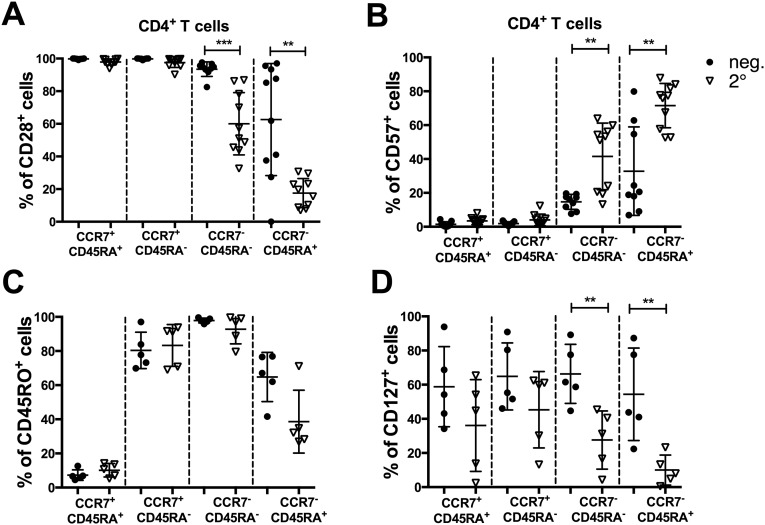
Perforin and granzyme B expression was also selectively enriched in CCR7−CD45RA+ CD4+ T cells in donors expressing the protective DRB1*0401 allele, compared with those expressing the susceptible DRB1*0801 allele (P = 0.02 and 0.007 for perforin and granzyme B, respectively; Fig. 3 H and I). Furthermore, granzyme B was also selectively up-regulated in the DENV-specific T cells, as identified by DENV-specific IFN-γ secretion (P = 0.02). Finally, it has been shown that highly differentiated CD4 cytotoxic T cells often coexpress CD8α (28). Accordingly, we tested for expression of this marker. As shown in Fig. 3_J_, up to 40% of the cells of CCR7−CD45RA+ express CD8α in donors that express DRB1*0401. Furthermore, CD8α was also up-regulated in DENV-specific cells compared with total CD4+ T cells (P = 0.001). Further characterization of these subsets in DENV-negative and -positive donors revealed a highly differentiated phenotype evidenced by down-regulation of CD28, CD45RO, and CD127, whereas CD57 expression was high (Fig. S2).
Fig. S2.
Further phenotypic characterization of CD4+ T-cell subsets. PBMCs from donors seronegative for DENV (neg; filled circles) and donors with neutralizing Ab patterns characteristic of multiple DENV infections (2°; open triangles) were stained with mAbs and analyzed by flow cytometry. CD4+ T cells were grouped according their expression of CCR7 and CD45RA. Expression levels for CD28-positive (A), CD57-positive (B), CD45RO-positive (C), and CD127-positive (D) cells in each subset were plotted. Statistical significance was measured by using the Mann–Whitney test. **P ≤ 0.01; ***P ≤ 0.001.
Because the T-box transcription factors T-bet and Eomesodermin (Eomes) are known to induce multiple cytolytic functions in CD4+ T cells, we next examined coexpression of these factors within CD4+ T-cell subsets (30, 31). Both transcription factors were coexpressed significantly higher in CCR7−CD45RA+ cells (P = 0.003; Fig. 3_K_) further suggesting the cytotoxic phenotype CD4 T-cell differentiation programming indicated by the other markers. These phenotypic traits of the DENV-specific CD4 T cells are largely in accordance with those of cytolytic CD4+ T cells found in latent infection with human cytomegalovirus (CMV). To compare the cytotoxic profiles we have tested multiple DRB1*0401 donors with a peptide pool containing CMV-specific peptides. CMV-specific IFN-γ+ responses have been detected in four of seven donors tested. The phenotype and cytotoxic profiles in these four donors have subsequently been analyzed (Fig. S3_A_). As expected, we saw the majority of CMV-specific responses originate from the effector memory subsets (Fig. S3 B and C). As seen with dengue specific cells also the majority of CMV-specific cells did not express the chemokine markers CXCR3, CCR4, or CCR6 (Fig. S3_D_). CMV-specific cells further expressed significantly more CD107, perforin, and granzyme compared with the total CD4 T cells. No difference has been observed in the expression of CD226 and TIGIT. In contrast to DENV-specific cells, CMV-specific cells did not significantly up-regulate CX3CR1, and IFN-γ+ CD4 cells coexpressed significantly less CD8α (Fig. S3 E_–_K).
Fig. S3.
Phenotypic characterization of CMV-specific CD4+ T-cell subsets. (A) IFN-γ production upon peptide stimulation with CMV-specific peptides was measured for donors expressing DRB1*0401 alleles (n = 7). (B) Representative staining of the memory CD4+ T cells subsets and IFN-γ–producing cells donor is shown. Memory subsets were defined as naïve T cells (CCR7+CD45RA+), central memory T cells (CCR7+CD45RA+), and the two effector memory subsets (CCR7−CD45RA− and CCR7−CD45RA+), according to their expression of these markers. (C and D) The relative distribution of IFN-γ response between the memory T cells subsets (C) and subsets defined by chemokine receptor expression is shown (D). (E_–_K) Expression of CD226 (E), TIGIT (F), CD107a (G), perforin (H), granzyme B (I), CD8α (J), and CX3CR1 (K) was compared between bulk CD4+ and IFN-γ–producing CD4+ T cells. Error bars represent mean ± SEM. Statistical significance was determined by using the one-tailed Mann–Whitney test.
DENV-Specific CD4+ T Cells Express CX3CR1 and Mediate Direct Cytotoxic Activity.
Given that the DENV-specific CD4 T cells did not express any of the CD4 T-cell chemokine markers from our conventional panel, we considered whether other receptors might be expressed. The CX3CR1 chemokine receptor has been implicated in the trafficking and adhesion of lymphocytes and lymphocyte survival and maintenance in inflamed tissues (32). We measured the expression of this chemokine receptor in secondary DENV infection samples. The fraction of CX3CR1+ effector memory cells was highest in CCR7−CD45RA+ cells (Fig. 4_A_). Similarly, whereas only ∼10% of total CD4+ T cells expressed CX3CR1, almost 50% of the DENV-specific IFN-γ–producing cells were positive for the expression of this chemokine receptor (P = 0.008; Fig. 4_C_).
We next set out to demonstrate that the DENV-specific HLA class II restricted CD4+ T cells are not only expressing a pattern of markers associated with cytotoxicity, but that they can indeed mediate cell–cell killing. Carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled target cells were pulsed with the pool of DENV epitopes, and the number of DRhi target cells recovered after overnight incubation with effector cells was measured. As shown in Fig. 4_D_, significant killing was observed when sorted CX3CR1+ T cells from DENV DRB1*0401 donors were used, with 12% (range 9–18%) killing observed at a 5:1 effector:target ratio. Data from DRB1*0802 individuals, showing a lower activity with these cells, further helps support the potential implication of a cytolytic mechanism (Fig. 4_D_). In summary, we demonstrate that dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells that displays an Eomes+ Tbet+ perforin+ granzyme B+ CD45RA+ CD4 CTL phenotype (Fig. 4_E_).
Discussion
These studies describe a human CD4+ T-cell subset than can directly function as effector cells by executing cytotoxicity in a peptide-specific and MHC class II-restricted manner. This subset is specifically expanded in donors with a history of DENV infection and, in particular, in those donors carrying an HLA allele associated with protection from severe DENV disease. Subpopulations of cytotoxic CD4+ T cells (CD4–CTL) associated with executing cytotoxic effector functions have been described, especially under conditions of chronic viral infections and antitumor reactivity (33–36). The functional and phenotypic traits of the DENV-specific CD4 T cells are largely in accordance with those of cytolytic CD4+ T cells found in latent infection with human CMV (HCMV) and HIV (37, 38). The fact that the phenotype of ex vivo cytotoxic CD4+ activity after exposure with an acute virus is similar to CD4 T cells in chronic infection points to general validity of this concept. In fact, T cells with granzyme B- and perforin-containing granules can be found at low frequencies in the circulation of most healthy individuals (37). This unexpected plasticity of CD4+ T cells results in the postthymic termination of the Th lineage fate and the functional differentiation of distinct MHC class II-restricted CD4+ CTL. The development of highly differentiated cytotoxic CD4 T cells appears to require multiple encounters with cognate antigen. Infection with one DENV serotype affords lifelong immunity against reinfection with the same serotype. However, the existence of four different serotypes allows for multiple encounters with conserved antigens between the serotypes. Accordingly, we demonstrate that we see a more distinct expansion of this cytotoxic CD4–CTL subset in donors experiencing multiple infections with different serotypes. This finding is further supported by the notion that in vitro stimulation of T-cell clones specific for DENV can result in the generation of cytotoxic CD4 T-cell clones (39–42).
Multiple organ system, such as the liver, and endothelial cell (EC) linings of blood vessels have been suggested to play an important role in the pathogenesis of DENV (43). The ligand for CX3CR1, CX3CL1/fractalkine, is a chemokine synthesized in ECs as a membrane protein. Membrane-bound fractalkine works as an adhesion molecule for these leukocytes, and the secreted form as a chemotactic factor (44). CX3CR1 expression allows migration from the bloodstream to peripheral tissue (32). This finding could potentially provide a mechanism for how DENV-specific CD4+ T cells are able to enter and remain in inflamed and infected tissue throughout the body to exert their prolonged effector functions.
MHC class II molecules are found on antigen-presenting cells (APCs) such as dendritic cells, phagocytes, ECs, and B cells. Antigens presented by MHC class II molecules are typically derived from extracellular proteins. However, it has been shown that autophagy promotes MHC class II presentation of peptides from intracellular source proteins (45). Interestingly, one of the mechanisms dengue virus uses to hijack host cell machinery to facilitate viral replication is by inducing autophagy, which is necessary for virus maturation and production of infectious virions (46, 47). A recent study has further provided evidence of a definitive link between Ab-enhanced DENV infection and autophagosome formation (48). Cytotoxic CD4+ T cells may contribute to protection by eliminating these DENV-infected, MHC II-expressing cells.
In summary, we present the characterization of defined HLA-restricted CD4+ T-cell responses resulting from natural infection with DENV in individuals from Sri Lanka where DENV is hyperepidemic. This subset is particularly expanded in donors carrying an allele associated with protection from severe DENV disease. We are aware that the evidence that these cells are protective is indirect because it is based on the association with a protective allele. Knowledge of the HLA alleles associated with increased susceptibility to severe disease has the potential to serve as a prognostic tool, allowing identification of individuals at increased risk for severe disease. A detailed characterization of the DENV-specific T-cell response will further contribute to the evaluation of vaccine candidates and may contribute to the definition of correlates of protection from severe disease.
Materials and Methods
Ethics Statement.
Blood samples were obtained from healthy adult blood donors from the National Blood Center, Ministry of Health, Colombo, Sri Lanka, in an anonymous fashion. Samples obtained were discarded buffy coats from routine blood donations at the National Blood Center and thus exempt from human subject review. The institutional review boards of both La Jolla Institute for Allergy and Immunology (LIAI) and the Medical Faculty, University of Colombo [serving as the NIH-approved Institutional Review Board (IRB) for Genetech] approved all protocols described in this study.
Human Blood Samples.
The 250 peripheral blood samples were obtained from healthy adult blood donors from the National Blood Center. Donors were of both sexes and between 18 and 60 y of age. Blood processing and HLA typing of both study populations were performed as described (18). PBMCs of donors were picked according their expression of one of the HLA alleles studied. The institutional review boards of both LIAI and the Medical Faculty, University of Colombo (serving as NIH-approved IRB for Genetech) approved all protocols described in this study.
Serology.
DENV seropositivity was determined by dengue IgG ELISA as described (49). Flow cytometry-based neutralization assays were performed for further characterization of seropositve donors, as described (50). Neutralization assays determined that the majority of donors have experienced infection with more than one serotype, further referred to as secondary infections. Donors that showed neutralization titers to only one of the serotypes were considered primary infections.
MHC Class II Binding Predictions and Peptide Selection.
To analyze DENV-specific HLA-restricted CD4 responses, we selected a set of DENV peptides predicted to bind HLA DRB1*0701, chosen because of its high prevalence in Sri Lanka (33% phenotypic frequency). Two additional alleles were selected because they are respectively associated with increased resistance (DRB1*0401) or susceptibility (DRB1*0802) to severe DENV clinical outcomes (9). The 15-mer peptides from all serotypes were predicted for their binding affinity to the selected MHC class II molecules. Epitope predictions for class II were performed for DENV1, 2, 3, and 4 for all isolates in the database by using the consensus prediction methods publicly available through the Immune Epitope Database and Analysis Resource (IEDB; www.iedb.org) (51, 52). For each allele, any peptide predicted to bind with high affinity (2% consensus threshold) and that was present in 30% or more of the isolates was synthesized. This approach resulted in the synthesis of 148 DRB1*0701, 132 DRB1*0401, and 142 DRB1*0802 peptides (Mimotopes). For screening studies, the class II peptides were combined into pools of ∼20 individual peptides, according to their predicted HLA restriction, resulting in seven pools per HLA allele. CMV-specific MHC class II peptides were selected through the IEDB.
MHC Purification and Binding Assays.
Purification of HLA class II MHC molecules by affinity chromatography and the performance of assays based on the inhibition of binding of a high-affinity radiolabeled peptide to quantitatively measure peptide binding were performed essentially as detailed elsewhere (53). Briefly, EBV-transformed homozygous cell lines were used as sources of MHC molecules. A high-affinity radiolabeled peptide (0.1–1 nM) was coincubated at room temperature or 37 °C with purified MHC in the presence of a mixture of protease inhibitors. After a 2-d incubation, MHC bound radioactivity was determined by capturing MHC/peptide complexes on Ab-coated Lumitrac 600 plates (Greiner Bio-one) and measuring bound cpm using the TopCount (Packard Instrument Co.) microscintillation counter. The concentration of peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated (IC50). As a positive control, the unlabeled version of the radiolabeled probe was also tested in each experiment.
In Vitro Expansion of DENV-Specific T Cells.
PBMCs were thawed, and CD4+ T cells were isolated by magnetic bead negative selection. CD45RA+ and CD45RA− cells were subsequently isolated by magnetic bead selection and cultured separately in RPMI 1640 (Omega Scientific) supplemented with 5% (vol/vol) human serum (Cellgro) at a density of 2 × 106 cells per mL in 24-well plates (BD Biosciences). Cells were cocultured with autologous APCs at a density of 1× 106 cells per mL and stimulated with DENV-specific pools (averaging 20 peptides per pool). Cells were kept at 37 °C in 5% CO2, and additional IL-2 (10U/mL; eBioscience) was added 4, 7, and 11 d after initial antigenic stimulation. On day 14, cells were harvested and screened for reactivity against individual DENV-specific peptides.
IFN-γ ELISPOT Assay.
After 14 d of in vitro expansion, 5 × 104 PBMCs were incubated in triplicate with 0.1 mL of complete RPMI 1640 in the presence of HLA-matched peptide pools and individual peptides (2 μg/mL). After a 20-h incubation at 37 °C, the cells were incubated with biotinylated IFN-γ mAb (mAb 7-B6-1 Mabtech) for 2 h and developed as described (18).
Flow Cytometry.
Detailed information of all mAbs used in this study is listed in Table S1. For ICS, PBMCs were cultured in the presence of HLA-matched peptide pools (10 μg/mL) and GolgiPlug containing brefeldin A (BD Biosciences) for 6 h and subsequently permeabilized, stained, and analyzed as described (18). Staining for intracellular nuclear proteins was performed by using the Foxp3/Transcription Factor Staining Buffer Set according to the manufacturer’s instructions (eBioscience).
Table S1.
mAbs used in this study
| Target | Color | Clone | Volume,* μL | Company |
|---|---|---|---|---|
| CD8 | V500 | RPA-T8 | 3 | BD Horizon |
| CD3 | Alexa Flour 700 | UCHT1 | 1 | BD Pharmingen |
| CCR4 (CD194) | PE-Cy7 | 1G1 | 3 | BD Pharmingen |
| CCR6 (CD196) | Biotin | 11A9 | 3 | BD Pharmingen |
| CXCR3 (CD183) | APC | 1C6/CXCR3 | 10 | BD Pharmingen |
| CD226 | FITC | DX11 | 5 | BD Pharmingen |
| CCR7 (CD197) | PerCP/Cy5.5 | G043H7 | 4 | Biolegend |
| Streptavidin | BV 605 | B175472 | 1 | Biolegend |
| CXCR5 (CD185) | FITC | J252D4 | 4 | Biolegend |
| CD28 | PE | CD28.2 | 1 | Biolegend |
| CX3CR1 | PE | 2A9-1 | 1 | Biolegend |
| CD4 | APC eflour 780 | RPA-T4 | 1 | eBioscience |
| CD45RA | eFlour 450 | HI100 | 2 | eBioscience |
| IFN-γ | FITC | 4S.B3 | 5 | eBioscience |
| CD107a | PE | eBioH4A3 | 5 | eBioscience |
| PD-1 (CD279) | PE | eBioJ105 | 1 | eBioscience |
| TIGIT | PE | MBSA43 | 5 | eBioscience |
| CD57 | PE | TB01 | 1 | eBioscience |
| EOMES | FITC | WD1928 | 5 | eBioscience |
| Tbet | PE | eBio4B10 | 5 | eBioscience |
| Perforin | FITC | dG9 | 1 | eBioscience |
| Granzyme B | PE | GB11 | 1 | eBioscience |
| HLA-DR | PE-Cy7 | L243 | 1 | eBioscience |
In Vitro Cytotoxicity Assay.
For the cytotoxicity assay, cells were surface-stained and sorted with the FACSAria III (BD Biosciences). Effector memory CD4 T cells (CD3+, CD4+, CCR7−, and CX3CR1+) and naïve CD4 T cells (CD3+, CD4+, CCR7+, and CD45RA+) were sorted as the effector cells and APCs (CD3-) sorted as targets. APCs were then stained with CFSE (Life Technologies) at a concentration of 5 μM for 20 min at room temperature and pulsed with peptide (10 μg/mL) for 60 min at 37 °C. The APCs and effectors were cocultured overnight at 37 °C. Cells were then harvested, stained for HLA-DR, and analyzed by flow cytometry.
Acknowledgments
We thank the National Blood Center, Ministry of Health, Colombo, Sri Lanka for providing buffy coat samples used in this study and the staff of Genetech Research Institute for processing the samples in a timely manner followed by storage and shipment to LIAI. This work was supported by National Institutes of Health Contracts Nr. HHSN272200900042C and HHSN27220140045C (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Alves MJ, et al. Clinical presentation and laboratory findings for the first autochthonous cases of dengue fever in Madeira Island, Portugal, October 2012. Euro Surveill. 2013;18(6):20398. [PubMed] [Google Scholar]
- 2.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman MG, Harris E. Dengue. Lancet. 2014;385(9966):453–465. doi: 10.1016/S0140-6736(14)60572-9. [DOI] [PubMed] [Google Scholar]
- 4.Halstead SB. Dengue. Lancet. 2007;370(9599):1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 5.Halstead SB. Immune enhancement of viral infection. Prog Allergy. 1982;31:301–364. [PubMed] [Google Scholar]
- 6.Halstead SB, Mahalingam S, Marovich MA, Ubol S, Mosser DM. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: Disease regulation by immune complexes. Lancet Infect Dis. 2010;10(10):712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 8.Zellweger RM, Prestwood TR, Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 2010;7(2):128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malavige GN, et al. HLA class I and class II associations in dengue viral infections in a Sri Lankan population. PLoS One. 2011;6(6):e20581. doi: 10.1371/journal.pone.0020581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens HA, et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens. 2002;60(4):309–318. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- 11.Loke H, et al. Strong HLA class I-restricted T cell responses in dengue hemorrhagic fever: A double-edged sword? J Infect Dis. 2001;184(11):1369–1373. doi: 10.1086/324320. [DOI] [PubMed] [Google Scholar]
- 12.Appanna R, Ponnampalavanar S, Lum Chai See L, Sekaran SD. Susceptible and protective HLA class 1 alleles against dengue fever and dengue hemorrhagic fever patients in a Malaysian population. PLoS One. 2010;5(9):e13029. doi: 10.1371/journal.pone.0013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen TP, et al. Protective and enhancing HLA alleles, HLA-DRB1*0901 and HLA-A*24, for severe forms of dengue virus infection, dengue hemorrhagic fever and dengue shock syndrome. PLoS Negl Trop Dis. 2008;2(10):e304. doi: 10.1371/journal.pntd.0000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falcón-Lezama JA, et al. HLA class I and II polymorphisms in Mexican Mestizo patients with dengue fever. Acta Trop. 2009;112(2):193–197. doi: 10.1016/j.actatropica.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 15.Sierra B, et al. HLA-A, -B, -C, and -DRB1 allele frequencies in Cuban individuals with antecedents of dengue 2 disease: Advantages of the Cuban population for HLA studies of dengue virus infection. Hum Immunol. 2007;68(6):531–540. doi: 10.1016/j.humimm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Greenbaum J, et al. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63(6):325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50(3-4):201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 18.Weiskopf D, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci USA. 2013;110(22):E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alagarasu K, et al. Association of HLA-DRB1 and TNF genotypes with dengue hemorrhagic fever. Hum Immunol. 2013;74(5):610–617. doi: 10.1016/j.humimm.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 20.Brown MG, Salas RA, Vickers IE, Heslop OD, Smikle MF. Dengue HLA associations in Jamaicans. West Indian Med J. 2011;60(2):126–131. [PubMed] [Google Scholar]
- 21.Cardozo DM, et al. Evidence of HLA-DQB1 contribution to susceptibility of dengue serotype 3 in dengue patients in Southern Brazil. J Trop Med. 2014;2014:968262. doi: 10.1155/2014/968262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaFleur C, et al. HLA-DR antigen frequencies in Mexican patients with dengue virus infection: HLA-DR4 as a possible genetic resistance factor for dengue hemorrhagic fever. Hum Immunol. 2002;63(11):1039–1044. doi: 10.1016/s0198-8859(02)00682-1. [DOI] [PubMed] [Google Scholar]
- 23.Southwood S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160(7):3363–3373. [PubMed] [Google Scholar]
- 24.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 25.Harari A, Vallelian F, Pantaleo G. Phenotypic heterogeneity of antigen-specific CD4 T cells under different conditions of antigen persistence and antigen load. Eur J Immunol. 2004;34(12):3525–3533. doi: 10.1002/eji.200425324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sallusto F, Lanzavecchia A. Heterogeneity of CD4+ memory T cells: Functional modules for tailored immunity. Eur J Immunol. 2009;39(8):2076–2082. doi: 10.1002/eji.200939722. [DOI] [PubMed] [Google Scholar]
- 27.Locci M, et al. International AIDS Vaccine Initiative Protocol C Principal Investigators Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheroutre H, Husain MM. CD4 CTL: Living up to the challenge. Semin Immunol. 2013;25(4):273–281. doi: 10.1016/j.smim.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mucida D, et al. Transcriptional reprogramming of mature CD4⁺ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat Immunol. 2013;14(3):281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6(12):1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 31.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302(5647):1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 32.Imai T, et al. Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell. 1997;91(4):521–530. doi: 10.1016/s0092-8674(00)80438-9. [DOI] [PubMed] [Google Scholar]
- 33.Marshall NB, Swain SL. Cytotoxic CD4 T cells in antiviral immunity. J Biomed Biotechnol. 2011;2011:954602. doi: 10.1155/2011/954602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quezada SA, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med. 2010;207(3):637–650. doi: 10.1084/jem.20091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van de Berg PJ, van Leeuwen EM, ten Berge IJ, van Lier R. Cytotoxic human CD4(+) T cells. Curr Opin Immunol. 2008;20(3):339–343. doi: 10.1016/j.coi.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 36.van Leeuwen EM, et al. Emergence of a CD4+CD28- granzyme B+, cytomegalovirus-specific T cell subset after recovery of primary cytomegalovirus infection. J Immunol. 2004;173(3):1834–1841. doi: 10.4049/jimmunol.173.3.1834. [DOI] [PubMed] [Google Scholar]
- 37.Appay V, et al. Characterization of CD4(+) CTLs ex vivo. J Immunol. 2002;168(11):5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 38.Suni MA, et al. CD4(+)CD8(dim) T lymphocytes exhibit enhanced cytokine expression, proliferation and cytotoxic activity in response to HCMV and HIV-1 antigens. Eur J Immunol. 2001;31(8):2512–2520. doi: 10.1002/1521-4141(200108)31:8<2512::aid-immu2512>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 39.Kurane I, et al. Flavivirus-cross-reactive, HLA-DR15-restricted epitope on NS3 recognized by human CD4+ CD8- cytotoxic T lymphocyte clones. J Gen Virol. 1995;76(Pt 9):2243–2249. doi: 10.1099/0022-1317-76-9-2243. [DOI] [PubMed] [Google Scholar]
- 40.Kurane I, Zeng L, Brinton MA, Ennis FA. Definition of an epitope on NS3 recognized by human CD4+ cytotoxic T lymphocyte clones cross-reactive for dengue virus types 2, 3, and 4. Virology. 1998;240(2):169–174. doi: 10.1006/viro.1997.8925. [DOI] [PubMed] [Google Scholar]
- 41.Mathew A, et al. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J Virol. 1998;72(5):3999–4004. doi: 10.1128/jvi.72.5.3999-4004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng L, Kurane I, Okamoto Y, Ennis FA, Brinton MA. Identification of amino acids involved in recognition by dengue virus NS3-specific, HLA-DR15-restricted cytotoxic CD4+ T-cell clones. J Virol. 1996;70(5):3108–3117. doi: 10.1128/jvi.70.5.3108-3117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martina BE, Koraka P, Osterhaus AD. Dengue virus pathogenesis: An integrated view. Clin Microbiol Rev. 2009;22(4):564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imaizumi T, Yoshida H, Satoh K. Regulation of CX3CL1/fractalkine expression in endothelial cells. J Atheroscler Thromb. 2004;11(1):15–21. doi: 10.5551/jat.11.15. [DOI] [PubMed] [Google Scholar]
- 45.Dengjel J, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102(22):7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8(5):422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mateo R, et al. Inhibition of cellular autophagy deranges dengue virion maturation. J Virol. 2013;87(3):1312–1321. doi: 10.1128/JVI.02177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang YT, et al. Autophagy facilitates antibody-enhanced dengue virus infection in human pre-basophil/mast cells. PLoS One. 2014;9(10):e110655. doi: 10.1371/journal.pone.0110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanakaratne N, et al. Severe dengue epidemics in Sri Lanka, 2003-2006. Emerg Infect Dis. 2009;15(2):192–199. doi: 10.3201/eid1502.080926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraus AA, Messer W, Haymore LB, de Silva AM. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol. 2007;45(11):3777–3780. doi: 10.1128/JCM.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang P, et al. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLOS Comput Biol. 2008;4(4):e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang P, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sidney J, et al. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr Protoc Immunol. 2013;Chapter 18:Unit 18.3. doi: 10.1002/0471142735.im1803s100. [DOI] [PMC free article] [PubMed] [Google Scholar]