Individuality and variation of personal regulomes in primary human T cells (original) (raw)
. Author manuscript; available in PMC: 2016 Jul 29.
Abstract
Here we survey variation and dynamics of active regulatory elements genome-wide using longitudinal samples from human individuals. We applied Assay of Transposase Accessible Chromatin with sequencing (ATAC-seq) to map chromatin accessibility in primary CD4+ T cells isolated from standard blood draws of 12 healthy volunteers over time, from cancer patients, and during T cell activation. Over 4,000 predicted regulatory elements (7.2%) showed reproducible variation in accessibility between individuals. Gender was the most significant attributable source of variation. ATAC-seq revealed previously undescribed elements that escape X chromosome inactivation and predicted gender-specific gene regulatory networks across autosomes, which coordinately affect genes with immune function. Noisy regulatory elements with personal variation in accessibility are significantly enriched for autoimmune disease loci. Over one third of regulome variation lacked genetic variation in cis, suggesting contributions from environmental or epigenetic factors. These results refine concepts of human individuality and provide a foundational reference for comparing disease-associated regulomes.
INTRODUCTION
Understanding the basis of individual variation is a central goal in genetics and epigenetics. The advent of global gene expression and chromatin mapping technologies has greatly increased our understanding of gene regulatory mechanisms (Degner et al., 2012; Kasowski et al., 2013; McVicker et al., 2013; Vernot et al., 2012; Whitney et al., 2003). However, prior methods often required tens of millions of cells. Investigators were forced to expand cells through artificial means such as immortalization or extensive ex vivo expansion— manipulations that can significantly alter the regulatory landscape. Hence, prior studies have focused on the impact of inherited genetic variation on gene expression or chromatin states (Degner et al., 2012; Kasowski et al., 2013; McVicker et al., 2013; Vernot et al., 2012), but the fidelity and variation of the human gene regulatory landscape in vivo are surprisingly not known.
ATAC-seq is a recently introduced and sensitive method to map open chromatin sites, predicted transcription factor finding, and nucleosome position from as few as 500 cells (Buenrostro et al., 2013; Lara-Astiaso et al., 2014; Lavin et al., 2014), even in single cells (Buenrostro et al., 2015; Cusanovich et al., 2015). Such a comprehensive molecular portrait of predicted gene regulatory events affords a “personal regulome”—a summary of gene regulatory events in a snap shot of time within a single individual. Although ATAC-seq provides a possible approach to interrogate primary human cell types and minute clinical samples, the feasibility and accuracy of large-scale applications have not been demonstrated. Here we generate and analyze 58 high-resolution personal regulomes of a single cell type—human CD4+ T cells—comprised of over 1.7 billion measurements. We develop methods to integrate diverse sources of genomic and epigenomic information to address the regulatory variation as a function of individuality, time, and disease (Figure 1a).
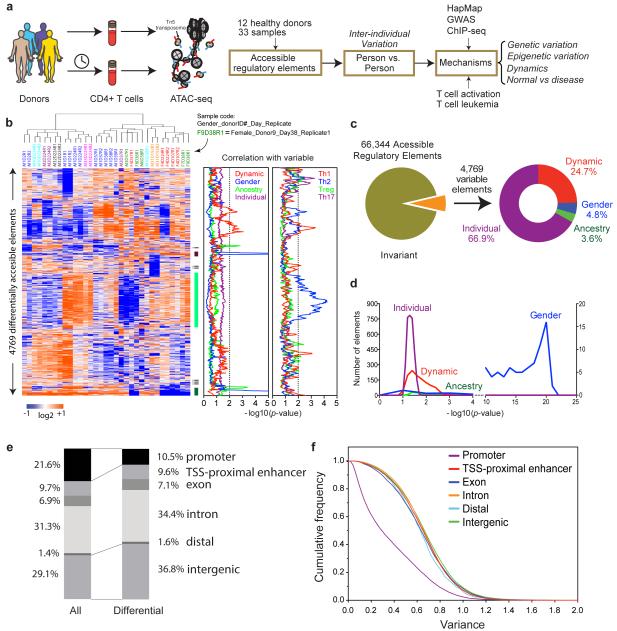
Figure 1. Landscape of individual variation in T cell regulome.
a, Schematic outline of study design.
b, Heatmap of regulatory elements with differential activity. Each column is a sample; each row is an element. Samples and elements are organized by two dimensional hierarchical clustering. Color scale indicates relative ATAC-seq signal as indicated. Top: Samples from the same donor are labeled with the same color, and sample id is coded with gender, donor ID, sample day, and replicate number. E.g. F9D38R1 indicates this sample is female, from donor 9 drawn at day 38 and is the first replicate. Middle: three clusters of differential accessible elements of interest. Right: correlation of cluster activity with demographic variables and T cell subtype signatures is shown in corresponding color. The dotted line indicates _p_=0.01, FDR <0.05. Cluster i and iii are associated with gender; cluster ii is associated with a Th2 signature.
c, Distribution of the variable regulatory element associated with attributable demographic features.
d, Significance of the association of the variable regulatory element activity with demographic features.
e, Distribution of genomic features of all and differential accessible elements, respectively.
f, Scale of variance of genomic features of all elements with accessibility.
RESULTS
Landscape and variation of personal regulomes in CD4+ T cells
We assessed the landscape and variation of chromatin accessibility in human CD4+ T cells in 33 samples provided by 12 healthy donors (Figure 1a). In this exploratory study, we wished to document dominant sources of regulome variation readily evident from small numbers of healthy individuals; other well-selected and larger populations are likely to reveal many other potential contributions to regulome variation. Most donors gave at least two independent samples days to months apart; one donor was sampled six times over seven months (Table S1). From each standard 5mL blood draw, we enriched at least 50,000 CD4+ T cells by negative selection without ex vivo expansion (avoiding potentially activating antibodies in positive selection), and performed ATAC-seq to map the location and accessibility of regulatory elements genome-wide (Supplementary Methods). CD4+ T cells include multiple subsets (including naïve, memory Th1, Th2, Th17, Treg, and Tfh cells); known frequency counts suggest that these major subsets would be sampled by our approach(Maecker et al., 2012). Each library was sequenced to obtain on average more than 30 million paired-end reads. We used ZINBA (Rashid et al., 2011) to identify focal peaks of chromatin accessibility that typify active regulatory elements, and sequence counts within accessibility peaks were subjected to quantile normalization to yield a quantitative portrait of active regulatory elements in each sample. Pearson correlation of replicates and Irreproducibility Discovery Rate (IDR) analysis (Landt et al., 2012) indicate high quality of the data and excellent reproducibility between replicates (Figure S1a-d). Systematic comparison of published ATAC-seq peaks to histone modification and DNase I hypersensitivity sequencing (DHS-seq) confirmed that ATAC-seq highlights active enhancers and promoters (Figure S1e-f).
To identify individual variation in T cell regulomes, we applied intrinsic analysis (Perou et al., 2000), a method that highlighted elements that varied in accessibility across individuals but not between repeat samples from the same individual (Figure 1b). Noisy measurements that vary regardless of donor identity are filtered out by this approach. We also used permutation analysis as further protection against noisy data, using signals in observed but not permuted data to estimate FDR (Supplementary Methods). The selected elements reveal complex but distinctive patterns of activity, such that replicate samples from the same donor at a same time were always clustered together in unsupervised hierarchical clustering (12 of 12 pairs). Correlation of the pattern of regulome activity with known subject variables revealed potential relationships with gender, self-reported ancestry, and other individual variation (p< 0.01, false discovery rate (FDR) <0.05, Figure 1b, c). As CD4+ cells are comprised of several subsets, the individual variation observed may reflect variations in cell subset composition in addition to gene-specific regulation. Comparison of chromatin accessibility data (by DNase I hypersensitivity) from purified T cell subsets, including Th1, Th2, Th17, and Treg cells, suggests that some of the regulome variation may be correlated with cell subsets (Figure 1b, right panel).
CD4+ T cells from healthy donors exhibited 66,344 accessible sites in our survey, inferred to be active regulatory elements (Figure 1c). Approximately 92.8% of these sites showed invariant activity across individuals and time, indicating a high degree of fidelity in the regulatory landscape. Nearly 7.2% of accessible sites (4769 elements) show significant variable activity (Z-score>1, FDR<0.1, and fold change>5). 24.7% of these elements varied in accessibility in the same individual over time, and hence may reflect dynamic regulation of the immune system (Figure 1c). The remainder elements showed stable inter-individual differences, which can be associated with known variables with different degrees of confidence (Figure 1d). Notably, gender is the most informative variable, being strongly associated (p<10−10) with differential activity of several hundred elements. In all cases, self-reported gender was concordant with biological sex as determined by chromosome complement (see below). In contrast, potentially regulatory impact from ancestry was less significant in this cohort, and differential activity attributable to individual or dynamic differences are weakly but broadly associated with thousands of elements. Many elements currently assigned to the “individual” category are idiosyncratic in their differential activity; many are “private” variations that are observed reproducibly in one donor but not across multiple individuals in our survey. Notably, although some of the dynamic and individual variation correlated with Th1 vs. Th2 signals, gender—the most significant variable—is not appreciably associated with known cell subsets (Figure 1b). The variable elements are enriched for distal regulatory elements and depleted for promoters, suggesting variation in long-range gene regulation (Figure 1e, f).
Gender-specific regulome in T cells
We focused next on gender-specific variation in the T cell regulome, because it emerged as the most significant attributable source of inter-individual variation and because well-studied mechanisms of dosage compensation provided a rich interpretative framework to understand our data. In mammals, males have XY sex chromosomes whereas females have XX. Dosage compensation occurs via random epigenetic silencing of one of the two female X chromosomes, termed X chromosome inactivation (XCI), that is heritable through somatic cell divisions over life (Flynn and Chang, 2014; Lee and Bartolomei, 2013). XCI is controlled by several long noncoding RNAs, including XIST, which is transcribed from and mediates the epigenetic silencing of the inactive X chromosome (Xi). The active X chromosome (Xa) and Xi thus harbor distinct chromatin modifications and gene expression patterns. A subset of X-linked genes escape from XCI in a tissue-specific fashion through poorly understood mechanisms, leading to differential X-linked expression in male vs. female cells.
Among regulatory elements with significant differential activity between male and female cells, a majority (but not all) mapped to the sex chromosomes (Figure 2a, b). As expected, only male samples showed signal for Y chromosome-linked elements, and female samples showed increased activity of X chromosome-linked elements. Gender-specific regulomes may arise from effect of sex-specific hormones, random X inactivation, X-linked escapee genes, or additional differences (Rubtsova et al., 2015). Sex hormone changes may impact all chromosomes, whereas the XCI effects are linked to the X chromosome.
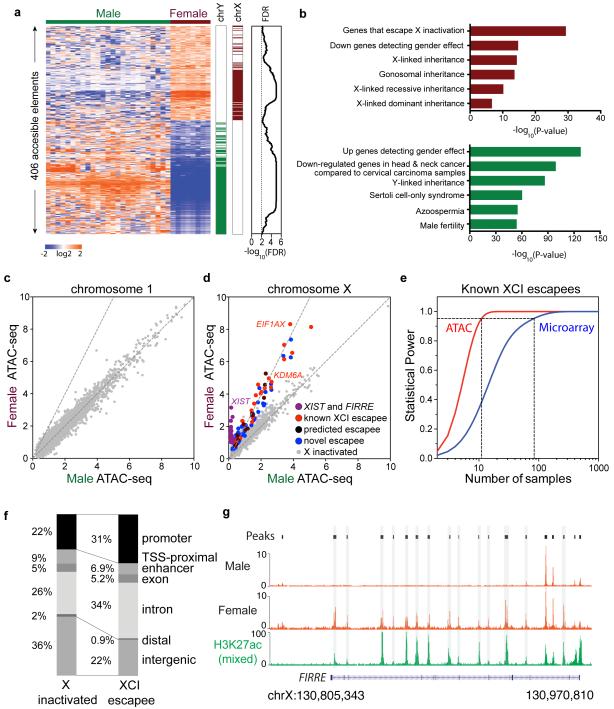
Figure 2. Regulatory variation in sex chromosomes.
a, Association of gender-specific regulatory activity with sex chromosomes. Left: heat map, color scale indicates relative ATAC-seq signal as indicated; Middle: bar graph indicates regulatory elements on chromosome X (dark red) and Y (dark green); Right: For each regulatory element, an FDR of significance estimated from random permutation.
b, Gene Ontology terms enriched in female (dark red) and male (dark green)-enriched regulatory elements.
c,d, Male vs. female ATAC-seq signal across an autosome (chr1) or X chromosome. Dotted lines indicate slopes of 1 and 2 respectively. Regulatory elements of X inactivated and of known, predicted, and novel to escape XCI were color coded as described in the figure. Scales represent average coverage per base. The X-axis and Y-axis indicate the average base coverage.
e, Statistical power of ATAC-seq (red) vs. microarray (blue) to detect known XCI escapee genes is shown. ATAC-seq requires 11 samples of each gender while microarray requires 81 samples of each gender to reach a power of 0.95 (dotted line, _p_=0.01).
f, Distribution of genomic features in elements that are X inactivated (left) or escape XCI (right).
g, Normalized male vs. female specific ATAC-seq signal at gene FIRRE locus. The enhancer associated histone modification H3K27ac in mixed gender samples is shown for comparison.
Comparative analysis of regulatory activity between males and females across autosomes (e.g. chromosome 1) revealed equivalent activity profiles at the chromosomal level, as evidenced by a slope of 1 in the two dimensional plot (Figure 2c). In contrast, comparison of male and females across the X chromosome readily revealed three types of elements: (i) elements that are dosage compensated and thus have equivalent activity between males and females (slope =1, gray dots in Figure 2d), (ii) active elements on the Xi and thus only active in female cells (slope = infinity; purple dots, e.g XIST), and (iii) elements that escape XCI and thus show a two-fold increase in activity in female over male cells (slope= 2; red, blue, and dark violet dots).
We identified 43 elements associated with 17 coding genes (e.g. EIF1AX and KDM6A) and 3 noncoding genes known to escape XCI, as well as elements associated with 7 escapees predicted by others (Zhang et al., 2013) and 12 XCI escapees that to our knowledge have not been reported previously (Figure 2d, Figure S2, Table S2). We analyzed the ImmVar dataset, which measured genome-wide mRNA levels of CD4+ T cells from 163 healthy male and 244 female donors(Ye et al., 2014). Gender-specific gene expression profiles validated 16 out of 17 known XCI genes and 6 of 7 novel XCI escapees predicted by ATAC-seq that had well-measured transcripts (Figures S3a). We note that escapee genes that were not validated tend to have lower ATAC-seq signals and mRNA expression compare to those that were validated (Figure 6), which may be below the detection confidence of array technology.
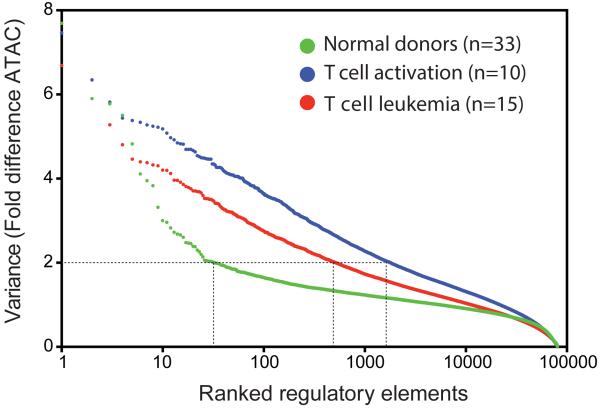
Figure 6. Scale of regulatory variation in as function of individuality versus disease.
Regulome variation in CD4+ T cells derived from normal donors (green), during T cell activation (blue), or in T cell leukemia (red) are ranked and compared. There are 32, 517, and 1800 regulatory elements in normal, T cell leukemia and T cell activation respectively whose variance are greater than 2.
Genes that escape XCI in females offered an opportunity to compare the accuracy and statistical power of ATAC-seq data vs. standard microarray data(Ye et al., 2014) (Figure 2e). Such genes are known to have a 2:1 expression ratio in females vs. males. Whereas ATAC-seq analysis accurately identified the two-fold ratio in female vs. males across a range of gene activity levels (Figure 2d), microarrays under estimated the difference (Figure S4). Using the known XCI escapees that are detected by both ATAC-seq and RNA microarray analysis (_P_-value<10−4, Student t-test) as positive controls, we performed a power analysis to determine the number of samples required to have 95% probability of detecting this true difference with significance level less than 0.01. ATAC-seq requires only 11 samples of each gender while mRNA microarray requires 81 samples of each gender to reach a power of 0.95 (Figure 2e), indicating ATAC-seq is over 7 times more sensitive than microarray. The ability of ATAC-seq to detect rare elements (e.g. Y-linked sequences in males) and accurately quantify one vs. two copies of X-linked activity suggests that our methods are sensitive and precise, and could achieve excellent statistical power with a small sample size.
The comparison of gender-specific regulomes also yielded unexpected insights that are not possible from gene expression measurements. For example, we observed that accessible elements that escape XCI are more likely to be found at promoters and introns of known escapee genes, but not at intergenic distal regions (Figure 2f). Moreover, our analysis revealed evidence of gender-specific regulatory landscapes of XCI escapees (Figure 2g). XCI escape has traditionally been considered simply a failure of Xi silencing; hence it was believed that the regulatory pattern on the Xa will simply be duplicated on the Xi for escapee genes. Indeed, the term “XCI escape” implies this preconceived notion; however, there is no direct evidence to support or refute this model for XCI escape. We identified Xi-specific regulatory elements on XCI escapees, which have signal only in female but not male cells (Figure 2d, purple dots). For instance, FIRRE is a recently described X-linked lncRNA that escapes XCI, and is involved in chromosome topological organization (Hacisuleyman et al., 2014; Yang et al., 2015). RNA in situ hybridization documented two equivalent RNA foci in female cells and one focus in male cells (Hacisuleyman et al., 2014). FIRRE contains a series of putative intronic enhancers embedded throughout its locus, as documented by the enhancer-associated modification histone H3 lysine 27 acetylation (H3K27ac) in a survey of mixed sex cells (Bernstein et al., 2012). Intriguingly, our data suggests that two FIRRE enhancers in intron 2 are active in male cells whereas over a dozen enhancers in introns 2-12 are active only in female cells (Figure 2g). This regulatory divergence implies gender-specific regulation, allele-specific regulation of FIRRE on Xa vs. Xi, or combinations of both strategies.
Extending the concept of allelic regulatory divergence, the selection pressure for dosage compensation is thought to be the maintenance of the expression level of homologous gene pairs on the X and Y-chromosomes (Bellott et al., 2014). Analysis of the regulatory landscape of XY homolog pairs shows that all 15 pairs examined showed divergence on par with or exceeding that of FIRRE alleles, indicating that the regulatory inputs into the sex-linked gene homologs are distinctive as a rule (Figure S5a-c). These results highlight the value of examining dosage compensation from the perspective of the personal regulome.
Gender-specific regulome of autosomes reveal propensity for autoimmunity
The dramatic chromatin accessibility differences between the genders on sex chromosomes prompted us to examine the scope of gender-specific differences on autosomes. Gender-specific regulomes for T cells have special relevance to human disease because of the strong epidemiological evidence indicating an association between female sex and autoimmunity (reviewed by (Rubtsova et al., 2015)). Four out of five patients with any type of autoimmune disease are female, and for some common diseases such as systemic lupus erythematosus, the preponderance of females to males is 9 to 1. It has been long postulated that epigenetic or regulatory differences between male and female immune cells may underlie autoimmune susceptibility (Rubtsova et al., 2015), but gender-specific differences are incompletely understood.
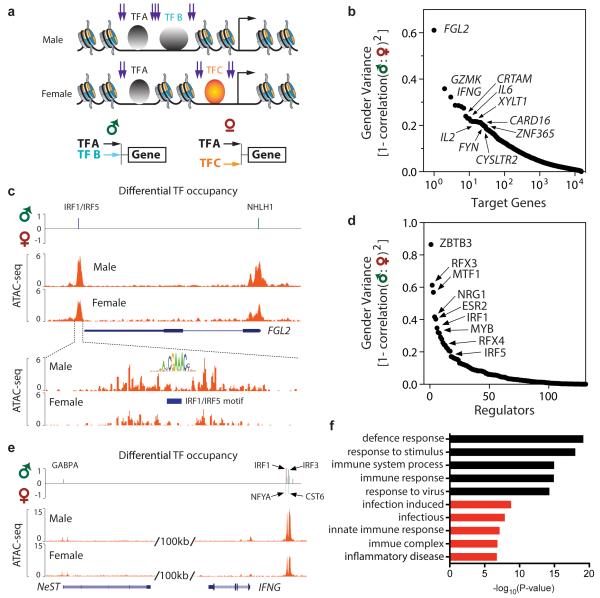
We sought to identify gender-specific differences in gene regulatory networks from ATAC-seq data (Figure 3). DNA transposition occurs preferentially at nucleosome free regions adjacent to TF binding sites, but the TF binding sites themselves are protected from transposition. Hence, the pattern of ATAC-seq reads can directly reveal the binding profiles of hundreds of TFs whose cognate motifs are known at once (Buenrostro et al., 2013). One caveat of this approach is that TF family members that bind similar motifs cannot be distinguished. To this end, for each ATAC-seq profile, we identified enriched transcription factor (TF) motifs and factor footprints associated with ATAC-seq peaks to generate a matrix of predicted TF to gene relationships(Sherwood et al., 2014); the inferred regulation is weighted by the distance of the element to the transcription start site of each gene (Supplementary Methods, Figure S5d,e). The end result is a set of active TF regulators and their cognate sites on each gene for each individual. We then compared male vs. female samples for differences in this predicted regulatory matrix (Figure 3a-d, Figure S6f).
Figure 3. Gender-specific T cell regulome across autosomes.
a, Schematic of strategy to construct gender-specific gene regulatory network. Three TFs, A-C, are depicted; B and C bind a target gene differentially depending on gender. Purple arrows depict expected ATAC-seq signal.
b. Rank ordered genes with predicted gender-specific regulatory variation across their cognate loci.
c, Genomic tracks of FGL2 locus showing ATAC-seq signal in male vs. female samples. Top: Differential TF occupancy, defined as the TF occupancy score in male samples minus female samples, is shown. Upward signal indicates greater occupancy in males; downward signal indicates greater occupancy in females. The identity of the TF with gender-specific signal is shown.
Bottom: Zoom in view to visualize the ATAC-seq footprint at an IRF motif.
d, Rank ordered TFs with with gender-specific variation in occupancy profiles.
e, Genomic track of NeST-IFNG locus; gender-specific signal displayed as in panel c.
f, Functional enrichment of top 1000 genes with gender-specific differences in ATAC-seq signal. Black: Gene Ontology terms. Red: Disease terms.
At the level of target genes, we found several hundred genes that show significant differences in their predicted regulatory network in male vs. female T cells (z-score≥ 2, Figure 3b, Table S3). The scale of variance—defined here as the pattern of regulation in one gender that differs from the regulatory pattern of the other gender (1 - Pearson correlation of male vs. female) —ranges from 0.13 to 0.61 for the top 100 genes; hence such gender-specific variation is likely more modulatory than deterministic. Ranking autosomal genes by gender-specific regulatory variance revealed that the top divergent genes include many genes with well-known and important function in immune function or development, including FGL2, GZMK, IFNG, CRTAM, CARD16, FYN, IL2, and IL6 (Figure 3b). Indeed, significant gender-specific differences in T cell production of IFN-gamma and IL-2 have been documented in healthy children (Wiegering et al., 2009), validating our unbiased approach. IFN-gamma is also known to be affected by sex hormones (Rubtsova et al., 2015). In addition, direct molecular counting via Nanostring nCounter analysis of GZMK, IL2, IL6 and NLRP2 transcripts indicates significant gender-divergent responses to T cell activation (Figure S3b), which also supports our discovery (Ye et al., 2014). In summary, among the top 500 predicted gender-specific genes, the mRNA levels of 30 were measured using Nanostring, of which 20 show significant differential expression between males vs. females (p <0.05, _t_-test).
FGL2 stands out because it is the number one gene in the entire genome for gender-specific variance in chromatin accessibility and because the variance is nearly twice that of the next most variable gene (Figure 3c). FGL2 shows greater promoter ATAC-seq signal in male than female T cells, and analysis of specific TF signals revealed that IRF family and NHLH1 transcription factors are bound in male, but not female, T cells (Figure 3c). Notably, FGL2 encodes a fibrinogen-like protein secreted by regulatory T cells and other cells that has immunosuppressive activity (Marsden et al., 2003). Mouse knockout showed Fgl2 is required for Treg function and prevention of spontaneous autoimmunity (Shalev et al., 2008). Similarly, CRTAM encodes a T cell adhesion molecule that is recently recognized to critically control the differentiation of CD4+ cells into inflammatory Th17 cells (Cortez et al., 2014). Thus, gender-specific regulation of FGL2 and CRTAM, which to our knowledge has not been reported, may contribute in part to gender-linked differences for autoimmune disease.
Extending this concept to the top 1000 differentially regulated genes, we note that these genes are significantly enriched for biological functions in defense response, response to virus, immune complex, and inflammatory disease (p<10−6 for each, FDR<0.05, hypergeometric test, Figure 3f). The regulatory elements of many well-expressed genes, such as house keeping genes, are surveyed in these experiments but did not show gender-specific differences. Thus, gender-specific regulation in T cells is focused on genes with coherent biological function that impact immune function and autoimmunity.
To understand the mechanisms of gender-specific regulome divergence, we examined TF regulators that may exhibit gender-specific activity. In contrast to the ~1000 target genes, we observed just a handful of TFs with gender-associated divergence (Figure 3d, Table S3). Among the most divergent is ESR2 (encoding estrogen receptor beta); its differential can be understood based on the female hormone estrogen and serve as a positive controls. The top divergent regulator maps to the cognate motif of ZBTB3, a little studied factor. ZBTB3 is poorly expressed in CD4+ cells; hence this motif may be recognized by another zinc finger family protein. Two notable gender-divergent regulators are IRF family members, encoding well-studied TF activated by interferon signaling and other signals in innate and adaptive immunity responses that can cooperate or compete with other TFs to exert regulatory effects (Ikushima et al., 2013). Thus, a large number of genes were differently regulated, but the differential regulation was associated with a small number of TF motifs that repeated showed differential activity across many genes. This result suggests that a small number of regulators may impact a large number of target genes to yield the observed male-female divergence in the regulome.
The IFNG locus emerged as a prime example of intersection of predicted regulatory divergence of both regulators and target genes (Figure 3b,e). IFNG is the third most divergent gene in our analysis. IFNG encodes interferon gamma and is a key regulator of immune response and Th1 cell differentiation. Multiple studies have documented gender-specific association of allelic variants at IFNG regulatory elements or IFNG protein levels with human disease. For example, IFNG variants are associated with multiple sclerosis (Kantarci et al., 2008) and asthma (Loisel et al., 2011) in males but not females, but the mechanisms are not known. NeST (also known as IFNG-AS1 or TMEVPG1) is located proximal to IFNG and encodes a long noncoding RNA that is required to program active chromatin state and promote expression of IFNG.(Gomez et al., 2013) NeST is convergently transcribed relative to IFNG, and a long isoform of NeST is transcribed through the IFNG promoter. NeST itself is induced by Th1 polarization (Hu et al., 2013), and murine NeST was first discovered as a genetic locus that controlled pathogen resistance and immune-mediated demyelinating disease (Bihl et al., 1999).
We found that human IFNG and NeST show gender-specific regulation (Figure 3e). A cluster of elements nearest to 5’ of IFNG is equally active in male vs. female cells, but high-resolution analysis indicated that IRF family members occupied these sites more strongly in males, but NF-YA and CST6 did so in females. Males also have higher TF occupancy of NeST. These results suggest that a positive regulatory loops comprising of NeST, IRF, and IFNG may differ in a gender-specific fashion. Consistent with this idea, careful genetic analyses showed that NeST locus mutation has stronger pathogenic impact in male than female mice (Bihl et al., 1999). The fact that our unbiased approach examining target genes and regulators independently nominated IFNG highlights interferon signaling as a major gender-specific regulatory feature in T cells. Our results support the considerable epidemiologic and genetic evidence of IFNG involvement in gender variation of immune function and autoimmunity, and enrich them by nominating specific TFs and lncRNA as potential players in this mechanism.
T cell chromatin accessibility variation is enriched at sites of causal genetic variants for autoimmune diseases
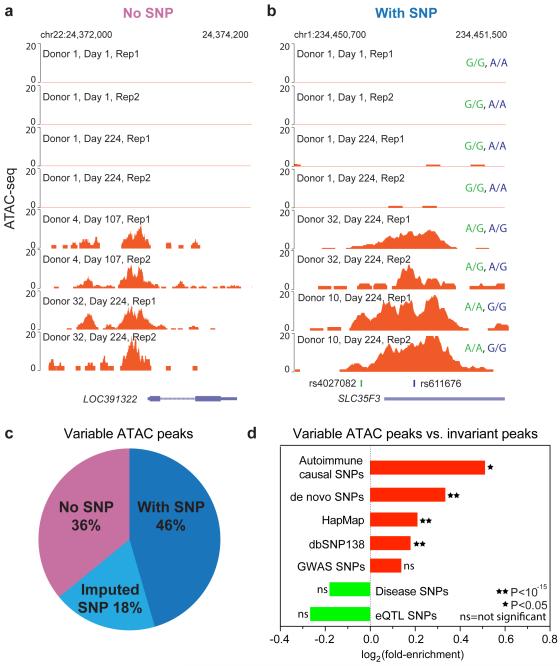
To understand how inherited DNA variation may underlie variations in personal regulomes, we intersected the set of variable regulatory elements with a previously curated set of single nucleotide polymorphisms (SNPs), particularly SNPs linked to human diseases or variation in chromatin state or gene expression (Boyle et al., 2012),(Farh et al., 2014). Prior work indicated that most dhsQTLs with strong effect size acted in cis, meaning that they affected the chromatin feature where the SNP itself is located(Degner et al., 2012). We identified the genotypes using sequencing information in ATAC-seq peaks, and validated them by standard SNP genotyping of a subset of donors (Figure 4a,b, Figure S6a,b). In addition, we imputed unmeasured SNPs by applying a standard imputation method IMPUTE2 (Howie et al., 2009), using the current version of 1,000 Genomes haplotypes as a reference. We found that 48% and 18% of the 4769 variably active elements overlapped detected or imputed SNPs, respectively (Figure 4c). Thus, more than a third (36%) of regulome variation occurs in the absence of genetic variation in cis. Prior data further indicate that only a small minority (<1%) of SNPs can significantly explain variation in chromatin accessibility (Degner et al., 2012). Additional contributions of personal regulome variation may include _trans-_acting genetic variants, environmental and epigenetic factors, and variations in cell subset composition.
Figure 4. Intersection of regulome variation with genetic variation.
a, Example of a reproducible regulome variation without underlying sequence variation.
b, Example of a reproducible regulome variation with underlying sequence variation; genotype of the two SNPs in each individual is indicated.
c, Regulome variation intersection with detected SNPs (dark blue) and imputed SNPs (light blue). Over a third of variable peaks do not intersect with SNPs (light purple).
d, Enrichment of the indicated classes of SNPs in variable ATAC-seq peaks vs. invariant peaks. Autoimmune causal SNPs showed highest enrichment.
We discovered that the intersection of personal regulome variation and genetic variation is highly relevant for human disease. We obtained SNP sets from the recently published set of causal autoimmune SNPs (Farh et al., 2014), GWAS and eQTLs SNPs from NCBI, HapMap, dbSNP138, disease SNPs (of all organ systems) from regulomeDB (Boyle et al., 2012) and de novo SNPs from the donors (Supplementary Methods). First, we asked whether ATAC-seq peaks of CD4+ T cells, representing active regulatory elements, are enriched in SNPs as a class compared to the remainder of the genome. Autoimmune casual SNPs are strongly enriched in CD4+ T cell ATAC-seq peaks (p <10−15, binomial test) (Figure S6c), consistent with the idea that causal genetic variants in autoimmune diseases impact immune cell enhancers(Farh et al., 2014). Second, we tested whether ATAC-seq peaks that show inter-individual variation are enriched for disease or eQTL SNPs compared to invariant open chromatin sites in CD4+ T cells. We found that autoimmune causal SNPs were most significantly enriched in variable peaks compare to invariant open chromatin sites (Figure 4d), including causal variants for type I diabetes, rheumatoid arthritis, lupus erythematosus, Crohn’s disease, and vitiligo (Table S4). As negative controls, generic SNP sets from GWAS of all diseases, or eQTLs (which are not T cell-specific) showed no significant enrichment. Collectively, these results illustrate the biological significance of the variable regulatory elements, and variability in chromatin accessibility across individuals emerges as a novel feature associated with locations of causal disease SNPs . Variable elements are by definition “noisy” and capable of being readily switched on or off—properties that may enable even a single nucleotide mutation to change their activity(Farh et al., 2014).
Regulome dynamics in T cell activation
Personal regulomes can also allow be interpreted with the help of in vitro experiments that create an interpretative framework. We measured changes in the chromatin accessibility caused by specific hypothesis-driven perturbations in vitro, and examined whether any of the same changes occurred in patient-derived samples in vivo. We illustrate this concept by assessing the regulome dynamics during T cell activation. We isolated CD4+ cells from donor 1 and stimulated them with PMA and ionomycin, collected cells at 0, 1, 2, 4 hours or 0 and 4 hours of unstimulated controls in duplicate, and performed ATAC-seq to map the regulatory elements genome-wide.
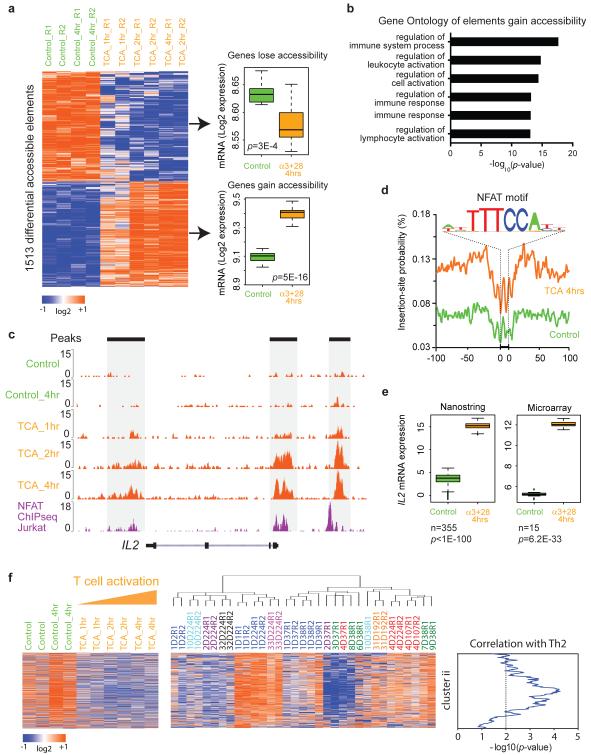
We identified 1513 regulatory elements that gained or lost accessibility upon T cell activation from 1 to 4 hours (Figure 5). 770 elements, mapping to 591 genes, gained accessibility (presumably induced) while 773 elements, mapping to 593 genes, lost accessibility (presumably repressed). By comparing genome-wide mRNA microarray data in normal human CD4+ T cells(Ye et al., 2014), we found that on average, genes that gained ATAC-seq signal showed significantly increased mRNA level upon activation (_p_=5×10−16), and conversely genes that lost ATAC-seq signal also had decreased mRNA expression (_p_= 3×10−4, Figure 5a). For gene loci that gain chromatin accessibility, 61 show increased mRNA level by more than 2-fold in mRNA expression while only 1 gene shows decreased mRNA level. Thus chromatin accessibility and gene expression are highly concordant, as anticipated. This finding further validates the accuracy of ATAC-seq and our mapping of regulatory elements to genes. As expected, regulatory elements that gain accessibility during the T cell activation show significant Gene Ontology enrichment in regulation of immune system processes, leukocyte activation and immune response (_p_-value<10−10, Figure 5b). T cell activation strongly activates a suite of genes, including interleukin 2 (IL2) in part through the inducible transcription factor NFAT(Northrop et al., 1994). ATAC-seq data visualized enhancer activation at IL2 locus, and identified inducible TF footprints of NFAT that is validated by published NFAT ChIP-seq (Figure 5c-e).
Figure 5. Regulome dynamics during T cell activation.
a, Left: heat map showing time course of regulatory elements with differential ATAC-seq activity during T cell activation. Each column is a sample; each row is an element. Samples and elements are organized by supervised hierarchical clustering. Color scale indicates relative ATAC-seq signal as indicated. Samples from untreated control and 4 hours control are colored green, and T cell activation at 1, 2, and 4 hours are colored orange. Right: box-plots of mRNA expression levels of the indicated genes in untreated control CD4+ T cells or after 4 hours activation with anti-CD3/CD28. Number of replicates = 15. _P_-value estimated from Student t-test.
b, Gene Ontology terms of regulatory elements gain in accessibility during T cell activation.
c, Dynamics of ATAC-seq signal at IL2 locus (orange track) during T cell activation (TCA) at the indicated times. NFAT ChIP-seq data in Jurkat cells (purple track) is shown for comparison.
d, Visualization of ATAC-seq footprint for transcription factor NFAT (motif shown) in control cells (green) vs. cells after T cell activation for 4 hours (orange). ATAC-seq signal across all NFAT binding sites in the genome were aligned on the motif and averaged.
e, Box-plots of mRNA expression levels of IL2 in untreated control (green) and 4 hours T cell activation with anti-CD3/CD28 (orange), obtained from Nanostring (left) and microarray (right). mRNA level from 355 or 15 healthy donors were measured by Nanostring or microarray, respectively. Log2 data are shown; _P_-value estimated from Student t-test.
f, Use of regulome signature of T cell activation to interpret individual variation. Regulatory element accessibility during T cell activation in vitro (left) vs. personal variation from healthy donors (right) are shown. Cluster ii from Figure 1b is found to exhibit coordinate deactivation during T cell activation and is also correlated with a Th2 signature. Donor sample dendrogram and demographic correlation are as in Figure 1b.
Next, we used the T cell activation regulome to interpret personal regulome variation. We discovered that a set of regulatory elements exhibiting inter-individual variation strongly corresponded to elements that are coordinately deactivated with T cell activation; this very same cluster is also associated with elevated Th2 gene signature (cluster ii, Figure 5f). Thus, a previously unassigned set of personal regulome variation may be related to the state of T cell activation in these donors. At a broader level, this new result illustrates the general concept that we can use ATAC-seq profiles from laboratory-based, well defined perturbations or cell populations to interpret the complex regulome patterns observed in clinical samples across real populations.
Regulome variation in the context of T cell activation and cancer
Finally, we explored the scope of regulome variation across individuals versus that with cell stimuli or a disease state. Although comparison of regulomes in disease vs. health may be a powerful approach to understand disease mechanism, an underlying assumption in cross-sectional studies (i.e. comparing different individuals who are healthy vs. sick at the same point in time) is that the disease-relevant variations significantly outnumber inter-individual variation—an assumption that needs to be tested. To this end, we performed ATAC-seq at 4 time points of human CD4+ T cells activated with ionomycin and phorbol ester (n=10 samples), as well as leukemic CD4+ T cells isolated from patients in the leukemic phase of CD4+ cutaneous T cell lymphoma (CTCL, n=15 samples). We found that the regulome dynamics in cancer and T cell activation are at least 10 to 100-fold greater than inter-individual variation (Figure 6). Notably, for the largest amplitude differences (>5 fold), the number of variable elements in these different states starts to converge, highlighting the importance of knowledge about inter-individual variation to interpret disease-associated data. T cell activation may exhibit greater regulome variation in this analysis because the cell population is temporally synchronized with respect to stimulation where as the leukemia samples are not. These results demonstrate the feasibility of using these data as foundational reference to which disease-associated regulomes can be compared. CTCL-specific regulome differences will be presented in detail elsewhere.
DISCUSSION
Here we evaluate the feasibility of using a recently introduced and sensitive genomic technology, ATAC-seq, to visualize the personal regulome from a standard blood draw, the most common source of human samples for clinical diagnostics. This work expands the number of primary human cell samples studied via ATAC-seq by twenty-fold, and we provide foundational data and methods to compare and visualize differences in personal regulome. We were able to enumerate the number, location, and potential sources of in vivo variation in chromatin accessibility on a genome-wide scale, providing many notable and unexpected findings. Our cohort was not designed to test many possible potential contributors of regulome variation, and lack of association here does not rule out their roles. Rather, these data serve as a starting point to understand potential sources of variation in chromatin accessibility in the population.
Our analyses suggest that sex is a major source of individual variation in the T cell regulome. We demonstrated the capacity of ATAC-seq to detect even two-fold differences in chromatin access, and many predicted regulome differences were validated by independent gene expression measurements. We identified X-linked chromatin accessibility sites that are more active in females vs. male T-cells, which nominated elements associated with escape from XCI in female cell. We further identified autosomal sites that have differential accessibility between sexes, which may related to differences in sex hormones, indirect regulation from sex-chromosome dosage, or other sex-related associated differences (Rubtsova et al., 2015). Variation between sexes has its root cause in genetics from the differential inheritance of sex chromosomes, which in turn leads to epigenetic differences in dosage compensation and organismal differences, such as different hormonal environments and life history events. These events are often linked to social or cultural roles that can also impact physiology. Our work adds to the increasing recognition of sexual dimorphism at the molecular level across many organs (Rinn et al., 2004). Building on the large body of epidemiologic and clinical data for gender differences in immune function and autoimmunity, our results nominated specific genes, transcription factors, and predicted regulatory circuits as potential drivers of gender differences for future studies. Similarly, we found that the majority of the variation in the regulatory landscape may go beyond genetic variation; such regulatory variation may arise from epigenetic differences from life history or environmental factors such as the microbiome that offers many opportunities for future investigations (Yurkovetskiy et al., 2013).
Finally, comparison of regulome variation in healthy donors vs. disease documented the feasibility of using the personal regulome approach to investigate disease biomarkers and mechanisms. The added value of regulome analysis over mRNA biomarkers may arise from the fact that casual SNPs associated with human diseases predominantly impact distal enhancers (Farh et al., 2014), and thus cannot be assessed by traditional mRNA measurements alone. Recent discovery of widespread allelic bias in enhancer-promoter interactions motivates the need for regulome analysis in addition to RNA measurements(Dixon et al., 2015), as exemplified by our detection of differential regulatory landscape of FIRRE in males vs. females. Furthermore, regulome analysis may directly interrogate chromatin or TF pathways that are direct drug targets. While useful, potential limitations of ATAC-seq for personal regulome prediction include Tn5 sequence bias, sensitivity as a function of sequencing depth, and cell type heterogeneity. Integration of additional epigenomic measures, such as direct TF binding or enhancer RNA synthesis, may improve enhancer activity prediction(Dogan et al., 2015). As more than one billion blood samples are obtained per year in the U.S alone (Becich, 2000), using ATAC-seq to monitor personal regulomes in health and disease offers many exciting possibilities.
METHODS SUMMARY
Enrichment of CD4+ cells from peripheral blood, ATAC-seq and primary data analysis were as described (Buenrostro et al., 2013). Differential peaks was defined via intrinsic analysis, with cutoff of Z-score>1, unlog2ed fold change >5 and a FDR<0.1. Gene ontology enrichment scores were obtained from GREAT. Statistical power analysis was performed using the “pwr.t2n.test” function in R. TF footprinting analysis was performed using PIQ v1.2 (Sherwood et al., 2014) with input motifs set from jasper. Donor DNA was genotyped using Illumina HumanOmni2.5-8+ v1.1 DNA Analysis BeedChip Kits. De novo mutation calling was performed using VarScan v2.2.8, and unmeasured SNPs were imputed by IMPUTE2. Enrichments of SNP sets were estimated by binomial test in R. Variance of each peak was calculated using “var” function in R. Genomic data are available at NIH GEO (GSE60682), and processed data in BAM format are available through NCBI SRA (SRP059154). Complete methods are available in Supplemental Information.
Supplementary Material
1
2
Highlights.
- ATAC-seq identifies personal gene regulatory landscapes of T cells over time.
- Sexual dimorphism in chromatin accessibility coordinately impact immune genes.
- Noisy enhancers are enriched for disease enhancers in autoimmunity.
- Foundational resource for comparison to human disease states or perturbations.
ACKNOWLEDGEMENTS
We thank all the donors for participating in the study, and M.M. Davis, K. Kirkegaard, J. Pritchard, P.J. Utz and A. Satpathy (Stanford) for discussion. Supported by NIH P50HG007735 (H.Y.C. and W.J.G.), and NIH U19AI110491, Stanford Cancer Center, Scleroderma Research Foundation (H.Y.C.), and Haas Family Foundation (Y.H.K. and H.Y.C.) H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
L.C.Z, P.G.G, and H.Y.C. conceived the project; K.Q and L.C.Z designed the experiment. L.C.Z, P.G.G., R.L., and M.L. performed the experiments; K.Q. and P.G.G. developed computational algorithms and conducted data analysis; L.C.Z., Y.H.K. and M.L. obtained the human samples; K. Q., W.J.G., and H.Y.C. wrote the paper with contributions from all authors.
References
- Becich MJ. Information management: moving from test results to clinical information. Clin Leadersh Manag Rev. 2000;14:296–300. [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihl F, Brahic M, Bureau JF. Two loci, Tmevp2 and Tmevp3, located on the telomeric region of chromosome 10, control the persistence of Theiler's virus in the central nervous system of mice. Genetics. 1999;152:385–392. doi: 10.1093/genetics/152.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015 doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez VS, Cervantes-Barragan L, Song C, Gilfillan S, McDonald KG, Tussiwand R, Edelson BT, Murakami Y, Murphy KM, Newberry RD, et al. CRTAM controls residency of gut CD4+CD8+ T cells in the steady state and maintenance of gut CD4+ Th17 during parasitic infection. J Exp Med. 2014;211:623–633. doi: 10.1084/jem.20130904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner JF, Pai AA, Pique-Regi R, Veyrieras JB, Gaffney DJ, Pickrell JK, De Leon S, Michelini K, Lewellen N, Crawford GE, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan N, Wu W, Morrissey CS, Chen KB, Stonestrom A, Long M, Keller CA, Cheng Y, Jain D, Visel A, et al. Occupancy by key transcription factors is a more accurate predictor of enhancer activity than histone modifications or chromatin accessibility. Epigenetics & chromatin. 2015;8:16. doi: 10.1186/s13072-015-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2014 doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14:1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Negishi H, Taniguchi T. The IRF Family Transcription Factors at the Interface of Innate and Adaptive Immune Responses. Cold Spring Harb Symp Quant Biol. 2013;78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- Kantarci OH, Hebrink DD, Schaefer-Klein J, Sun Y, Achenbach S, Atkinson EJ, Heggarty S, Cotleur AC, de Andrade M, Vandenbroeck K, et al. Interferon gamma allelic variants: sex-biased multiple sclerosis susceptibility and gene expression. Arch Neurol. 2008;65:349–357. doi: 10.1001/archneurol.2007.66. [DOI] [PubMed] [Google Scholar]
- Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, Zaugg JB, Kundaje A, Liu Y, Boyle AP, Zhang QC, Zakharia F, Spacek DV, et al. Extensive Variation in Chromatin States Across Humans. Science. 2013 doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Loisel DA, Tan Z, Tisler CJ, Evans MD, Gangnon RE, Jackson DJ, Gern JE, Lemanske RF, Jr., Ober C. IFNG genotype and sex interact to influence the risk of childhood asthma. J Allergy Clin Immunol. 2011;128:524–531. doi: 10.1016/j.jaci.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nature reviews Immunology. 2012;12:191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden PA, Ning Q, Fung LS, Luo X, Chen Y, Mendicino M, Ghanekar A, Scott JA, Miller T, Chan CW, et al. The Fgl2/fibroleukin prothrombinase contributes to immunologically mediated thrombosis in experimental and human viral hepatitis. The Journal of clinical investigation. 2003;112:58–66. doi: 10.1172/JCI18114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, Lewellen N, Myrthil M, Gilad Y, Pritchard JK. Identification of Genetic Variants That Affect Histone Modifications in Human Cells. Science. 2013 doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop JP, Ho SN, Chen L, Thomas DJ, Timmerman LA, Nolan GP, Admon A, Crabtree GR. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome biology. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Rozowsky JS, Laurenzi IJ, Petersen PH, Zou K, Zhong W, Gerstein M, Snyder M. Major molecular differences between mammalian sexes are involved in drug metabolism and renal function. Dev Cell. 2004;6:791–800. doi: 10.1016/j.devcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Rubtsova K, Marrack P, Rubtsov AV. Sexual dimorphism in autoimmunity. The Journal of clinical investigation. 2015;125:2187–2193. doi: 10.1172/JCI78082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev I, Liu H, Koscik C, Bartczak A, Javadi M, Wong KM, Maknojia A, He W, Liu MF, Diao J, et al. Targeted deletion of fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. J Immunol. 2008;180:249–260. doi: 10.4049/jimmunol.180.1.249. [DOI] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O'Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Stergachis AB, Maurano MT, Vierstra J, Neph S, Thurman RE, Stamatoyannopoulos JA, Akey JM. Personal and population genomics of human regulatory variation. Genome Res. 2012;22:1689–1697. doi: 10.1101/gr.134890.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegering V, Eyrich M, Wunder C, Gunther H, Schlegel PG, Winkler B. Age-related changes in intracellular cytokine expression in healthy children. Eur Cytokine Netw. 2009;20:75–80. doi: 10.1684/ecn.2009.0149. [DOI] [PubMed] [Google Scholar]
- Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, Moore JM, Filippova GN, Xu J, Liu Y, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome biology. 2015;16:52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CJ, Feng T, Kwon HK, Raj T, Wilson MT, Asinovski N, McCabe C, Lee MH, Frohlich I, Paik HI, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Castillo-Morales A, Jiang M, Zhu Y, Hu L, Urrutia AO, Kong X, Hurst LD. Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol Biol Evol. 2013;30:2588–2601. doi: 10.1093/molbev/mst148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2