Antibiotic susceptibility profile of bacilli isolated from the skin of healthy humans (original) (raw)
Abstract
In the present work, twelve bacilli were isolated from four different regions of human skin from Bela population of Nagpur district, India. The isolated bacilli were identified by their morphological, cultural and biochemical characteristics. Seven isolates were Gram negative rods, out of which five were belong to genus_Pseudomonas_. Three among the five Gram positive isolates were identified as Dermabactor and the remaining two_Bacillus_. Their antimicrobial susceptibility profile was determined by Kirby-Bauer disc diffusion method. The isolates showed resistance to several currently used broad-spectrum antibiotics. The Dermabactor_genus was resistant to vancomycin, although it was earlier reported to be susceptible. Imipenem was found to be the most effective antibiotic for_Pseudomonas while nalidixic acid, ampicillin and tetracycline were ineffective. Isolates of Bacillus displayed resistance to the extended spectrum antibiotics cephalosporin and ceftazidime. Imipenem, carbenicillin and ticarcillin were found to be the most effective antibiotics as all the investigated isolates were susceptible to them. Antibiotic resistance may be due to the overuse or misuse of antibiotics during the treatment, or following constant exposure to antibiotic-containing cosmetic formulations.
Key words: antibiotic resistance, identification, microflora, nosocomial, opportunistic
Introduction
Human skin is considered to be a critical barrier between the human body and its outer environment. It prevents loss of moisture and restricts the entry of pathogenic organism (Bojar and Holland, 2002). Hence it seems to be an excellent ecosystem that harbors varying microbial communities (Kong and Segre, 2012). These microbial communities are distributed across the human skin and live in physiologically diverse and topographically distinct niches, depending on the environmental conditions specific to distinct region of the skin (Grice et al., 2009; Oh et al., 2012). The normal flora generally coexists asymptomatically with host, but can cause infection whenever the host immune system gets compromised or skin is damaged. Microflora causing such infection are said to be opportunistic pathogens.
The contribution of Gram negative bacilli as normal microflora of the human skin is quite small as compared to their extraordinary numbers in the human gut (Lockhart et al., 2007). This may be attributed to desiccation considered a major factor preventing the colonization and multiplication of Gram negative organisms on skin (Elsner, 2006). Gram negative organisms such as Pseudomonas areuginosa, Pasteurella multocida, Vibrio vulnificus are not considered typical resident of human skin microflora since they may cause cutaneous infections (Larson et al., 1986a, 1986b).
Bacteria can infect human skin when they gain access to the human host. This happens when the host has the broken skin or mucus membrane or is immunocompromised (Hogenova et al., 2004). The treatment of infections then depends on use of different antibiotics treatments. It is plausible that this may account for phenomenon of emergence of resistance of bacteria to various antimicrobial agents (Patil and Chopade, 2001). The development of antimicrobial resistance reflects an evolutionary process in response to the antimicrobial therapy that may select physiologically or genetically competent strain capable of surviving high dose of antimicrobial agents (Zhang Li and Liu, 2007). The acquired resistance may be transfer in the form of resistance genes to other species via horizontal gene transfer (Pardesi et al., 2007; Barak et al., 2005). In the recent times, information is increasing on cosmetic products contain antibiotics may again drive the development of antibiotic resistance among the normal microflora of human skin (Hegstad et al., 2010). In a related study antibiotic resistant strains of human microflora were isolated from peoples who did not have prior contact with hospital environment, and from doctors who were closely connected with hospital. It was shown that isolated strain from the doctors, who were not under any kind of antibiotic treatment, were five times more multidrug resistant to antibiotics used in hospital environment (Kwaszewska et al., 2009; Berlau et al., 1999).
Although, studies have reported the role of skin bacilli in nosocomial infections, but bacilli are still under-explored with respect to their antibiotic resistance profile. Antibiotic-resistant flora from human skin can potentially transmit the antibiotic resistance gene pool to other known pathogens of human skin. Hence, the need for routine examination of antibiotic susceptibility of isolates from human skin is important. In this study, isolation of pure culture of bacilli from human skin samples was carried out. The isolated bacilli were identified using morphological, cultural and biochemical characteristics, after which antibiotic resistance profile studied, to gain better understanding about the antibiotic resistance of skin microflora. The finding from this is expected to guide definitive antibiotic therapy.
Material and Methods
Sample collection from human skin
Samples were collected from twelve healthy people of Bela village from Nagpur district. The people were within the the age group of 18 to 45 years and with no observable symptomatic disease. The use of cosmetics was not considered as selection criteria as the study population was from a village and used very little or no cosmetics. The samples were collected by rubbing sterile cotton swab on skin and transferred to tube containing nutrient broth. Samples were collected from four different regions of healthy human skin, making a total of twelve samples (Table 1). Moist regions of the skin that favor the growth and colonization of bacteria were selected for sample collection. Each sample was coded by a number and an alphabet corresponding to the sites selected.
Table 1. Regions of skin from different people depicting the name of sample.
| Skin Region | Sample |
|---|---|
| Inner Elbow | 9IE, 4IE, 15IE, 10IE, 7IE2 |
| Philtrum | 10J, 7J, 14J, 12JY |
| Cheek | 5CT, 14C |
| Forehead | 14FH |
Identification of bacilli from collected samples
Isolation of pure culture from the samples was carried out by streak plate method. The isolates were identified by their morphological, cultural and biochemical characteristics. The pure cultures were characterized using different methods of microscopic observation including Gram staining, motility. The cultural characteristics on solid media include observation of colony size, margin, surface, color, elevation. Nutrient agar was used for the study. The biochemical characterization include sugar test, IMViC, and demonstration of enzyme activity like starch hydrolysis, tributyrin hydrolysis, oxidase, catalase, nitrate reduction and urease.
Antibiotic susceptibility testing
The antibiotics used along with their abbreviations (as per CLSI) and concentration are mentioned as follows; Amoxicillin AMR (30 μg/disc), Ampicillin AMP (25 μg/disc), Carbenicillin CAR (100 μg/disc), Ceftazidime CAZ (30 μg/disc), Cephalothin CEF (30 μg/disc), Ciprofloxacin CIP (30 μg/disc), Cloxacillin CLX (30 μg/disc), Gentamycin GEN (30 μg/disc), Imipenum IPM (10 μg/disc), Kanamycin KAN (30 μg/disc), Methicillin MET (30 μg/disc), Nalidixic acid NAL (30 μg/disc), Piperacillin PIP (100 μg/disc), Roxithromycin ROX (30 μg/disc), Streptomycin STR (10 μg/disc), Tetracycline TET (30 μg/disc), Ticarcillin TIC (30 μg/disc), Tobramycin TOB (30 μg/disc), Vancomycin VAN (100 μg/disc). The antibiotic discs (6 mm diameter were purchased from Hi-media Laboratory Ltd., Mumbai (India). The Kirby-Bauer method of antibiotic sensitivity testing is a relatively simple, reliable and rapid test was used in this study. The effectiveness of antimicrobial in sensitivity testing is based on the size of zone of inhibition that surrounds a disc that has been impregnated with a specific concentration of an agent. The Kirby-Bauer method is a standardized system for antimicrobial resistance profiling. Plates were prepared with Mueller Hinton agar. Inoculation was carried out by spread plate method. Isolated bacilli were grown in nutrient broth up to 1.0 OD and 100 μL of each was spread uniformly by spreader on agar surface. Various antibiotic discs then placed on the agar plate and the plates were incubated at 37 °C for 24 h. After 24 h incubation, plates were observed for zone of inhibition around the antibiotic discs. The zone of inhibition for different antibiotics was then measured. The antibiotics used for the antibiotic resistance profiling were selected on the basis of their class as described along with their abbreviation and concentration (μg/disc). The spectrum of activity was also considered for selection of antibiotics. For example, broad spectrum tetracyclines, quinolones and third generation cephalosporins are active against both Gram negative and Gram positive organisms, while narrow spectrum antibacterial drugs have limited activity.
Amplification of 16S rRNA and lipase A genes
Nutrient agar is generally used for bacterial growth but can also support the growth of many microorganisms other than bacteria which can confound the outcome of a result. In order to eliminate this possibility, PCR amplification with eubacteria-specific 16S rRNA universal primers was carried out. Since these primers are known to be highly specific for eubacteria, amplification with these primers could confirm the isolates as bacteria. Isolation of genomic DNA was carried out from all the isolates. Cell lysis occurs due to the action of SDS (Sodium dodecyl sulphate) which destabilizes the plasma membrane. Upon centrifugation, cell debris along with trapped RNA, proteins are separated. Resulting supernatant contains genomic DNA. This was followed by the amplification of 16S rRNA gene with universal primers (FP Bac AGAGTTTTATCCTGGTCAG, RP univ592r ACCGCGGCKGCTGGC). 1.5 μL of template DNA was used for amlification of genomic DNA for 40 cycles (denaturation 94 °C-10 min; denaturation, 94 °C- 1 min; anneling 54.9 °C- 1 min; polymerization 72 °C- 10 min). Amplification was also carried out for Lipase A gene to validate the results obtained on tributyrin agar plates for lipid hydrolysis (Figure 1) (Lindh et al., 2005). 1.5 μL of genomic DNA was used for amplification with Lipase A primers (FP L1 ATGGTTCACGGTATTGGA GG, RP L2 CTGCTGTAAATGGATGGAGG) for 40 cycle with condition of denaturation 94 °C-10 min; denaturation, 94 °C- 1 min; anneling 54.6 °C- 1 min; polymerization 72 °C- 10 min. The PCR product length for 16S rRNA and Lipase A gene was 540 bp and 371 bp respectively.
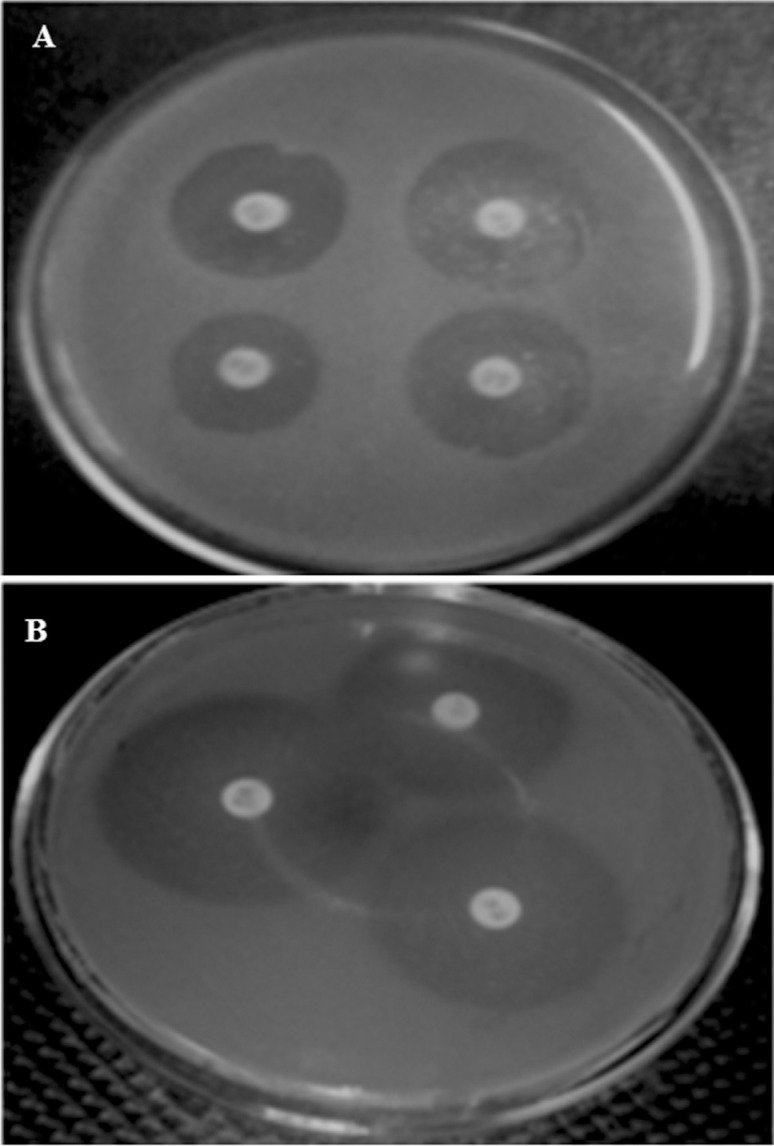
Figure 1. Representative result for antibiotic susceptibility profile. Zone of inhibition obtained on Mueller Hinton (MH) agar for (A) 10IE isolate and (B) 7IE2 isolate.
Results
Isolation and identification of bacilli from skin samples
The pure cultures of bacteria from 12 skin samples were obtained by streak plate method. The isolated pure cultures were then identified based on their morphological, cultural and a biochemical characteristic with reference to Bergey's manual of determinative bacteriology (Brockman, 1986). The results are summarized in Table 2. From twelve isolated pure cultures, seven were found to be Gram negative while others were Gram positive The characteristics observed were then compared with Bergey's manual of determinative bacteriology and the isolates were categorized to genus level and to species level wherever possible.
Table 2. Identification chart for the isolates based on the Bergey's manual of Determinative Bacteriology.
| Characteristics | P. stutzeri 9IE | P. mendocina 4IE | P. stutzeri 15IE | P. mendocina 14FH | P. mendocina 14C | Aeromonas spp. 14J | Proteus spp. 7J | B. licheniformis 5CT | Sulfobacillus 10IE | Dermabactor spp. 10J | Dermabactor spp. 12JY | Dermabactor spp. 7IE2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram staining | - | - | - | - | - | - | - | + | + | + | + | + |
| Endospore | + | + | ||||||||||
| Motility | + | + | + | + | + | + | + | + | - | - | - | - |
| Catalyse | + | + | + | + | + | + | + | + | + | + | + | + |
| Oxidase | + | - | + | + | + | + | + | + | + | - | - | - |
| Glucose fermentation | + | + | + | - | - | - | - | + | + | + | + | - |
| Lactose fermentation | - | - | - | - | - | - | - | - | + | - | - | - |
| Mannitol fermentation | - | - | - | - | - | - | - | + | + | + | - | - |
| IMViC I | - | - | - | - | - | - | - | - | - | - | - | - |
| M | - | - | - | - | - | - | - | - | - | - | - | - |
| VP | - | - | - | - | - | - | - | - | - | - | - | - |
| C | + | - | + | - | - | - | - | + | - | + | - | - |
| Starch Agar | + | - | + | - | - | + | - | + | + | - | - | - |
| Tributyrin Agar | + | - | + | + | + | + | - | + | - | + | - | - |
| Urease | - | - | - | + | + | - | + | + | + | - | - | + |
| Nitrate reduction | + | + | + | - | - | + | - | + | + | - | - | - |
Antibiotic resistance profile of identified bacilli
After identification, the antibiotic susceptibility of all isolates was determined. The results for the antibiotic susceptibility are shown in Table 3 and 4. The seven Gram negative isolates were tested for 19 antibiotics. They were found to be resistant to streptomycin, nalidixic acid and ampicillin, while three of the isolates were resistant to cephalothin and tetracycline. It was also found that Imipenem, carbenicillin and ticarciliin were the most effective antibiotic to which all Gram negative isolates were susceptible. The five Gram positive isolates were found to be highly resistant to ceftazidime antibiotic. Two isolates were also resistant to cephlothin and vancomycin, while all Gram positive isolates were susceptible to Imipenem and methicillin.
Table 3. Antimicrobial susceptibility pattern for Gram negative isolates.
| Antibiotics | Susceptibility of isolates to antibiotics with diameter of inhibition (mm) shown in bracket | ||||||
|---|---|---|---|---|---|---|---|
| P. stutzeri (9IE) | P. luteolla | P. stutzeri | Proteus spp. | Aeromonas spp. | P. mendocina | P. mendocina | |
| GEN | R(16) | S(22) | S(20) | I(17) | I(18) | I(16) | I(15) |
| KAN | R(13) | I(16) | S(22) | S(18) | S(18) | S(18) | S(18) |
| TOB | S(21) | R(16) | I(18) | I(16) | I(16) | I(15) | I(16) |
| STR | S(11) | R(15) | S(21) | S(22) | S(18) | R(00) | R(00) |
| NAL | R(11) | R(00) | I(14) | R(09) | I(18) | R(06) | R(08) |
| CIP | R(24) | S(28) | S(34) | I(28) | S(33) | R(08) | I(18) |
| IPM | S(36) | S(32) | S(30) | S(29) | S(32) | S(28) | S(26) |
| PIP | S(20) | S(22) | S(20) | S(35) | R(16) | S(18) | S(18) |
| TIC | S(24) | S(22) | S(22) | S(22) | S(19) | S(28) | S(35) |
| CAR | S(24) | S(28) | S(21) | S(15) | S(21) | S(30) | S(28) |
| VAN | R(00) | I(22) | S(22) | S(22) | R(16) | S(18) | S(19) |
| AMP | R(07) | R(11) | R(18) | S(21) | R(09) | R(25) | R(18) |
| MET | S(22) | R(15) | I(20) | S(28) | S(25) | R(15) | S(30) |
| CAZ | S(24) | S(22) | S(22) | R(08) | R(00) | I(12) | S(20) |
| CEF | R(00) | S(30) | R(00) | S(20) | R(00) | S(22) | S(21) |
| TET | R(16) | R(17) | R(15) | S(19) | I(15) | I(28) | S(21) |
Table 4. Antimicrobial susceptibility pattern for Gram positive isolates.
| Antibiotics | Susceptibility of isolates to antibiotics with zone of inhibition (mm) shown in bracket | ||||
|---|---|---|---|---|---|
| Sulfobacillius | Dermabactor spp. | Dermabactor spp. | Dermabactor spp. | B. licheniformis | |
| GEN | S(19) | R(12) | R(13) | I(14) | S(20) |
| KAN | S(19) | I(16) | S(20) | R(08) | S(20) |
| TOB | I(16) | R(15) | I(16) | R(10) | S(18) |
| STR | S(20) | S(15) | S(22) | R(00) | S(18) |
| NAL | I(16) | I(15) | R(00) | R(11) | I(16) |
| CIP | S(35) | S(31) | R(08) | I(18) | S(33) |
| IPM | S(40) | S(37) | S(30) | S(40) | S(40) |
| PIB | S(24) | R(17) | S(21) | S(27) | R(11) |
| TIC | S(25) | S(19) | S(28) | S(40) | R(12) |
| CAR | S(22) | S(20) | S(32) | S(40) | R(09) |
| VAN | S(19) | R(13) | R(09) | S(20) | S(17) |
| TET | S(27) | R(16) | S(26) | S(30) | R(15) |
| MET | S(22) | I(19) | S(29) | S(29) | I(15) |
| CLX | I(20) | I(18) | S(31) | S(30) | R(06) |
| CAZ | S(19) | R(00) | R(00) | R(00) | R(10) |
| ROX | R(13) | S(33) | S(26) | I(17) | R(00) |
| CEF | S(27) | S(38) | R(00) | S(22) | R(00) |
| TET | S(26) | S(28) | R(13) | S(30) | I(18) |
Six isolates from inner elbow region of different persons were found to be resistant to nalidixic acid, ampicillin and tetracycline. Out of which three isolates were highly resistant to cephalothin. Among the 19 antibiotics Imipenem, ticarcillin, carbenicillin and piperacillin were the most effective to which all isolate were susceptible. Four isolates from philtrum region were found to be highly resistant to ceftazidime and two of them were also resistant to nalidixic acid and vancomycin while the other were also resistant to streptomycin, tobramycin and piperacillin . All isolates showed susceptibility to Imipenem, carbenicillin and ticarcillin. Figure 2 shows the zone of inhibition obtained around antibiotic disc for two isolates 10IE and 7IE2 after 24 h of incubation.
Figure 2. Representative result for lipid hydrolysis on tributyrin agar.
Amlification of 16S rRNA and Lipase A genes
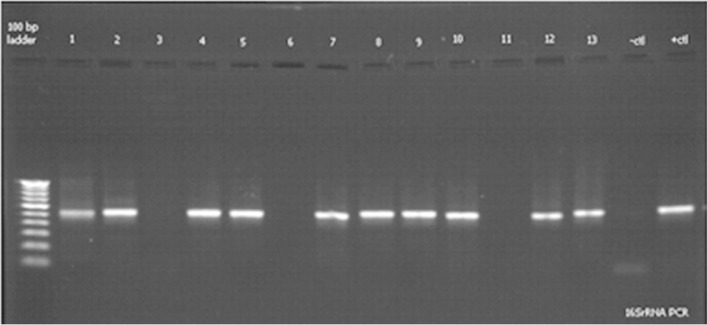
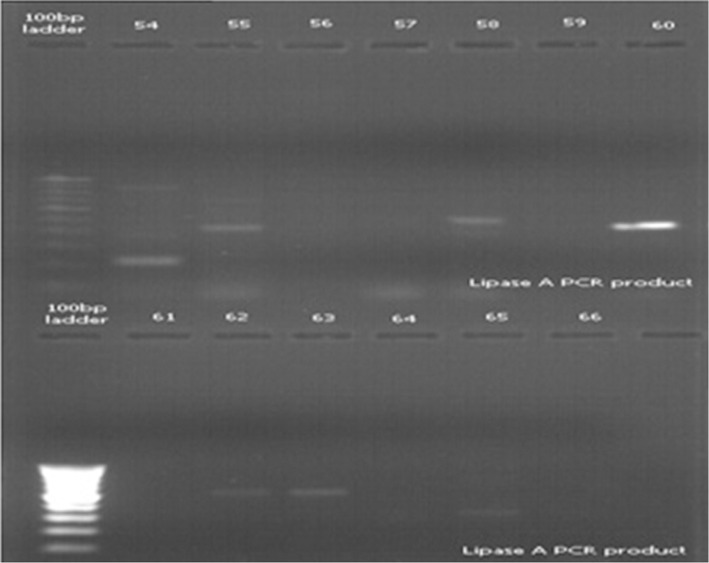
Microorganism other than bacteria, like yeasts, may form colonies on nutrient agar. Hence, PCR amplification with eubacterial 16S rRNA-specific primers was carried out, as 16S rRNA gene is highly conserved across the bacterial kingdom. The 16S rRNA gene of all isolates was partially amplified with 16S rRNA universal primers and PCR products (540 bp) obtained were run on 1.2% agarose gel. Amplification of the gene for 16s rRNA was successfully carried out for confirming that the isolates were bacterial species (Figure 3). Study of lipid hydrolysis is included in the biochemical characterization of isolates. Lipid hydrolysis was studied on tributyrin agar. Seven isolates were found positive for lipid hydrolysis (Figure 1). Lipase A primers was used for validating the result obtained for lipid hydrolysis on tributyrin agar. Lipid hydrolysis is a significant virulence factor for skin pathogenesis. Results of amplification with Lipase A primers confirmed the presence of this virulence factor and validate the results obtained on tributyrin agar for lipid hydrolysis as shown inFigure 4.
Figure 3. Agarose gel showing amplification of 16S rRNA genes from different isolates. Lane 1- 9IE, 2- 4IE, 3- 15IE, 4- 7IE2, 5- 7J, 6- 10IE, 7- 10J, 8- 14J, 9- 14FH, 10- 14C, 11- 12JY, 12- 13C, 13- 5CT.
Figure 4. Agarose gel amplification of Lipase A gene from different isolates. Lane 54- 9IE, 55- 4IE, 56- 15IE, 57- 7IE2, 58- 7J, 59- 10IE, 60- 10J, 61- 14J, 62- 14FH, 63- 14C, 64- 12JY, 65- 13C, 66- 5CT.
Discussion
An enhanced understanding of the skin microbiome is necessary to gain insight into microbial involvement in human skin disorder, and to enable novel promicrobial and antimicrobial therapeutic approaches for their treatment. Hence it is important to know the antibiotic susceptibility of isolates from human skin (Todar, 2012; Wilson, 2008). The significance of coryneforms in opportunistic infections is growing especially in immuncoompromised patients. Coryneforms have been described as the main etiologic factors of opportunistic infections (Kazmierczak_et al_., 2005). For instance Dermabactor is a relatively new genus and D. hominis is a relatively new species.D. hominis has been assigned in to Coryneform group 3 and group 5 respectively. Little has been learnt about its epidemiology except that it is found among the human cutaneous flora (Funke_et al_., 1996; Kazmierczak_et al_., 2005). In previous studies Dermabactor was found to be resistant to aminoglycoside, fluroquinolones, macrolides and lincosamide. In present work three isolates of Dermabactor displayed high resistance to ceftazidime, and two of them also showed resistance to aminoglycoside (tobramycin) and quinolones (nalidixic acid) (Funke et al., 1996). Dermabactor, which was reported to be susceptible to glycopeptides (vancomycin) earlier, displayed resistance to vancomycin in this study. In another study Southern African strain of B. anthracis was studied for its susceptibility to newly developed antibiotics (Frean et al., 2003). In previous study lack of activity of extended spectrum cephalosporin against B. _anthracis_was reported (Paavilainen et al., 2000). In this study one isolate of Bacillus displayed resistance to cephalosporin and ceftazidime. In another prime study, Pseudomonas stutzeri shown susceptibility to nalidixic acid and resistance to ampicillin and streptomycin (Hentges et al., 1985; Lalucat et al., 2006). In present work isolates of Pseudomonas displayed resistance to nalidixic acid, ampicilln and streptomycin, while they showed susceptibility to ceftazidime, piperacillin and ticarcillin. Imipenem was found to be the most effective antibacterial for Pseudomonas in agreement with previous studies.
On comparing the results from the present study with previous studies, it can be considered that the antibiotic resistance has increased in skin isolates. Antibiotics to which isolates were found susceptible in previous study were ineffective in present work. Isolates from skin of healthy humans showed resistant to board spectrum of antibiotics. Skin colonizing normal microbial flora generally resides peacefully without harming host, but may cause infection upon physical injury or in case of immunocompromised hosts. The presence of high antimicrobial resistance in skin microbiome can be a cause for concern as there is probability of horizontal gene transfer of the antibiotic resistance gene pool to skin pathogens. This can become a serious challenge in clinical therapy. It can be concluded that this increasing antibiotic resistance may be a result of uncontrolled utilization of antibiotics. Therefore skin of healthy human can be considered to be one of the most important reservoirs for microorganism causing clinically acquired infections. The study has reported that cosmetic formulations also contain antibiotics and drive antibiotic resistance in skin microflora (Horner et al., 2012). The use of cosmetics even though not considered for selecting individuals in present work but still, it may be considered as a contributing factor for development of antibiotic resistance in skin microflora. Untreated wastewater from antibiotic industry may aid in developing the reservoir of antibiotic resistance gene pool in environmental bacteria. These resistance genes may be transfer to human microbiome including pathogens (Cabello et al., 2013; Li et al., 2010). In addition to this, detergents like quaternary ammonium salt can also be responsible for development of cross resistance against antibiotics (Hegstad et al., 2010).
References
- Barak O, Treat JR, James WD. Antimicrobial peptides: Effect of innate immunity in the skin. Adv Dematol. 2005;2:357–374. doi: 10.1016/j.yadr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Berlau J, Aucken H, Malnick H, et al. Distribution of Acinetobactor species of skin of healthy human. Eur J Clin Microbiol Infect. 1999;18:179–183. doi: 10.1007/s100960050254. [DOI] [PubMed] [Google Scholar]
- Bojar KA, Holland KT. The human cutaneous microbiota and factors controlling colonization. World J Microbiol Biotechnol. 2002;18:889–903. [Google Scholar]
- Brockman ER. In: Bergey's Manual of Systematic Bacteriology. Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Vol. 3. Williams & Wilkins; Baltimore: 1986. [Google Scholar]
- Cabello F, Godfrey H, Tomova A, et al. Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ Microbiol. 2013;15:1917–1942. doi: 10.1111/1462-2920.12134. [DOI] [PubMed] [Google Scholar]
- Elsner P. Antimicrobial and the skin physiological and pathological flora. Dermatol. 2006;33:35–41. doi: 10.1159/000093929. [DOI] [PubMed] [Google Scholar]
- Frean J, Klugman KP, Arntzen L, et al. Susceptibility of bacillus to eleven antimicrobial agents including novel fluroquinolones and ketolides. J Antimicrob Chemother. 2003;52:297–299. doi: 10.1093/jac/dkg364. [DOI] [PubMed] [Google Scholar]
- Funke G, Verena PU, Graevenitz AV. Antimicrobial Susceptibility Patterns of Some Recently Established Coryneform Bacteria. Antimicrob Agents Chemoter. 1996;40:2874–2878. doi: 10.1128/aac.40.12.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:593–595. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegstad K, Langsrud S, Lunestad BT, et al. Does the wide use of quaternary ammonium compounds enhance the selection and spread of antibacterial resistance and thus threaten our health. Microbiol Drug Resist. 2010;16:91–104. doi: 10.1089/mdr.2009.0120. [DOI] [PubMed] [Google Scholar]
- Hentges DJ, Stein AJ, Casey SW, et al. Protective role of intestinal flora against Pseudomonas aeruginosa in mice: influence of antibiotics on colonization resistance. Infect Immun. 1985;47:118–122. doi: 10.1128/iai.47.1.118-122.1985. 2856912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenova HT, Stepankova R, Hudcovic T, et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93:97–108. doi: 10.1016/j.imlet.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Horner C, Mawer D, Wilcox M. Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J Antimicrob Chemother. 2012;67:2547–2559. doi: 10.1093/jac/dks284. [DOI] [PubMed] [Google Scholar]
- Kazmierczak AK, Kwaszewska JK, Szewczyk EM. Opportunistic Coryneform Organism-Resident of Human Skin. Pol J Micribiol. 2005;54:27–37. [PubMed] [Google Scholar]
- Kazmierczak AK, Szewczyk EM. Bacteria forming a resident flora of the skin as a potential source of opportunistic infections. Pol J Micribiol. 2004;53:249–255. [PubMed] [Google Scholar]
- Kenneth Todar. Online Textbook of Bacteriology. University of Winconsin, Department of Bacteriology; 2012. [Google Scholar]
- Kong HH, Segre JA. Skin Microbiome Looking Back to move forward. J Invest Dermatol. 2012;132:933–939. doi: 10.1038/jid.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaszewska AK, Sobis-Glinkowska M, Szewczyk EM. Infuence of contact with hospital environment on sensitivity to antibiotic to lipophilic strains of Corynebacterium residing on human skin. Med Dosw Mikrobiol. 2009;61:359–366. [PubMed] [Google Scholar]
- Lalucat J, Bennasar A, Bosch R, et al. Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev. 2006;70:510–547. doi: 10.1128/MMBR.00047-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, McGinley KJ, Foglia AR, et al. Composition and Antimicrobial Resistance of Skin Flora in Hospitalized and Healthy Adults. J Clin Microbiol. 1986;23:604–608. doi: 10.1128/jcm.23.3.604-608.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, McGinley KJ, Foglia AR, et al. Skin colonization with antibiotic-Resistance (JK Group) and Antibiotic-Sensetive Lipophilic diptheroids in Hospitalized and Normal Adults. Jpn J Infect Dis. 1986;153:701–706. doi: 10.1093/infdis/153.4.701. [DOI] [PubMed] [Google Scholar]
- Li D, Yu T, Zhang Y, et al. Antibiotic Resistance Characteristics of Environmental Bacteria froman Oxytetracycline Production Wastewater Treatment Plant and the Receiving River. Appl Environ Mcrobiol. 2010;76:3444–3451. doi: 10.1128/AEM.02964-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhang GA, Liu Y. Analysis of predominant bacteria from burn infections and their resistance to antibiotics in recent years. Chin J Burns. 2007;23:255–259. [PubMed] [Google Scholar]
- Lindh JM, Terenius O, Faye I. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Environ Microbiol. 2005;71:7217–7223. doi: 10.1128/AEM.71.11.7217-7223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Abramson MA, Beekmann SE, et al. Antimicrobial resistance among Gram negative bacilli causing infection in intensive care unit in United State between 1993 and 2004. J Clin Microbiol. 2007;45:3352–3359. doi: 10.1128/JCM.01284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Conlan S, Polley EC, et al. Shift in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4:1–11. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavilainen T, Osterblad M, Leistevuo T, et al. Screening for antimicrobial resistance in normal bacterial flora of the skin using the replica plating method. Eur J Clin Microbial Infect. 2000;12:956–959. doi: 10.1007/s100960000406. [DOI] [PubMed] [Google Scholar]
- Pardesi KR, Yavankar SP, Chopade BA. Plasmid distribution and antimicrobial susceptibility of Acinetobactor genospecies from healthy tribal population of western India. Indian J Med Res. 2007;125:79–88. [PubMed] [Google Scholar]
- Patil JR, Chopade BA. Distribution and in vitro antimicrobial susceptibility of Acinetobactor species on the skin of healthy humans. Natl Med J India. 2001;14:204–208. [PubMed] [Google Scholar]
- Wilson M. Bacteriology of Humans: An Ecological Perspective. Wiley-Blackwell; Hoboken: 2008. 360 p [Google Scholar]