In vivo gene editing in dystrophic mouse muscle and muscle stem cells (original) (raw)
. Author manuscript; available in PMC: 2016 Jul 22.
Published in final edited form as: Science. 2015 Dec 31;351(6271):407–411. doi: 10.1126/science.aad5177
Abstract
Frame-disrupting mutations in the DMD gene, encoding dystrophin, compromise myofiber integrity and drive muscle deterioration in Duchenne muscular dystrophy (DMD). Removing one or more exons from the mutated transcript can produce an in-frame mRNA and a truncated but still functional protein. In this study, we develop and test a direct gene editing approach to induce exon deletion and recover dystrophin expression in the mdx mouse model of DMD. Delivery by adeno-associated virus (AAV) of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 endonucleases coupled with paired guide RNAs flanking the mutated Dmd_exon23 resulted in excision of intervening DNA and restored Dystrophin reading frame in myofibers, cardiomyocytes and muscle stem cells following local or systemic delivery. AAV-Dmd CRISPR-treatment partially recovered muscle functional deficiencies and generated a pool of endogenously corrected myogenic precursors in_mdx mouse muscle.
Duchenne muscular dystrophy (DMD) is a progressive muscle degenerative disease caused by point mutations, deletions, or duplications in the DMD gene that cause genetic frame-shift or loss of protein expression (1). Efforts under development to reverse the pathological consequences of DYSTROPHIN deficiency in DMD aim to restore its biological function through viral-mediated delivery of genes encoding shortened forms of the protein, upregulation of compensatory proteins, or interference with the splicing machinery to “skip” mutation-carrying exons in the mRNA and produce a truncated, but still functional protein (reviewed in (2)).
The potential efficacy of exon-skipping strategies is supported by the relatively mild disease course of Becker Muscular Dystrophy (BMD) patients with in-frame deletions in_DMD_ (3, 4), and by the capacity of antisense oligonucleotides (AONs), which mask splice donor or acceptor sequences of mutated exons in dystrophin mRNA, to restore biologically active DYSTROPHIN protein in mice (5, 6) and humans (7,8). Yet, limitations remain for the use of AONs, including variable efficiencies of tissue uptake, depending on AON chemistry, a requirement for repeated AON injection to maintain effective skipping, and the potential for AON-associated toxicities ((9, 10) and Supplementary Text).
Here, we sought to address these limitations by developing a one-time, multisystemic approach based on the genome-editing capabilities of the CRISPR/Cas9 system. This system, coopted originally from Streptococcus pyogenes (Sp), couples a DNA double strand endonuclease with short “guide RNAs” (gRNAs) that provide target specificity to any site in the genome that also contains an adjacent `NGG' protospacer adjacent motif (PAM) (11–14), thereby enabling targeted gene disruption, replacement, and modification.
To apply CRISPR/Cas9 for exon deletion in DMD, we first established a reporter system for CRISPR activity by “repurposing” the existing Ai9 mouse reporter allele, which encodes the fluorescent tdTomato protein downstream of a ubiquitous CAGGS promoter and “floxed” STOP cassette (15, 16) (Fig. S1A). Exposure to SpCas9, together with paired gRNAs targeting near the Ai9 loxP sites (hereafter Ai9 gRNAs), resulted in excision of intervening DNA and expression of tdTomato (Fig. S1A,B,E). We next designed and tested paired gRNAs (hereafter Dmd_23 gRNAs, Fig. S1C) directed 5' and 3' of mouse_Dmd exon23, which in mdx mice carries a nonsense mutation that destabilizes Dmd mRNA and disrupts DYSTROPHIN expression (17). Finally, we coupled the paired _Dmd_23 and Ai9 gRNAs using a two plasmid system that links expression of the CRISPR activity reporter (tdTomato) to genome editing events at the Dmd locus (Fig. S1D). _In vitro_transfection of primary satellite cells from mdx mice carrying the Ai9 allele (hereafter mdx;Ai9 mice) with SpCas9 + Ai9-_Dmd_23 coupled gRNAs induced gene editing at both the Ai9 locus, demonstrated by tdTomato expression (Fig. S1E), and _Dmd_locus, detected by genomic PCR (Fig. 1A) and confirmed by amplicon sequencing (Fig. S1F).Dmd editing was not detected in mdx;Ai9 cells receiving Ai9 gRNAs alone (Fig. 1A), although tdTomato expression was equivalently induced (Fig. S1E).
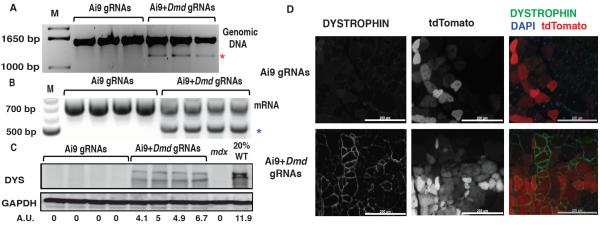
Figure 1. DYSTROPHIN expression in CRISPR-modified dystrophic satellite cells.
(A) Detection of exon23 excision by genomic PCR in myotubes derived from satellite cells transfected with SpCas9 and Ai9 gRNAs (left lanes) or coupled Ai9-_Dmd_23 gRNAs (right lanes). Unedited genomic product, 1572bp; gene-edited product (red asterisk), 1189bp. M, molecular weight marker. (B) RT-PCR detection of exon23-deleted mRNA. Unedited RT-PCR product: 738bp; exon23-deleted product (blue asterisk): 525bp. (C) Western blot detecting DYSTROPHIN in myotubes derived from gene-edited satellite cells. A.U.: Arbitrary Unit, normalized to GAPDH (loading control). DYS: DYSTROPHIN, WT: wild type (D) DYSTROPHIN immunofluorescence in mdx muscles transplanted with satellite cells transfected ex vivo with SpCas9 + Ai9 gRNAs (top) or SpCas9 + Ai9-_Dmd_23 coupled gRNAs (bottom). For merge: Green: DYSTROPHIN; Red: tdTomato; Blue: DAPI (nuclei). Scale bar: 200 μm. See also Fig. S1I.
To confirm that CRISPR-mediated Dmd editing results in irreversible genomic modification and production of exon-deleted mRNA and protein, primary satellite cells from mdx;Ai9 mice were co-transfected with SpCas9 + Ai9 or Ai9-Dmd_23 gRNAs, FACSorted based on tdTomato expression, expanded_in vitro (18), and differentiated to myotubes. RT-PCR (Fig. 1B) and amplicon sequencing (Fig. S1G) from these myotubes detected exon23-deleted Dmd mRNA in cells receiving Ai9-_Dmd_23 coupled gRNAs, but not in cells receiving only Ai9 gRNAs. Taqman analysis (9) further indicated that exon23-deleted transcripts represented 24–47% of total Dmd mRNA in cells receiving Ai9-_Dmd_23 coupled gRNAs, whereas exon23 deletion was undetectable with Ai9 gRNAs alone (Fig. S1H). DYSTROPHIN protein expression was also restored in CRISPR-modified mdx;Ai9 cells, as detected by Western blot of in vitro differentiated myotubes (Fig. 1C) and immunostaining of muscle sections from mdx mice transplanted with gene-edited mdx;Ai9 satellite cells (Fig. 1D and S1I). These data demonstrate that CRISPR/Cas9 can direct sequence-specific modification of disease alleles in primary muscle stem cells that retain muscle engraftment capacity.
We next adapted CRISPR for delivery via adeno-associated virus (AAV), employing the smaller Cas9 ortholog from Staphylococcus aureus (SaCas9), which can be packaged in AAV and programmed to target any locus in the genome containing a “NNGRR” PAM sequence (19). We generated Sa gRNAs targeting Ai9 and introduced several base modifications into the gRNA scaffold to enhance gene targeting by SaCas9 (Fig S2A–C). Using this modified scaffold, we tested _Dmd_23 Sa gRNAs (Fig. S2D) and produced AAVs encoding SaCas9 and Ai9 Sa gRNAs or Dmd_23 Sa gRNAs in a dual (Fig. S3A) or single (Fig. S3B) vector system. Comparison of exon23 excision efficiencies in transduced mdx myotubes demonstrated more efficient excision by dual AAV-CRISPR (Fig. S3C,D), as compared to single vector AAVs. Therefore, to test the potential for_in vivoDmd targeting by CRISPR/Cas9, we pseudotyped dual AAVs (AAV-SaCas9 + AAV-Ai9 gRNAs; hereafter, AAV-Ai9 CRISPR) to serotype 9, which exhibits robust transduction of mouse skeletal and cardiac muscle (20), and injected these AAVs into the tibialis anterior (TA) muscles of mdx;Ai9 mice (7.5E+11 vg each). Four weeks later, muscles were harvested to assess genome-editing events. TdTomato fluorescence was detected in muscles injected with AAV-Ai9 CRISPR, but not in muscles injected with vehicle alone (Fig. S4A). Co-delivery of AAV9-SaCas9 + AAV9-Dmd_23 gRNAs (hereafter AAV-Dmd CRISPR) likewise yielded robust and specific modification of the_Dmd locus in TA muscles in vivo. Genomic PCR (Fig. 2A) and Sanger sequencing (Fig. S4B) demonstrated exon23 excision in muscles injected with AAV-Dmd CRISPR, but not AAV-Ai9 CRISPR. Next-generation sequencing indicated minimal activity at the predicted highest-ranking genomic off-target sites (Fig. S12). RT-PCR (Fig. 2B) and sequencing (Fig. S4C) further confirmed the presence of exon23-deleted Dmd mRNA in muscles receiving AAV-_Dmd_CRISPR, with an average exon23-excision rate of 39%±1.8% (Fig. S3E). In vivo_CRISPR-mediated targeting of Dmd exon23 restored DYSTROPHIN expression in skeletal muscle, as detected by Western blot (Fig. 2C), immunofluorescence (Fig. 2D) and capillary immunoassay (Fig. S5A). Other pathological hallmarks of dystrophy were also restored in AAV-Dmd CRISPR injected muscles, including sarcolemmal localization of the multimeric dystrophin-glycoprotein complex and neuronal nitric-oxide synthase (Figs. S6 and S7). In contrast, DYSTROPHIN expression was undetectable by Western (Fig. 2C) and present only on rare revertant fibers in_mdx;Ai9 mice receiving control AAV-Ai9 CRISPR (Fig. 2D; (21)). Finally, to evaluate the functional consequences of CRISPR-mediated induction of exon23-deleted DYSTROPHIN, we subjected a subset of mdx;Ai9 mice injected intramuscularly with AAV-Dmd CRISPR to in situ muscle force assessment. Muscles receiving AAV-Dmd CRISPR showed significantly increased specific force (Fig. 2E) and attenuated force drop after eccentric damage (Fig. 2F), as compared to contralateral, vehicle-injected muscles and also AAV-Ai9 CRISPR injected muscles. In contrast, differences in specific force (Fig. 2E) and force drop (Fig. 2F) for AAV-Ai9 CRISPR injected mice were not significantly different between the virus-injected and vehicle-injected muscles.
Figure 2. AAV-CRISPR enables in vivo excision of Dmd exon23 and restores DYSTROPHIN expression in adult dystrophic muscle.
(A,B) Detection of exon23 excision in TA muscles from mdx;Ai9 mice injected intramuscularly with AAV-Ai9 CRISPR (left lanes) or AAV-Dmd_CRISPR (right lanes) by genomic PCR (A; unedited product 1012bp; exon-excised product 470bp) and RT-PCR (B). Asterisks mark gene-edited bands. M, molecular weight marker. (C) Western blot detecting DYSTROPHIN in muscles injected with AAV-Ai9 CRISPR (left) or AAV-Dmd CRISPR (right), with relative signal intensity determined by densitometry at bottom. A.U.: Arbitrary Unit, normalized to GAPDH. (D) Representative immunofluorescence images for DYSTROPHIN (green) and DAPI (blue) in_mdx;Ai9 muscles injected with AAV-Ai9 (left) or AAV-Dmd (right) CRISPR. Scale bar: 500 μm. (E,F) Muscle specific force (E) and decrease in force after eccentric damage (F) for wild type mice injected with vehicle (n=9), mdx;Ai9 mice injected with AAV-_Dmd_CRISPR in the right TA and vehicle in the left TA (n=12), mdx;Ai9 mice injected with AAV-Ai9 CRISPR in the right TA and vehicle in the left TA (n=12). *P<0.05, **P<0.01, n.s., not significant, One-Way ANOVA with Newman-Keuls multiple comparisons test.
We next evaluated the potential for multisystemic gene editing in vivo using AAV-CRISPR. Dual AAV-Ai9 CRISPR vectors (1.5E+12 vg each) were co-injected intraperitoneally into mdx;Ai9 mice at postnatal day 3 (P3). Three weeks later, widespread tdTomato expression was detected in all cardiac and skeletal muscles analyzed (Fig. S8A). Parallel injections of mdx;Ai9 mice with AAV-Dmd CRISPR revealed exon23-deleted transcripts in multiple skeletal muscles and cardiac muscle, with targeting levels varying from 3–18% in different muscle groups (Fig. 3A and S3F). Exon23 was not excised in animals receiving AAV-Ai9 CRISPR instead (Fig. 3A, S3F and S8B). Finally, Western blot (Fig. 3B and S8C), immunofluorescence (Fig. 3C), and capillary immunoassay (Fig. S5B) confirmed that DYSTROPHIN protein was largely absent in muscles of control mdx;Ai9 mice receiving AAV-Ai9 CRISPR and restored in mice receiving AAV-Dmd CRISPR. Similar systemic dissemination of AAV and excision of exon23 in multiple organs were seen in two adult mice injected intravenously with AAV-Dmd CRISPR at 6 weeks of age (Fig. S9).
Figure 3. Systemic dissemination of AAV-CRISPR targets Dmd exon23 and restores DYSTROPHIN in dystrophic cardiac and skeletal muscles.
(A) Exon23-deleted transcripts in muscles quantified by Taqman. Data plotted for individual mice (n=7 receiving AAV-Dmd CRISPR (blue) and n=3 receiving AAV-Ai9 CRISPR (red)) and overlaid with mean ± SEM. (B) Western blots detecting DYSTROPHIN in the indicated muscles of mdx;Ai9 mice receiving systemic AAV-CRISPR. Right lanes correspond to muscles from 7 different mice injected intraperitoneally with AAV-Dmd CRISPR. Relative signal intensity, determined by densitometry, presented as A.U.: Arbitrary Unit normalized to GAPDH. (C) Representative immunofluorescence staining for DYSTROPHIN (green) in_mdx;Ai9_ mice injected with AAV-Ai9 (top) or AAV-Dmd(bottom) CRISPR. Blue: DAPI (nuclei). Scale bar: 200 μm.
Dystrophic pathology and other muscle injuries activate muscle stem cells (also known as satellite cells), leading to regenerative responses that add new nuclei to damaged fibers ((2) and see Supplementary Text). To evaluate AAV-CRISPR gene editing in satellite cells in vivo, we crossed_mdx;Ai9_ mice with Pax7-ZsGreen animals, in which satellite cells are specifically marked by green fluorescence (22), and injected these animals intramuscularly or systemically with AAV9 encoding Cre (hereafter, AAV-Cre) or Ai9-CRISPR components. Muscles were harvested 2 weeks later (Fig. 4A), and analyzed by FACS. tdTomato expression was apparent in Pax7-ZsGreen+ satellite cells following local or systemic delivery of AAV-Cre or AAV-Ai9 CRISPR (Fig. 4B and S10A–C), although excision rates were lower for AAV-Ai9 CRISPR than for AAV-Cre. In vitro differentiation of ZsGreen+ satellite cells from mice receiving intramuscular or systemic AAV-Cre or AAV-Ai9 CRISPR produced tdTomato+ myotubes, demonstrating preservation of myogenic potential in AAV-transduced and gene-edited satellite cells (Fig. 4Cand S10D). TdTomato+ gene–edited satellite cells also engrafted recipient mdx muscle and contributed to in vivo muscle regeneration after transplantation (Fig. S10E).
Figure 4. Satellite cells in dystrophic muscles are transduced and targeted with systemically disseminated AAV-CRISPR.
(A) Experimental design. (B) Percent of ZsGreen+ satellite cells expressing tdTomato+ after intraperitoneal injection of_Pax7-ZsGreen_+/−_;mdx;Ai9_ mice. Individual data points overlaid with mean ± SD; vehicle (n=3), AAV-Cre (n=4) AAV Ai9 CRISPR (n=5). (C) Representative immunofluorescence of myotubes differentiated from FACSorted satellite cells from mice injected intraperitoneally with vehicle, AAV-Cre, or AAV-Ai9 CRISPR. Green: Myosin heavy chain (MHC); Red: tdTomato. Blue: DAPI (nuclei) Scale bar: 200 μm. (D) Exon23-deleted DMD mRNA in satellite-cell derived myotubes from mice previously injected intraperitoneally with AAV-Dmd CRISPR (right lanes), compared to control AAV-Ai9 CRISPR (left lanes).
We next analyzed Dmd editing in Pax7-ZsGreen+ satellite cells following intramuscular or systemic delivery of AAV-Dmd CRISPR or AAV-Ai9 CRISPR. Satellite cells were isolated by FACS, expanded and differentiated in vitro (Fig. 4A). RT-PCR revealed a truncated_Dmd_ transcript of the expected size and sequence for gene-edited_Dmd_ in satellite cell-derived myotubes from many of the AAV-Dmd CRISPR injected muscles, but none of the AAV-Ai9 CRISPR injected muscles (Fig. 4D and S10F,H,I). Quantification of exon23-excision revealed variable efficiencies (Fig. S10G,J), likely reflecting targeting of only a subset of endogenous satellite cells that may be variably represented among the isolated and cultured cells. Finally, genomic PCR and amplicon sequencing confirmed targeted excision at the_Dmd_ locus in satellite cell-derived myotubes (Fig. S10K), and capillary immunoassay analysis revealed restored DYSTROPHIN expression (Fig. S10L). As expected, injection of AAV-Dmd CRISPR did not induce tdTomato expression in satellite cells or myofibers of _mdx;Ai9_mice (Fig. S11).
In summary, this study provides proof-of-concept evidence supporting the efficacy of_in vivo_ genome editing to correct disruptive mutations in DMD in a relevant dystrophic mouse model. We show that programmable CRISPR complexes can be delivered locally and systemically to terminally differentiated skeletal muscle fibers and cardiomyocytes, as well as muscle satellite cells, in neonatal and adult mice, where they mediate targeted gene modification, restore DYSTROPHIN expression and partially recover functional deficiencies of dystrophic muscle. As prior studies in mice and humans indicate that DYSTROPHIN levels as low as 3–15% of wild-type are sufficient to ameliorate pathologic symptoms in the heart and skeletal muscle (23–26), and levels as low as 30% can suppress the dystrophic phenotype altogether (27) and protect dystrophic muscle from damage (28), the restoration of DYSTROPHIN achieved here by one-time administration of AAV-Dmd CRISPR clearly encourages further evaluation and optimization of this system as a new candidate modality for the treatment of DMD (see Supplementary Text).
Supplementary Material
supplement
Acknowledgments
We thank the HSCI/HSCRB Flow Cytometry Core, the SERI/MEEI Gene Transfer Vector Core, the Parker lab at Harvard and J. Goldstein for technical assistance. Work was funded in part by grants from HHMI and NIH (1DP2OD004345, 5U01HL100402, 5PN2EY018244) to A.J.W. M.T. is an Albert J. Ryan fellow. F.A.R. is a Junior Fellow at the Harvard Society of Fellows. W.X.Y. was supported by T32GM007753 from the National Institute of General Medical Sciences (NIGMS). F.Z. is a New York Stem Cell Foundation Robertson Investigator and supported by NIMH (5DP1-MH100706) and NIDDK (5R01DK097768-03), a Waterman Award from the National Science Foundation, the Keck, New York Stem Cell, Damon Runyon, Searle Scholars, Merkin, and Vallee Foundations, and B. Metcalfe. W.L.C. is supported by the National Science Scholarship from the Agency for Science, Technology and Research (A*STAR), Singapore. G.M.C. is supported for this work by NHGRI CEGS P50 HG005550. Content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or the National Institutes of Health. M.T., A.J.W., W.L.C. and G.M.C. are inventors on a patent application (PCT/US15/63181) filed by Harvard University related to in vivo genetic modifications and gene editing in muscle. G.M.C. is an inventor on issued patents (US9023649, US9074199) filed by Harvard University related to CRISPR. L.H.V. is an inventor on a patent application (US2007036760) filed by the University of Pennsylvania related to AAV capsid sequences. F.Z., L.C., F.A.R. and W.Y. are inventors on patents and patent applications (8,865,406, 8,906,616 and accepted EP 2898075, from International patent application WO 2014/093635) filed by the Broad Institute related to SaCas9 optimized components and systems. A.J.W. is an advisor for Fate Therapeutics. G.M.C and F.Z. are founders and scientific advisors of Editas Medicine and F.Z. is a scientific advisor for Horizon Discovery. G.M.C. has equity in Caribou/Intellia, Egenesis and Editas (for full disclosure list, see: http://arep.med.harvard.edu/gmc/tech.html). L.H.V. is co-founder, shareholder, member of the Scientific Advisory Board, and consultant of GenSight Biologics, a consultant to Novartis and Eleven Bio, and received honoraria and consulting fees from Regeneron Pharmaceuticals and Cowen, Jefferies and Sectoral. AAV9 vector sequences are available through an MTA from the University of Pennsylvania. Sa Cas9 plasmids are openly available through a UBMTA from Addgene.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Supplementary Materials: www.sciencemag.org Materials and Methods Supplementary Text Figures S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12 Tables S1, S2, S3, S4 References (28–54)
References and Notes
- 1.Koenig M, et al. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Tabebordbar M, Wang ET, Wagers AJ. Annu Rev Pathol. 2013;8:441–475. doi: 10.1146/annurev-pathol-011811-132450. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura A, et al. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2008;15:757–763. doi: 10.1016/j.jocn.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Taglia A, et al. Acta Myol. 2015;34:9–13. [PMC free article] [PubMed] [Google Scholar]
- 5.Lu QL, et al. Nat Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- 6.Echigoya Y, et al. Mol Ther Nucleic Acids. 2015;4:e225. doi: 10.1038/mtna.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Deutekom JC, et al. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 8.Kinali M, et al. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goyenvalle A, et al. Nat Med. 2015;21:270–275. doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 10.Vila MC, et al. Skeletal muscle. 2015;5:1–12. [Google Scholar]
- 11.Cong L, et al. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mali P, et al. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran FA, et al. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, et al. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madisen L, et al. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Materials and methods are available as supplementary materials on Science Online
- 17.Sicinski P, et al. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 18.Xu C, et al. Cell. 2013;155:909–921. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran FA, et al. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 21.Lu QL, et al. J Cell Biol. 2000;148:985–996. doi: 10.1083/jcb.148.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosnakovski D, et al. Stem Cells. 2008;26:3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Putten M, et al. J Mol Cell Cardiol. 2014;69:17–23. doi: 10.1016/j.yjmcc.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 24.van Putten M, et al. FASEB J. 2013;27:2484–2495. doi: 10.1096/fj.12-224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Putten M, et al. PLoS One. 2012;7:e31937. doi: 10.1371/journal.pone.0031937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long C, et al. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neri M, et al. Neuromuscul Disord. 2007;17:913–918. doi: 10.1016/j.nmd.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Xu L, et al. Mol Ther. 2015 Oct 9; doi: 10.1038/mt.2015.192. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplement