Ligand Activation of ERRα by Cholesterol Mediates Statin and Bisphosphonate Effects (original) (raw)
. Author manuscript; available in PMC: 2017 Mar 8.
Published in final edited form as: Cell Metab. 2016 Jan 14;23(3):479–491. doi: 10.1016/j.cmet.2015.12.010
SUMMARY
Nuclear receptors (NRs) are key regulators of gene expression and physiology. Nearly half of all human NRs lack endogenous ligands including estrogen-related receptor α (ERRα). ERRα has important roles in cancer, metabolism and skeletal homeostasis. Affinity chromatography of tissue lipidomes with the ERRα ligand-binding domain (LBD) and subsequent transcriptional assays identified cholesterol as an endogenous ERRα agonist. Perturbation of cholesterol biosynthesis or inhibition of ERRα revealed the interdependence of cholesterol and ERRα. In bone, the effects of cholesterol, statin and bisphosphonate on osteoclastogenesis require ERRα; and consequently, cholesterol-induced bone loss or bisphosphonate osteoprotection are lost in ERRα knockout mice. Furthermore, statin induction of muscle toxicity and cholesterol suppression of macrophage cytokine secretion are impaired by loss or inhibition of ERRα. These findings reveal a key step in ERRα regulation and explain the actions of two highly prescribed drugs, statins and bisphosphonates.
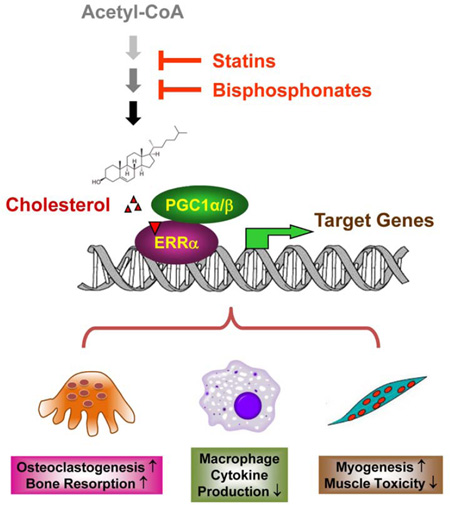
Graphical abstract
INTRODUCTION
Estrogen-related receptors (ERRs) are a family of nuclear receptors that consist of ERRα, ERRβ and ERRγ (Giguere, 2002; Giguere et al., 1988). ERRs control pathophysiological processes, such as cancer and skeletal homeostasis, by regulating the expression of specific target genes. ERRα is the most studied member of this subgroup and an orphan receptor because it lacks an endogenous (or natural) ligand. The identification of an endogenous nuclear receptor ligand is crucial to understand how the receptor is regulated. This information is of fundamental biological interest. Moreover, if the nuclear receptor is involved in diseases, knowledge on how to manipulate the levels of a ligand can be used to develop novel therapeutics. For instance, knowing that estrogen, a product of the aromatase enzyme, activates the estrogen receptor α (ERα), a nuclear receptor, led to the development of aromatase inhibitors to treat ERα-driven cancer by inhibiting estrogen production (Johnston and Dowsett, 2003).
ERRα was originally identified based on its homology to ERα (Giguere et al., 1988). Analysis of the individual domains reveals that the DNA-binding domains (DBDs) of ERRα and ERα are 70% homologous, but their ligand-binding domains (LBDs) are only 36% similar, explaining why ERα ligands do not activate ERRα. 17β-estradiol, estrone and estriol are all ERα regulators (Ascenzi et al., 2006), but these steroid hormones have no effect on ERRα function (Horard and Vanacker, 2003). The absence of an endogenous or dietary lipid ligand for ERRα has led to the hypothesis that ERRα is a ligand-less receptor (Kallen et al., 2004). This idea is bolstered by constitutive activity of the ERRα in the absence of an added ligand and structural biology that revealed that the ERRα ligand-binding pocket is almost completely occluded.
ERRα modulates energy metabolism (Giguere, 2008; Luo et al., 2003); ERRα deletion or inhibition confers resistance to obesity and insulin resistance (Luo et al., 2003; Patch et al., 2011; Sladek et al., 1997). ERRα also regulates skeletal remodeling by controlling osteoclastogenesis, a key cellular differentiation process essential for bone resorption (Gallet and Vanacker, 2010; Wan, 2010; Wei et al., 2010); ERRα deletion impairs osteoclast differentiation and bone resorption leading to increased bone mass (Wei et al., 2010). Furthermore, ERRα is also a critical regulator of multiple cancers (Stein and McDonnell, 2006; Suzuki et al., 2004).
The importance of ERRα in human health has led to tremendous interest in this protein as a novel therapeutic target. A number of synthetic small-molecule ERRα antagonists have been developed. These compounds bind to the ERRα ligand-binding domain (LBD) and induce a conformational shift in the ERRα-LBD that interferes with co-activator binding to inhibit transcription. ERRα antagonists have been found to induce cancer cell death (Wu et al., 2009), inhibit tumor growth (Duellman et al., 2010), and improve insulin sensitivity and glucose tolerance (Patch et al., 2011). More generally, the discovery of these antagonists indicates that ERRα has a functional small-molecule binding pocket, renewing the idea that ERRα has an endogenous ligand.
The identification of endogenous nuclear receptor ligands is typically accomplished using functional screens to find metabolites that regulate nuclear receptor transcriptional activity. While successful, cell-based assays with natural metabolites have certain drawbacks that could potentially lead to false negatives. Recent work, for example, has demonstrated the importance of cellular proteins in the transport (Ayers et al., 2007) and metabolism (Chakravarthy et al., 2009) of endogenous ligands. To obviate the need to consider these variables, we decided on using a lipidomic strategy to identify candidate endogenous ligand(s) for ERRα.
RESULTS
Cholesterol enrichment and binding to the ERRα LBD
We used an affinity chromatography approach to determine whether the ERRα-LBD binds to any endogenous lipids (Figure 1A). This method was previously used to enrich known endogenous PPARγ ligands from the lipidome (Kim et al., 2011a), validating the compatibility of this approach with nuclear receptors. Recombinant N-terminal 6xHis-tagged ERRα-LBD was expressed, purified and immobilized onto IMAC Sepharose 6 Fast Flow beads (resin). Lipidomes were prepared for affinity chromatography by extracting mouse brain with chloroform/methanol/water, isolating the organic fraction, and concentrating this fraction containing all lipids. Brain was selected because of the robust expression of ERRα (Bookout et al., 2006). The lipids were then dissolved in DMSO and mixed with buffer to afford a lipidome sample that is compatible with affinity chromatography. Resin loaded with ERRα-LBD was incubated with the lipidome. After incubation, the buffer was filtered away from the resin, and the resin was washed with buffer to remove weak interactions. The 6xHis-tagged ERRα-LBD is then eluted from the beads through the addition of buffer containing imidazole. Performing the exact steps with IMAC Sepharose 6 Fast Flow beads lacking any protein afforded the control sample for these experiments.
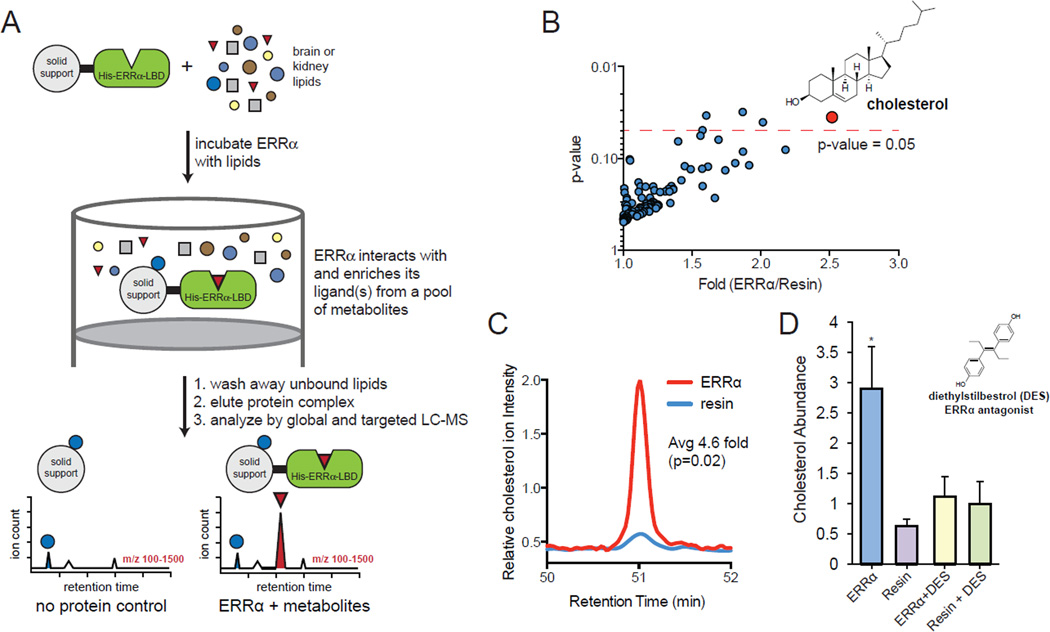
Figure 1. The ERRα-LBD selectively enriches cholesterol from the lipidome.
(A) A schematic diagram of the workflow for the identification of HIS-ERRα-LBD binders. HIS-ERRα-LBD is immobilized on a solid support and incubated with lipids from brain or kidney. After washing away unbound lipids, the protein complex is eluted, and eluent is analyzed by LC-MS. Lipids that bind HIS-ERRα-LBD are only present in the ERRα sample, but not in the no protein (resin only) control. (B) HIS-ERRα-LBD was incubated with a pool of lipids from brain, and bound lipids were analyzed by untargeted LC-MS. Cholesterol was the only lipid that was strongly and consistently enriched. (C) Cholesterol binding is also demonstrated by a targeted MS/MS method designed for sensitive and specific identification of sterol HIS-ERRα-LBD binding. A representative LC-MS chromatogram is shown. (D) Cholesterol binding could be blocked by the ERRα synthetic antagonist diethylstilbestrol (DES). In the absence of DES, HIS-ERRα-LBD strongly enriched cholesterol from a pool of brain sterols relative to a resin only sample. When DES was added to the sterol mixture, HIS-ERRα-LBD could no longer enrich cholesterol. Error bars, SD.
See also Figure S1, Figure S2 and Table S1.
The ERRα-LBD sample and the control sample were analyzed by untargeted liquid chromatography-mass spectrometry lipidomics. These lipidomics datasets were then compared using the XCMS software package (Smith et al., 2006), which quantifies the levels of metabolites in two samples to reveal any differences in lipid concentrations. XCMS identified a single ion that was significantly enriched by the beads bound to ERRα-LBD (p<0.05 and >2-fold), and this ion belongs to cholesterol (Figure 1B). This data indicate that the ERRα-LBD binds cholesterol. We have previously performed this affinity chromatography experiment with three different proteins (FABP, CRABP, StarD3) (Tagore et al., 2008) and three different nuclear receptors (PPARγ, PPARα and Nurr77) (Kim et al., 2011b; Vinayavekhin and Saghatelian, 2011). StarD3, a known cholesterol-binding protein, was the only one of these proteins or nuclear receptors to enrich cholesterol, demonstrating that this experiment is not biased towards cholesterol enrichment to support a specific interaction between ERRα-LBD and cholesterol.
We wanted to test the specificity of the ERRa-cholesterol interaction by repeating the affinity chromatography with tissue sterols. This required fractionation of the sterols away from other lipids using a validated method (McDonald et al., 2007). This sterol fraction from brain or kidney was then used for affinity chromatography with resin loaded with ERRa-LBD as described above. In addition, we used a targeted LC-MS method that is specific for sterols (i.e. cholesterol, oxysterols, etc.) (McDonald et al., 2007) to increase our detection sensitivity for lower abundance sterols. We measured 19 different sterols with this approach and only detected the enrichment of cholesterol (4.6- and 6.7-fold for the brain and kidney samples, respectively) (Figure 1C, S1A, Table S1). This data is consistent with the lipidomics experiment. Next we performed a competitive binding assay with diethylstilbestrol (DES), a synthetic ERRα antagonist that binds to the ERRα lipid-binding pocket (Tremblay et al., 2001), to test the specificity of the cholesterol interaction with the ERRα-LBD. DES was added to the brain lipidome before affinity chromatography with the ERRα-LBD beads. DES prevented enrichment of cholesterol (Figure 1D) by binding to the ERRα-LBD (Figure S1B), which indicates that cholesterol binds to the ERRα-LBD lipid-binding pocket.
To complement these affinity chromatography experiments, we further characterized the biochemical interaction between cholesterol and ERRα-LBD using several secondary assays. First, circular dichroism (CD) spectroscopy showed that cholesterol, as well as synthetic ERRα antagonists DES and XCT790, all induced conformational changes in ERRα-LBD; whereas estradiol, which does not bind to ERRα-LBD, did not (Figure S2). This change in conformation implies that all of these molecules can bind to the ERRα-LBD and the fact that estradiol, a lipid with similar biophysical properties, does not cause a change, supports that this binding is more than a simple hydrophobic interaction.
Second, computational docking of cholesterol into the ERRα ligand-binding pocket suggested that cholesterol bound with its hydroxyl group facing into the ligand-binding pocket (Figure 2A). We tested this model using a fluorescence polarization experiment with dye labeled cholesterol derivatives and site-directed mutagenesis. Fluorescence polarization indicated that 25-[N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)methyl]amino]-27-norcholesterol (25-NBD-cholesterol), a cholesterol derivative with a dye extending from carbon 25 of the norcholesterol, bound ERRα-LBD with a dissociation constant of 9 µM. According to the structural model (Figure 2A), carbon 25 is outside of the ERRα-LBD lipid-binding pocket and therefore should not interfere with binding, which is what we observed. By contrast, 6-NBD-cholesterol, which would place the NBD directly inside the binding pocket, did not bind the ERRα-LBD at all (Figure 2B). Furthermore, cholesterol, but not estradiol, was able to compete with 25-NBD-cholesterol for binding to ERRα-LBD (Figure 2C).
Figure 2. Characterization of cholesterol binding to the ERRα-LBD.
(A) Molecular docking simulations suggested that cholesterol bound to ERRα in the ligand binding pocket with the hydroxyl group facing into the ligand binding pocket. In this model, cholesterol makes important interactions with three residues: E235, F232, and L228 (highlighted in blue). (B) Fluorescent cholesterol derivatives demonstrated that cholesterol bound with its hydroxyl group facing into the ligand binding pocket. Labeling at the alkyl but not hydroxyl group with an NDB fluorophore permitted ERRα-cholesterol binding. (C) Competition fluorescence polarization assay with increasing amount of cholesterol and estradiol indicates that only cholesterol was able to compete with 25-NBD-cholesterol in binding to ERRα-LBD. (D) Cholesterol binding quenches the intrinsic tryptophan fluorescence intensity of ERRα-LBD, while estradiol, which is known to not bind ERRα, has no effect on ERRα tryptophan fluorescence. (E) A E235A, F232A, L228A ERRα triple mutant had impaired cholesterol binding ability relative to wild type ERRα. Error bars, SD.
See also Figure S3.
Third, to ensure that the NBD is not contributing to the binding, we performed a tryptophan fluorescence spectroscopy assay with cholesterol. In this experiment, binding of cholesterol to the ERRα-LBD quenches intrinsic tryptophan fluorescence of the ERRα-LBD, to afford a label-free method for testing binding. In contrast, estradiol did not quench the intrinsic tryptophan fluorescence of ERRα-LBD (Figure 2D).
Lastly, the model predicts a hydrogen bond between E235 and the cholesterol hydroxyl group, and F232 and L228 also appear to make important hydrophobic contacts with cholesterol (Figure 2A). Site-directed mutagenesis of ERRα-LBD at E235A, F232A and L228A generated a mutant ERRα-LBD that appeared folded properly by CD (Figure S3), but displayed a greatly reduced cholesterol binding (Figure 2E), indicating that disruption of the lipid-binding pocket interferes with cholesterol binding. Together, these experiments provide strong evidence of cholesterol binding to the ERRα-LBD, but do not indicate whether this binding is functional.
Cholesterol increases ERRα transcriptional activity
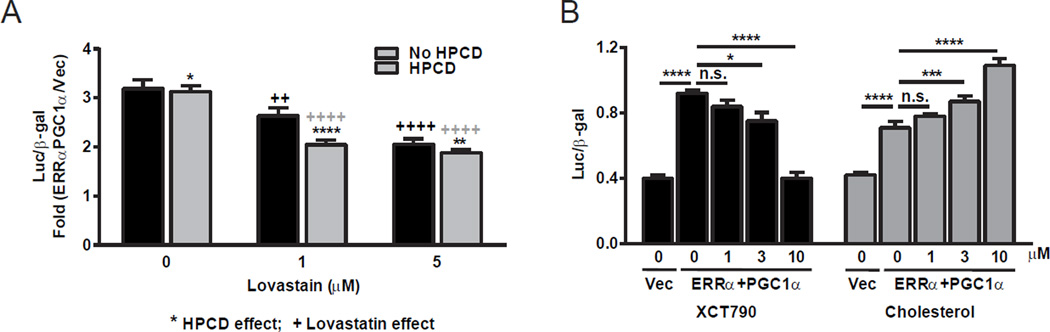
To determine whether cholesterol is a functional endogenous ERRα ligand in cells, we next examined the impact of modulating the cholesterol biosynthetic pathways on ERRα transcriptional activity. Conventional luciferase transactivation strategies, where a ligand is added to detect an increase in transcriptional activity, are complicated by the presence of abundant cholesterol in the media and cells. Rather than introducing cholesterol, we decided that a more prudent strategy would be to measure ERRα activity upon sterol depletion. We lowered intracellular sterol levels in three ways: the use of lipid free serum, the addition of the cholesterol-binding hydoxypropyl cyclodextrin (HPCD), and the addition of the lovastatin. The individual addition of HPCD or statin was able to attenuate ERRα transactivation (Figure 3A), indicating that sterol levels positively correlate with ERRα activity, as expected for an agonistic metabolic pathway that activates ERRα. Moreover, when combined, these compounds also had an additive effect on the lowering of ERRα activity (Figure 3A). Importantly, adding back cholesterol to these HPCD- and statin-treated samples rescued ERRα activity in a dose-dependent manner, indicating that cholesterol or a downstream sterol, but not a precursor of the cholesterol pathway, is involved in ERRα activity (Figure 3B). As a positive control, the synthetic ERRα antagonist/inverse agonist XCT790 further decreased ERRα transactivation (Figure 3B). As negative controls, these effects were not observed for ERRβ, ERRγ or other orphan nuclear receptors such as Nur77 (Figure S4). Importantly, the effects of cholesterol depletion and cholesterol add-back on ERRα activation of transcription were only observed in the presence of ERRα coactivators such as PGC1α (Figure 3A–B); in the absence of PGC1α, neither ERRα nor cholesterol could enhance the transcription (not shown), indicating that cholesterol activation of ERRα function is PGC1α-dependent. These results suggest that cholesterol is a functional ERRα agonist.
Figure 3. Cholesterol increases ERRα transcriptional activity.
ERRα transcriptional activity was analyzed using transient transfection and reporter assays. (A) Cholesterol depletion by lovastatin and/or the cholesterol binder hydroxypropyl-beta-cyclodextrin (HPCD) results in lower ERRα transcriptional activity. Results are shown as fold induction by ERRα+PGC1α transfection compared to vector (vec) transfection control (n=6). * indicates HPCD effect; + indicates lovastain effect. (B) The addition of cholesterol (blue bars) back to cholesterol-depleted cells dose-dependently rescues ERRα activity; whereas XCT790 (brown bars), a known ERRα antagonist and thus a control for this assay, further suppressed ERRα activity dose-dependently (n=6). CV-1 cells were transfected with expression vectors for ERRα and PGC1α co-activator as well as ERRE-luc and CMV-βgal reporters. For cholesterol depletion, the cells were treated with lovastatin at indicated concentration and HPCD (10mM) for 4hrs in DMEM containing 10% NCLPPS (newborn calf lipoprotein poor serum), and then with lovastatin in DMEM containing 10% NCLPPS for another 20hrs. For cholesterol additions to cells, cholesterol-depleted cells were treated with cholesterol or vehicle during the 20hrs of lovastatin treatment, or with XCT790 as a control. Error bars, SD.
See also Figure S4.
ERRα mediates statin-induced muscle toxicity
On the basis of these findings, we investigated whether the cholesterol pathway and ERRα are functionally dependent. We examined skeletal muscle, a well-known ERRα-regulated tissue. Statins are the most efficient drugs for the treatment of hypercholesterolemia that is generally considered safe and well tolerated. However, the most severe adverse effect of statins is myotoxicity, for which there is still no clear pathophysiology or effective treatment (Echaniz-Laguna et al., 2010; Sathasivam, 2012; Tomaszewski et al., 2011). It has been reported that statins induce myopathy by inhibiting myotube differentiation and inducing the expression of the muscle atrophy gene atrogin-1 (Fbxo32) (Hanai et al., 2007). Thus, we examined whether the effects of statin on muscle are ERRα-dependent in vitro and in vivo. Treatment of C2C12 myotube differentiation cultures with either lovastatin or simvastatin significantly suppressed myotube formation, decreased the expression of myotube markers, and increased the expression of Fbxo32 (Figure 4A–C). Treatment with the ERRα antagonist XCT790 exhibited similar effects (Figure 4A–C). This indicates that either a reduction of endogenous ERRα agonists by sterol depletion or an inhibition of ERRα activity by XCT790 leads to myotube defects. The effects of statins were largely rescued by adding back cholesterol to the cultures, but this rescue was absent when ERRα function was blocked by XCT790 (Figure 4A–C), indicating that regulation by cholesterol is ERRα-dependent. These observations may extend beyond C2C12 cells since statin can induce toxicity in many cell types.
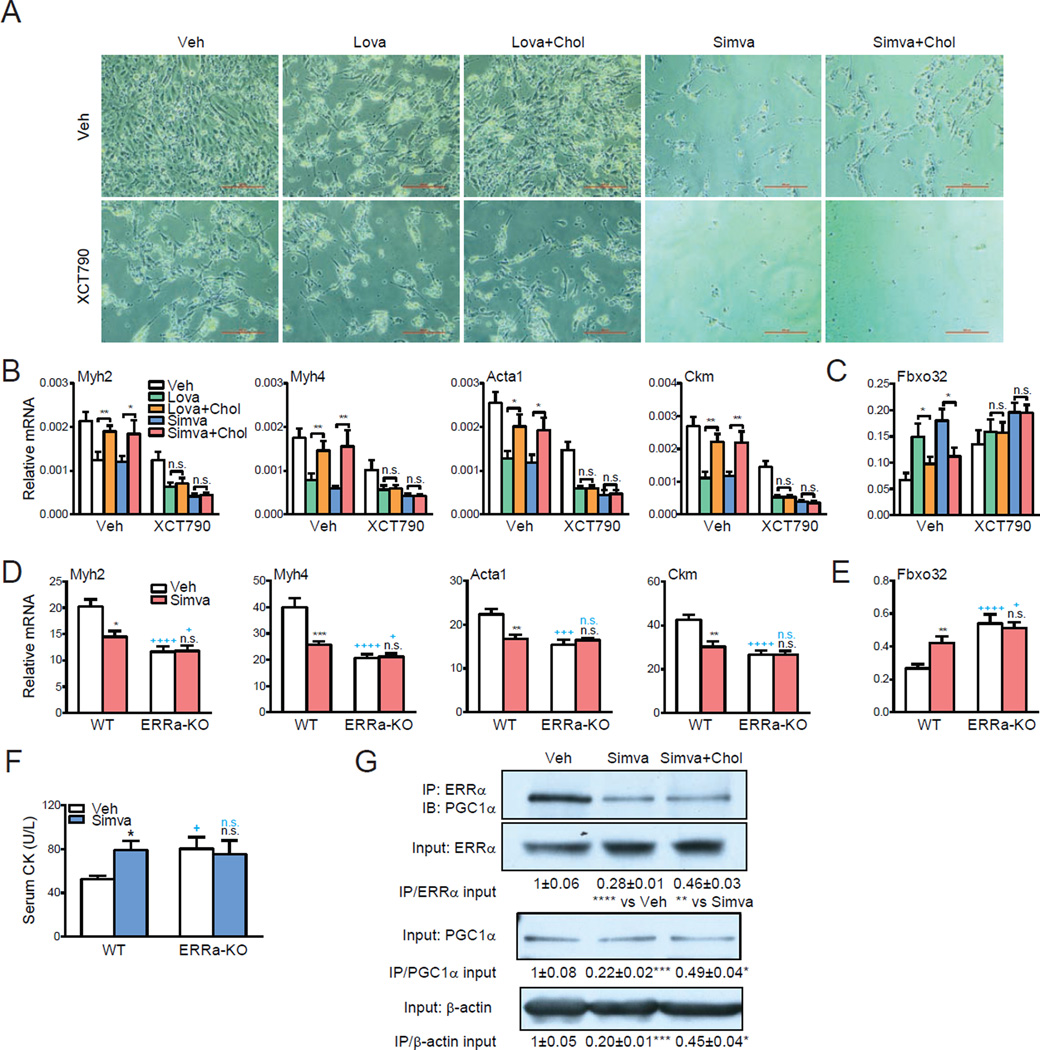
Figure 4. ERRα mediates statin-induced muscle toxicity.
(A–B) Effects of statins, cholesterol and XCT790 on C2C12 myotube differentiation. Lovastatin (Lova), simvastatin (Simva), cholesterol and XCT790 were added at 10µM. (A) Representative images of C2C12 differentiation cultures on day 5. Scale bar, 25µm. (B) mRNA expression of myotube differentiation markers on day 5 (n=3). (C) mRNA expression of the muscle atrophy gene atrogin-1 (Fbxo32) on day 5 (n=3). (D–F) Statin-induced muscle toxicity is abolished in ERRαKO mice. ERRαKO mice or WT controls (6 month old, females, n=6) were treated with simvastatin (Simva) for 2 weeks at 20mg/kg/day. (D) mRNA expression of myotube differentiation markers in quadriceps muscle (n=6). (E) mRNA expression of the muscle atrophy gene atrogin-1 (Fbxo32) in quadriceps muscle (n=6). (F) Serum muscle damage marker creatine kinase (CK, n=6). Black * and n.s. compare statin with vehicle (Veh) control; blue + and n.s. compare ERRαKO with WT control. (G) Co-immunoprecipitation analysis of the effects of statin and cholesterol on ERRα and PGC1α interaction. C2C12 myotube differentiation cultures were treated with vehicle, simvastatin or simvastatin+cholesterol at 10µM for 24 hr. Average results for the IP/Input ratio from 3 independent experiments are shown. Error bars, SD.
See also Figure S5 and S7.
We next treated ERRα-KO mice or WT controls with simvastatin (20mg/kg/day) or vehicle control for 2 weeks. Both simvastatin-treated WT mice and vehicle-treated ERRα-KO mice exhibited myopathy compared with vehicle-treated WT controls, shown by the decreased expression of myotube markers (Figure 4D) and increased expression of Fbxo32 in skeletal muscles (Figure 4E), as well as the elevated creatine kinase levels in serum, a biomarker for muscle damage (Echaniz-Laguna et al., 2010) (Figure 4F). Importantly, the myopathy-inducing effects of statin were completely abolished in ERRα-KO mice (Figure 4D–F). In line with these observations, ERRα association with its coactivator PGC1α was inhibited by statin, which was partially rescued by cholesterol in C2C12 myotube differentiation cultures (Figure 4G), suggesting that cholesterol binding to ERRα causes a conformational change in the LBD and the recruitment of coactivator to enhance ERRα-mediated transcription. Together, these findings indicate that physiological regulation by cholesterol and pharmacological regulation by statins are ERRα-dependent, supporting that the cholesterol pathway exerts a functional control of ERRα activity.
ERRα mediates cholesterol, statin and bisphosphonate regulation of osteoclastogenesis
To further investigate the functional role of cholesterol as an endogenous ERRα ligand, we examined osteoclastogenesis, a cellular differentiation process crucial for the physiological regulation of bone remodeling and skeletal regeneration, which also contributes to diseases such as osteoporosis and cancer bone metastases when overactive. Upon activation by receptor activator of nuclear factor kappa-B ligand (RANKL), osteoclasts are differentiated from monocyte/macrophage precursor cells in the blood circulation and bone marrow (Novack and Teitelbaum, 2008). We have recently shown that ERRα promotes osteoclast differentiation and activity, as a result, ERRα knockout mice exhibit decreased bone resorption and high bone mass (Wei et al., 2010). Interestingly, both statins and nitrogen-containing bisphosphonates suppress osteoclast function; and both inhibit the cholesterol synthesis pathway, by blocking the HMG-CoA reductase and farnesyl diphosphate synthase (FPPS), respectively (Russell, 2011; Toledano and Partridge, 2000). However, the molecular target for how exactly the cholesterol synthesis pathway impacts osteoclastogenesis is still an open question, although interference with protein prenylation by bisphosphonates has been suggested (Russell, 2011).
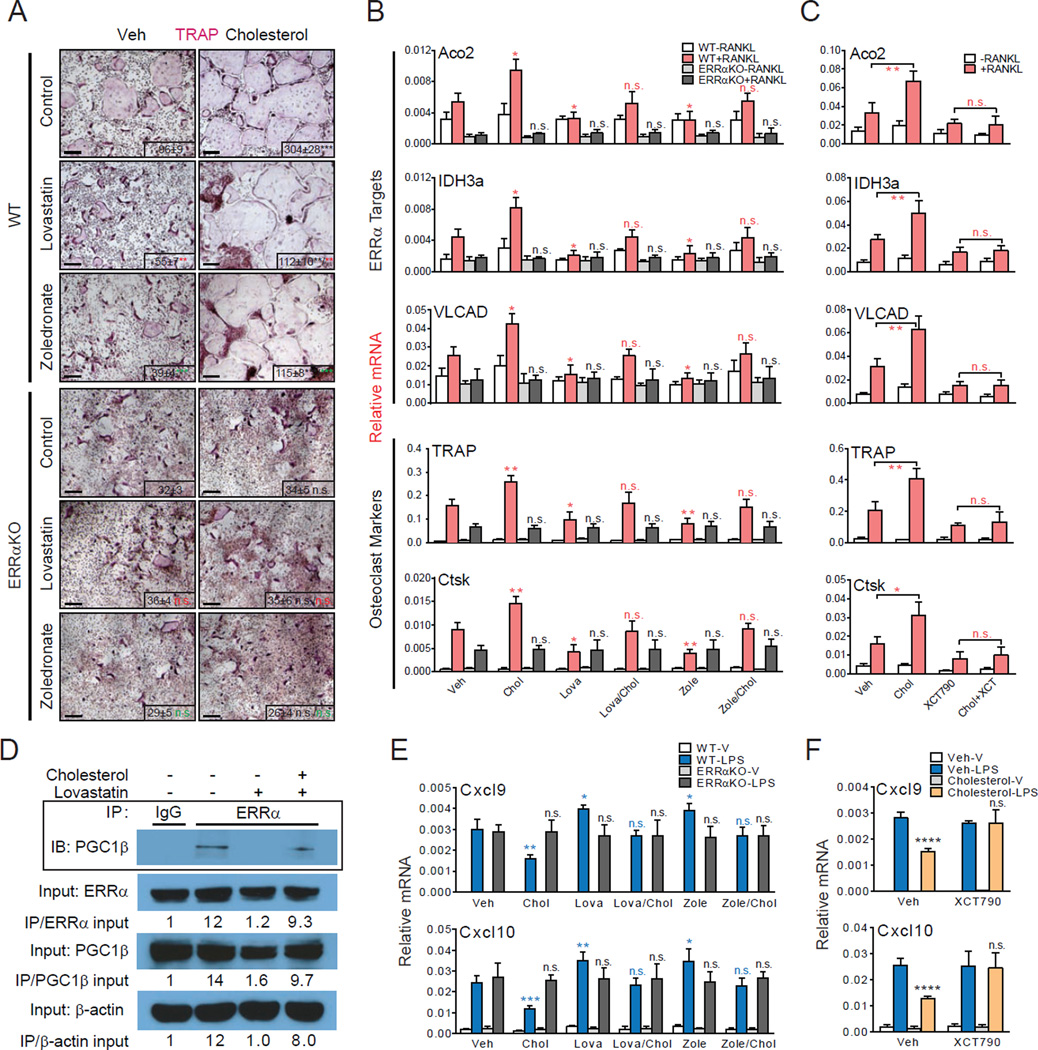
To test the hypothesis that cholesterol, statins and bisphosphonates regulate osteoclast differentiation via ERRα, we performed ex vivo bone marrow osteoclast differentiation assays. We found that osteoclastogenesis from WT bone marrow cells was dependent on the presence or absence of cholesterol. Cholesterol addition enhanced osteoclast formation, whereas cholesterol depletion by treatment with zoledronate (a clinically used nitrogen-containing bisphosphonates) or lovastatin inhibited osteoclast formation (Figure 5A). Cholesterol adding back to zoledronate- or lovastatin-treated cells was able to rescue osteoclast differentiation, indicating cholesterol or a downstream sterol, rather than cholesterol precursors in the biosynthetic pathway, was responsible for the changes in differentiation (Figure 5A). Gene expression analysis showed that cholesterol up-regulated the expression of both ERRα target genes that stimulate mitochondrial biogenesis and osteoclast activity, such as Aco2, IDH3a and VLCAD (Wei et al., 2010), and osteoclast differentiation markers such as TRAP (tartrate-resistant acid phosphatase) and Ctsk (Cathepsin K) (Figure 5B).
Figure 5. ERRα mediates cholesterol stimulation of osteoclastogenesis ex vivo.
(A–B) Cholesterol promotes whereas statin and bisphosphonate inhibit osteoclastogenesis in WT but not ERRαKO bone marrow osteoclast differentiation cultures. (A) Representative images of TRAP-stained differentiation cultures showing the number and size of mature osteoclasts. Mature osteoclasts were identified as multinucleated (>3 nuclei) TRAP+ (purple) cells. Scale bar, 25µm. Number of osteoclast/well is shown as insets; black * or n.s. compare cholesterol vs. veh, red * or n.s. compare lovastatin vs. control, green * or n.s. compare zoledronate vs. control. (B) Quantification of the mRNA expression of ERRα target genes and osteoclast differentiation markers. P-values compare each treatment condition with vehicle control in the same genotype. (C) Cholesterol induction of ERRα target genes and osteoclast differentiation markers in WT bone marrow osteoclast differentiation cultures was abolished by the treatment of an ERRα antagonist XCT790. Chol, cholesterol (10µM); lova, lovastatin (10µM); zole, zoledronate (20µM); ctrl, control; XCT, XCT790 (10µM). (D) Co-immunoprecipitation analysis of the effects of statin and cholesterol on ERRα and PGC1β interaction. Bone marrow osteoclast differentiation cultures were treated with RANKL for 6 days in the presence of lovastatin (10nM), cholesterol (2µM) or vehicle control. Representative results from three independent experiments are shown. (E) Cholesterol inhibits whereas statin and bisphosphonate enhance cxcl9 and cxcl10 expression in WT but not ERRαKO macrophages. Macrophages were differentiated from bone marrow cells of WT or ERRαKO mice with 20ng/ml MCSF for 8 days, in the presence of compound or vehicle, and then treated with 200µg/ml LPS for 6hr at the end. P-values compare each treatment condition with vehicle control in the same genotype. (F) Cholesterol suppression of cxcl9 and cxcl10 in WT macrophages was abolished by XCT790. Chol, cholesterol (10µM); lova, lovastatin (10µM); zole, zoledronate (20µM); ctrl, control; XCT, XCT790 (10µM). Error bars, SD.
See also Figure S5 and S6.
Similar experiments using ERRα knockout (ERRαKO) bone marrow osteoclast differentiation cultures reveal that the effects of cholesterol on osteoclastogenesis were completely abolished by ERRα deletion. In the absence of ERRα, osteoclast differentiation was neither enhanced by cholesterol nor suppressed by lovastatin or zoledronate (Figure 5A). Consistent with this observation, cholesterol, lovastatin or zoledronate no longer altered the expression of ERRα target genes or osteoclast differentiation markers (Figure 5B). In addition to genetic loss-of-function by ERRα deletion, biochemical inhibition with a synthetic ERRα antagonist XCT790 also prevented the effects of cholesterol on WT osteoclast differentiation (Figure 5C). The suppressive effects of XCT790 on ERRα target genes and osteoclast differentiation makers were absent in ERRαKO cultures, indicating that XCT790 mainly targeted ERRα (Figure S5A). These data indicate that cholesterol enhances osteoclastogenesis in an ERRα-dependent manner.
In osteoclasts, PGC1β is the ERRα coactivator, whereas PGC1α is not expressed (Wei et al., 2010). Once again, ERRα association with PGC1β was inhibited by statin, which was largely rescued by cholesterol add-back in osteoclast differentiation cultures (Figure 5D). Expression of ERRα and PGC1 was unaffected by statin or cholesterol under these conditions (Figure S5B–C). In conjunction with biochemical data demonstrating cholesterol binding to ERRα LBD and cellular reporter assays demonstrating cholesterol stimulation of ERRα transcriptional activity, these results suggest that cholesterol promotes osteoclastogenesis by functioning as an ERRα agonist, whereas statins and bisphosphonates suppresses osteoclastogenesis by reducing the bioavailability of the endogenous ERRα agonist cholesterol.
These clear phenotypic outcomes in cells provided us an opportunity to pin down the altered sterol species that modulate ERRα activity. Specifically, we measured sterol levels under different conditions during osteoclastogenesis using the highly sensitive sterol LC-MS measurements. Any sterol that activates ERRα must 1) have higher levels in cholesterol-treated cells, 2) have lower levels in lovastatin- and zoledronate-treated cells, and 3) have a level rescued back to normal control when cholesterol is added back to lovastatin- and zoledronate-treated cells. Although many sterols were detected and measured, cholesterol was the only sterol fitting all 3 criteria whose cellular levels correlated with changes in ERRα activity in response to statin and bisphosphonate (Figure S6). In combination with the biochemical characterization of ERRα-LBD as a cholesterol-binding protein, these cellular data supports the notion that cholesterol is an endogenous ERRα agonist. Although there may be additional, undetected or unknown, metabolites downstream of cholesterol, these results indicate that cholesterol is the major sterol in the cholesterol pathway that modulates ERRα activity.
ERRα activity. ERRα mediates cholesterol, statin and bisphosphonate regulation of macrophage
To examine the functional association between ERRα and cholesterol in the myeloid lineage under a different biological context, we next investigated whether the recently reported anti-inflammatory role of cholesterol in macrophages was also ERRα-dependent. Cholesterol has been shown to inhibit macrophage expression of chemokines Cxcl9 and Cxcl10 (Spann et al., 2012). We found that the LPS-induced Cxcl9 and Cxcl10 expression was suppressed by cholesterol but exacerbated by lovastatin or zoledronate in WT macrophages (Figure 5E). These effects were once again abolished in ERRαKO macrophages (Figure 5E). Furthermore, cholesterol inhibition of Cxcl9 and Cxcl10 expression was also absent in WT macrophages treated with XCT790 (Figure 5F). Similar regulation was observed for other inflammatory markers such as IL-1β and MMP9 (Figure S5D–E). In contrast, cholesterol regulation of SREBP and LXR target genes were intact in ERRαKO macrophages (Figure S5F). These data not only indicate that the chemokine-suppressive effect of cholesterol in macrophages is ERRα-mediated but also reveal a novel anti-inflammatory role for ERRα in a cholesterol rich environment. Importantly, these results provide further evidence that cholesterol is a functional ERRα agonist in another biological process.
ERRα mediates cholesterol regulation of bone resorption in vivo
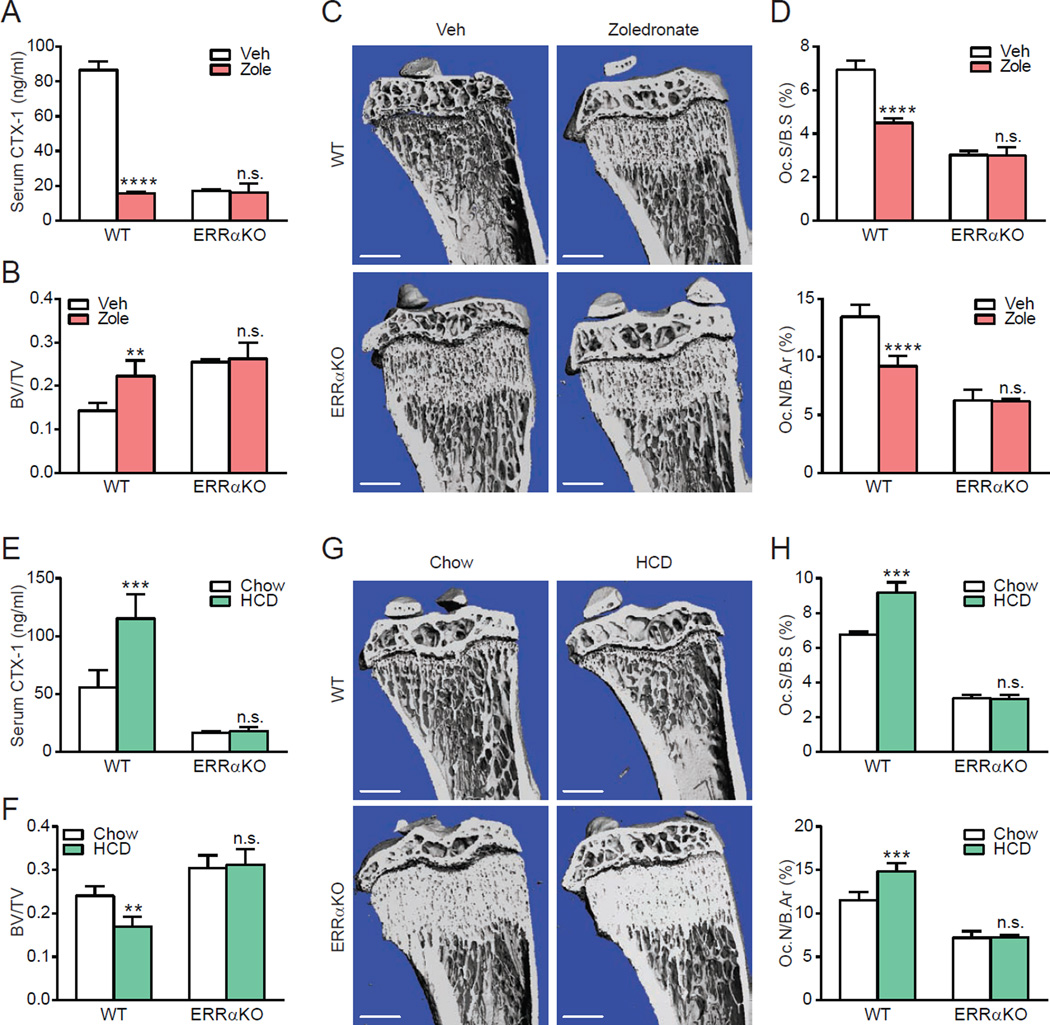
To gain further insight to whether the functional relationship between cholesterol and ERRα extends to physiology and pharmacology, we next performed in vivo studies in the context of bone resorption and skeletal remodeling. Epidemiological and pharmacological studies in human show that cholesterol-lowering drugs such as bisphosphonates and statins are bone protective (Walsh et al., 2012), whereas hypercholesterolemia is associated with bone loss (Orozco, 2004; Tarakida et al., 2011). As a loss-of-cholesterol-function approach, we treated WT or ERRαKO mice with zoledronate, which targets bone more efficiently than statins, and compared them to vehicle-treated mice 4 weeks later. ELISA analysis showed that zoledronate significantly decreased the serum levels of the bone resorption marker C-terminal telopeptide fragments of the type I collagen (CTX-1) in WT mice; in contrast, this effect was completely abolished in the ERRαKO mice (Figure 6A). Moreover, CTX-1 was reduced to a similar extent by both cholesterol ligand depletion in the zoledronate-treated mice (−82%) and by receptor deletion in the ERRαKO mice (−80%) (Figure 6A). In line with these results, microCT analysis of the proximal tibiae shows that zoledronate significantly increased bone mass in WT mice but not in ERRαKO mice (Figure 6B–C). Furthermore, bone histomorphometry analysis showed that zoledronate significantly reduced osteoclast surface and osteoclast number in WT mice but not in ERRαKO mice (Figure 6D). Serum cholesterol was reduced to a similar extent in both WT and ERRαKO mice (Figure S7A).
Figure 6. ERRα mediates cholesterol stimulation of bone resorption in vivo.
(A–D) Osteoprotective effects of zoledronate were abolished in ERRαKO mice. WT or ERRαKO mice (6 week old male, n=4) were treated with a single i.v. injection of zoledronate at 0.54 mg/kg or PBS vehicle control and analyzed 4 weeks later. (A) Serum levels of the bone resorption marker CTX-1. (B–C) MicroCT analysis of tibiae. (B) Trabecular bone volume/tissue volume ratio (BV/TV). (C) Representative CT images of the entire proximal tibia (scale bar, 1mm). (D) Bone histomorphometry analysis of osteoclast surface/bone surface (Oc.S/BS) (top) and osteoclast number/bone area (Oc.N/B.Ar) (bottom). (E–H) Bone loss induced by high cholesterol diet (HCD) feeding was abolished in ERRαKO mice. WT or ERRαKO mice (7 week old female, n=4) were fed with a HCD or chow control diet for 4 weeks. (E) Serum levels of the bone resorption marker CTX-1. (F–G) MicroCT analysis of tibiae. (F) Trabecular BV/TV. (G) Representative CT images of the entire proximal tibia (scale bar, 1mm). (H) Bone histomorphometry analysis of Oc.S/BS (top) and Oc.N/B.Ar (bottom). Error bars, SD.
See also Figure S7.
Complementarily, as a gain-of-cholesterol-function approach, we fed WT or ERRαKO mice with a high-cholesterol-diet (HCD) for 4 weeks and compared them to chow-diet fed control mice. ELISA analysis showed that HCD feeding significantly elevated serum CTX-1 levels by 107% in WT mice, once again, this effect was completely abolished in the ERRαKO mice (Figure 6E). In agreement, microCT analysis of the proximal tibiae shows that HCD feeding significantly decreased bone mass in WT mice but not in ERRαKO mice (Figure 6F–G). Furthermore, bone histomorphometry analysis showed that HCD feeding significantly increased osteoclast surface and osteoclast number in WT mice but not in ERRαKO mice (Figure 6H). In accordance with these observations, the expression of previously described ERRα target genes in osteoclast and muscle (Huss et al., 2004; Wei et al., 2010) was significantly increased by HCD feeding in an ERRα-dependent manner (Figure S7B–C). These in vivo studies indicate that the physiological function of cholesterol and the pharmacological actions of cholesterol-lowering drugs require ERRα, further supporting the notion that cholesterol is a functional endogenous ERRα agonist.
DISCUSSION
Here we identify and characterize cholesterol as a natural ERRα ligand using an integrated strategy that combines analytical chemistry, cell biology and animal physiology. The classic nuclear receptors are known as the receptors for steroids such as estrogen, androgen, glucocorticoid and progesterone, which are derivatives of cholesterol. Though cholesterol is not thought of a signaling molecule, cholesterol levels affect the activity of important transcription factors such as SREBPs via SCAP (Brown and Goldstein, 1997). Our binding model does not exclude other sterols with modifications outside of the binding site, and therefore it is possible that other sterols can also bind ERRα. Two pieces of our data support cholesterol as the major endogenous sterol ligand. First, the unbiased affinity chromatography experiments only enriched cholesterol. Other sterols should have been enriched if they could bind to ERRα in this mixture. Second, cholesterol was the only sterol with levels that correlated with the transcriptional activity of ERRα under all cellular treatment conditions we tried (Figure S6). As debates about the endogenous ligands of other nuclear receptors demonstrate, it is not possible to rule out additional ligands for ERRα, but our data provides the first evidence for cholesterol and the cholesterol biosynthetic pathway in the regulation of ERRα activity and biology.
Our results show that full length endogenous ERRα recruits PGC1 coactivators upon cholesterol addition but dissociates from PGC1 coactivators upon cholesterol depletion (Figure 4G, 5D), supporting a functional binding of cholesterol to ERRα in cells. Our data show that cholesterol enhances ERRα interaction with PGC1β in osteoclasts (Figure 5D), although the effects of cholesterol to increase ERRα interaction with PGC1α in myocytes (Figure 4G) were less discernible due to the limitation of statin-induced toxicity in these cells.
Moreover, computational docking of cholesterol into the ERRα ligand-binding pocket, fluorescence cholesterol labeling and site-directed mutagenesis suggest that cholesterol binds with its hydroxyl group facing into the ligand-binding pocket. Because the ligand-binding pockets of ERRα and other orphan nuclear receptors can adopt alternative conformations to bind molecules, these docking studies with the apo ligand-binding-domain, although informative, should be interpreted with caution.
Previous studies indicate that ERRα functions differently in many ways from classic nuclear receptors, and our identification of cholesterol as an endogenous ERRα agonist provides mechanisms for its unique property. First, the ubiquity of cholesterol explains why ERRα is constitutively active. Second, the ability of cholesterol to recruit coactivators PGC1α/β to ERRα explains why ERRα activity is highly dependent on PGC1α/β. Third, because cholesterol is essential for cellular function, cholesterol depletion can only be partially achieved (Figure S6); therefore, the effects of cholesterol depletion and cholesterol add-back on ERRα transcriptional outputs in the reporter assays were modest yet highly significant (Figure 3). These findings reveal ERRα as a metabolic-sensing nuclear receptor and a homeostatic rheostat, and distinguishing it from steroid receptors that respond to acute and steep rise in hormonal levels.
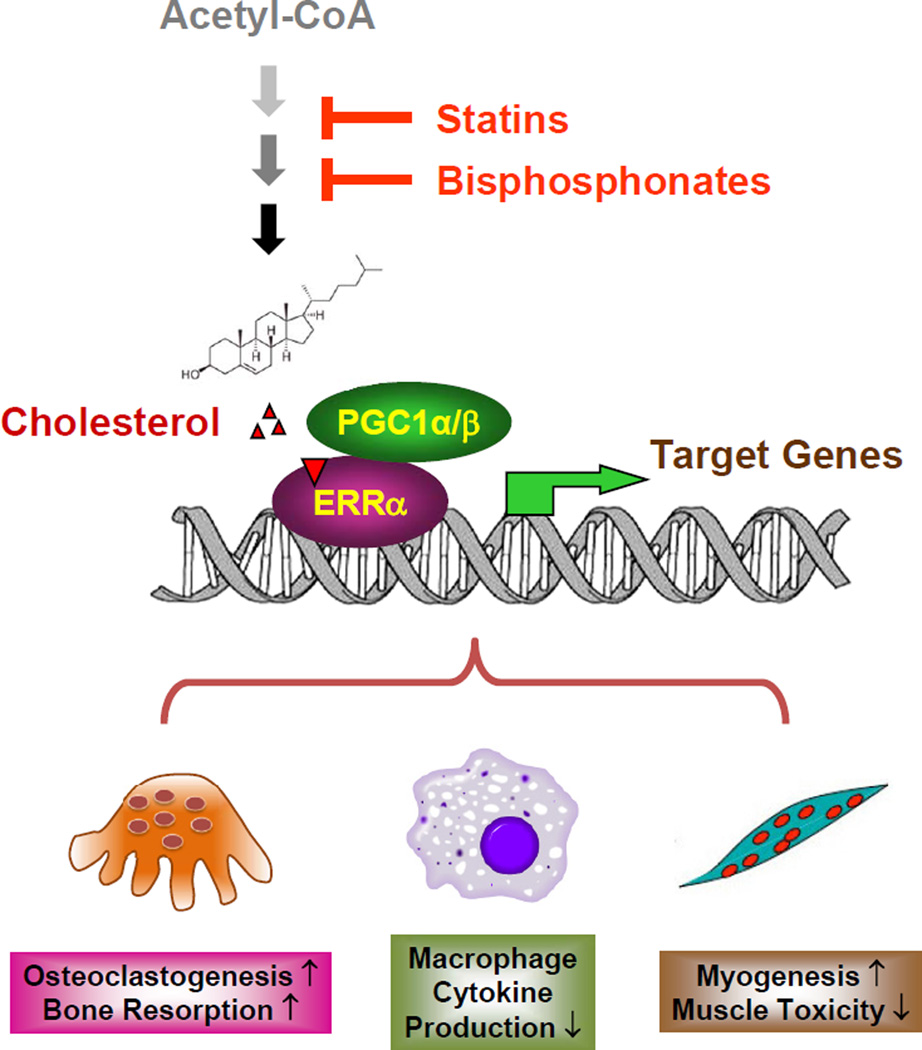
This discovery also elucidates a key mechanistic understanding for how ERRα serves as a critical metabolic and cholesterol sensor: cholesterol promotes osteoclastogenesis and myogenesis, as well as suppresses macrophage cytokine production, by activating its receptor ERRα; and ERRα regulates these cellular processes by responding to a rise or fall in cellular cholesterol level (Figure. 7). In macrophages, it is possible that cholesterol activation of ERRα may induce a transcriptional repressor for Cxcl9 and Cxcl10. Moreover, our findings illuminate new mechanistic insights for how clinically used drugs such as statins and bisphosphonates actually act by identifying ERRα as an essential mediator: both statin-induced muscle toxicity and bisphosphonate-induced osteoprotection result from a reduction of cholesterol synthesis and consequently decreased ERRα activity (Figure. 7). Particularly, our results question the long-standing hypothesis that the inhibition of osteoclast activity by bisphosphonates is solely due to the blockade of protein prenylation. Our results show that receptor loss by ERRα deletion and ligand depletion by statin (Figure 4) or bisphosphonate (Figure 6) led to a similar degree of serum CTX-1 reduction and serum CK induction; moreover, in the absence of the receptor ERRα, these effects by ligand depletion were completely abolished. Together, these findings suggest that the pharmacological actions of statins and bisphosphonates are ERRα-dependent.
Figure 7. A schematic diagram of how ERRα mediates the effects of cholesterol, statins and bisphosphonates.
Cholesterol serves as a natural ERRα agonist to recruit coactivators PGC1α/β and increase ERRα transcriptional activity, thereby promoting osteoclastogenesis and myogenesis but inhibiting macrophage cytokine production. A reduction in cholesterol synthesis by statins or bisphosphonates decreases ERRα transcriptional activity, thereby mediating the statin-induced muscle toxicity and bisphosphonate suppression of bone resorption.
In support of a negative impact of cholesterol on bone mass, epidemiological and pharmacological studies show that cholesterol-lowering drugs such as bisphosphonates and statins are bone protective in human (Walsh et al., 2012), whereas hypercholesterolemia is associated with bone loss (Orozco, 2004; Tarakida et al., 2011). Rare genetic disorders of cholesterol biosynthesis, however, have been associated with higher bone density. Desmosterolosis is caused by a mutation in the gene encoding 24-dehydrocholesterol reductase, which causes a marked accumulation of desmosterol but normal or slightly reduced cholesterol (Andersson et al., 2002; FitzPatrick et al., 1998; Schaaf et al., 2011). This is distinct from statins, which inhibit HMG CoA reductase and cause a generalized reduction in sterols including cholesterol and desmosterol. Thus, the osteosclerosis observed in 1 out of 3 desmosterolosis patients identified to date may be caused by an elevated desmosterol rather than decreased cholesterol. Antley-Bixler syndrome is caused by either mutations in the FGFR2 gene or mutations in the cytochrome p450 oxydoreductase (POR) gene. The latter results in a broad spectrum abnormality in steroidogenesis and hormonal disorders (Fluck et al., 2004), which may explain the craniosysnostosis phenotype.
Our conclusion of the ERRα-dependency of cholesterol regulation is supported by the consistency between ERRα-KO genetic model and XCT790 ERRα antagonist chemical model. Moreover, the effects of XCT790 we observed were completely abolished in ERRα-KO cultures (Figure S5A), demonstrating that ERRα is the major target of XCT790 in our system. Interestingly, a recent report (Eskiocak et al., 2014) showed that XCT790 can function as a mitochondrial uncoupler independent of ERRα in a melanoma cell line using siRNA ERRα knockdown; however, this in vitro observation was highly dependent on the cell lines, thus a context-specific effect that may not be extendable to generalized physiological settings.
The impairment of cholesterol, statin and bisphosphonate regulation in ERRα-KO mice was not due to a saturation of the phenotype. ERRα deletion or inhibition reduced baseline osteoclast and myotube differentiation, but did not completely abolish the ability to differentiate; for example, ERRα-KO osteoclast cultures indeed still showed induction of differentiation markers TRAP and Ctsk upon RANKL treatment (which is dependent on its receptor RANK and modulated by ERRα) (Figure 5B, bottom); only the induction of ERRα target genes were completely abolished (Figure 5B, top). Moreover, previous studies from our lab and others have shown that many other genetic defects can cause much more severe osteopetrotic phenotype, with a BV/TV ratio reaching 0.8 (Kong et al., 1999; Wei et al., 2011; Xing et al., 2013), more than 2-fold higher than the BV/TV of ~0.3 in ERRα-KO mice, indicating that the osteoclast and bone phenotype in ERRα-KO mice is by no means saturated. In addition, glucocorticoid-induced myopathy was intact in ERRα-KO, demonstrating that ERRα-KO mice remain sensitive to ERRα-independent stimuli (Figure S7D). Therefore, the inability of ERRα-KO cells and mice to respond to statin, bisphosphonate and cholesterol support a specific requirement of ERRα in the regulation by the cholesterol pathway.
Interestingly, there is a crosstalk between ERRα and ERα, including recognition of the estrogen response element (ERE) by ERRα. In ER negative (_ER_-) breast cancer, the presence of ERRα is a negative prognostic factor as it compensates for the loss of ERα in addition to triggering the expression of ERα-independent genes (Suzuki et al., 2004). Thus, the function of cholesterol as an ERRα agonist may provide molecular basis and mechanistic insight into clinical studies suggesting that statins can be used to treat or prevent _ER_- breast cancer (Cuzick et al., 2011; Kumar et al., 2008). The presence or absence of ERRα in _ER_- breast cancer may be a prognostic factor for the effectiveness of cholesterol-lowering treatments as anti-cancer therapeutics. Although it remains to be seen whether it is universally effective in the clinical settings to modulate ERRα transactivation through therapeutic regulation of cholesterol, as well as to modulate cholesterol function through ERRα-targeting small molecules, the deorphanization of ERRα using a widely applicable approach opens a new frontier in nuclear receptor biology.
EXPERIMENTAL PROCEDURES
Biochemical Analyses
6xHis-ERRα-LBD plasmid was obtained courtesy of Dino Moras. 6xHis-ERRα-LBD was transformed into Rosetta cells and expressed and purified as previously described (Greschik et al., 2008). Lipids were extracted into chloroform from brain or kidney by dounce homogenization. Global lipid pull-downs were performed as previously described (Kim et al., 2011a), but with larger concentrations of protein and lipid. Centrifuge columns (Pierce #89868) were loaded with 10µl IMAC sepharose 6 Fast Flow beads (GE). Beads were washed with water (2 × 500µl) and the wash was removed by centrifugation. 100µl 0.2M nickel sulfate was added to each column and the column was incubated for 30 min. Nickel sulfate was then eluted, and columns were washed with water (3 × 100µl) and protein buffer (20mM Tris, 200mM NaCl, pH 8.0) (3 × 200µl). 200µl of 25µM or 50µM HIS-ERRα-LBD or protein buffer containing no protein was incubated with resin for 2 hours. After incubation, protein was eluted and beads were washed with protein buffer containing 50mM imidazole (3 × 200µl). To prepare the lipid mixture, brain lipid extracts were dissolved in DMSO at 40µl/mg, vortexed, and then sonicated in a sonicator bath to ensure solubilization of the lipids. 12% lipid mixture was prepared by adding the DMSO lipid stock to protein buffer at 12% by volume. This mixture was vortexed and sonicated in a sonicator bath, and then 100µl was added to column and incubated for 30 min. The lipid mixture was then eluted and columns were washed with protein buffer containing 50mM imidazole (3 × 100µl). Protein-lipid conjugate was then eluted with protein buffer containing 250mM imidazole (3 × 100µl), and 80µl were injected on Agilent 6220 LC-MS TOF in negative or positive mode. For mass spectrometry, capillary voltage was 3500 V, fragmentor voltage was 100 V; nebulizer gas was 45 psi; drying gas temp was 350°C with a flow of 10 L/min; m/z range was from 100–1500 Daltons. This procedure was done in triplicate.
Targeted sterol analysis was performed using a MRM mode on an Agilent 6140 QQQ LC-MS/MS. Docking simulations were performed using autodock vina (Trott and Olson, 2010). Circular dichroism experiments were performed on a Jasco J-710 spectrophotometer. Tryptophan fluorescence quenching experiments and fluorescence polarization experiments were performed on a spectramax M5 spectrophotometer.
Cellular and In Vivo Analyses
Transfection and luciferase reporter assay were performed in CV-1 cells as described (Wei et al., 2010). Bone marrow osteoclast differentiation assays and bone analyses (microCT, ELISA and histomorphometry) were performed as described (Wei et al., 2010). C2C12 cells were cultured in DMEM+10% FBS, and differentiated into myotubes in DMEM+2% horse serum. ERRα knockout mice on a C57BL6/J background were previously described (Luo et al., 2003; Wei et al., 2010). Simvastatin was administered at 20mg/kg/day for 14 days. Zoledronate was administered by a single intravenous injection at 0.54 mg/kg. High cholesterol diet (2% cholesterol, 0.5% NaCholate) and normal chow diet control were from Harlan Teklad. All mouse experiments were conducted using littermates. Sample size estimate was based on power analyses performed using SAS 9.3 TS X64_7PRO platform at the UT Southwestern Biostatistics Core. With the observed group differences and the relatively small variation of the in vivo measurements, a sample size of four per group (n=4) will provide >90% power at type I error rate of 0.05 (two-sided test), and a sample size of three per group (n=3) will provide >80% power at type I error rate of 0.05 (two-sided test). All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center.
Statistical Analyses
All statistical analyses were performed with Student's t-Test and presented as mean ± standard deviation (s.d.) unless stated otherwise. The p values were designated as: *, p<0.05; **, p<0.01; ***, p<0.005; ****, p<0.001; n.s. non-significant (p>0.05).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Russell DeBose-Boyd and Marc Schumacher (UT Southwestern) for reagents and advice on cholesterol depletion; David Mangelsdorf (UT Southwestern) for ERRα and PGC1α plasmids; Vincent Giguère (McGill University) for ERRαKO mice; David Mangelsdorf, David Russell, Joseph Goldstein, Michael Brown and Russell DeBose-Boyd for critical reading of the manuscript and constructive suggestions. This work was in part supported by the UT Southwestern Endowed Scholar Startup Fund (Y.W.), NIH (RO1DK089113, Y.W.), The Welch Foundation (I-1751, Y.W.), Mary Kay Foundation (#073.14, Y.W.) and Searle Foundation (08-SSP-108, A.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
W.W., A.G.S., A.S. and Y.W. conceived the project and designed the experiments. A.G.S. conducted the lipidomic, mass spectrometry, biochemistry, and mutagenesis experiments and data analyses. W.W. conducted most of the cellular and animal experiments and data analyses. Xueqian W. conducted transfection and reporter assays. Xunde W. assisted with coIP. S.C. and Q.C. assisted with biochemical and mass spectrometry analyses. A.S. and Y.W. wrote the manuscript.
The authors declare that they have no financial conflict of interest.
References
- Andersson HC, Kratz L, Kelley R. Desmosterolosis presenting with multiple congenital anomalies and profound developmental delay. American journal of medical genetics. 2002;113:315–319. doi: 10.1002/ajmg.b.10873. [DOI] [PubMed] [Google Scholar]
- Ascenzi P, Bocedi A, Marino M. Structure-function relationship of estrogen receptor alpha and beta: impact on human health. Mol Aspects Med. 2006;27:299–402. doi: 10.1016/j.mam.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Ayers SD, Nedrow KL, Gillilan RE, Noy N. Continuous nucleocytoplasmic shuttling underlies transcriptional activation of PPARgamma by FABP4. Biochemistry. 2007;46:6744–6752. doi: 10.1021/bi700047a. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, DeCensi A, Arun B, Brown PH, Castiglione M, Dunn B, Forbes JF, Glaus A, Howell A, von Minckwitz G, et al. Preventive therapy for breast cancer: a consensus statement. The lancet oncology. 2011;12:496–503. doi: 10.1016/S1470-2045(11)70030-4. [DOI] [PubMed] [Google Scholar]
- Duellman SJ, Calaoagan JM, Sato BG, Fine R, Klebansky B, Chao WR, Hobbs P, Collins N, Sambucetti L, Laderoute KR. A novel steroidal inhibitor of estrogen-related receptor alpha (ERR alpha) Biochemical pharmacology. 2010;80:819–826. doi: 10.1016/j.bcp.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echaniz-Laguna A, Mohr M, Tranchant C. Neuromuscular symptoms and elevated creatine kinase after statin withdrawal. N Engl J Med. 2010;362:564–565. doi: 10.1056/NEJMc0908215. [DOI] [PubMed] [Google Scholar]
- Eskiocak B, Ali A, White MA. The estrogen-related receptor alpha inverse agonist XCT 790 is a nanomolar mitochondrial uncoupler. Biochemistry. 2014;53:4839–4846. doi: 10.1021/bi500737n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzPatrick DR, Keeling JW, Evans MJ, Kan AE, Bell JE, Porteous ME, Mills K, Winter RM, Clayton PT. Clinical phenotype of desmosterolosis. American journal of medical genetics. 1998;75:145–152. [PubMed] [Google Scholar]
- Fluck CE, Tajima T, Pandey AV, Arlt W, Okuhara K, Verge CF, Jabs EW, Mendonca BB, Fujieda K, Miller WL. Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat Genet. 2004;36:228–230. doi: 10.1038/ng1300. [DOI] [PubMed] [Google Scholar]
- Gallet M, Vanacker JM. ERR receptors as potential targets in osteoporosis. Trends in endocrinology and metabolism: TEM. 2010;21:637–641. doi: 10.1016/j.tem.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Giguere V. To ERR in the estrogen pathway. Trends in endocrinology and metabolism: TEM. 2002;13:220–225. doi: 10.1016/s1043-2760(02)00592-1. [DOI] [PubMed] [Google Scholar]
- Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocrine reviews. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- Greschik H, Althage M, Flaig R, Sato Y, Chavant V, Peluso-Iltis C, Choulier L, Cronet P, Rochel N, Schule R, et al. Communication between the ERRalpha homodimer interface and the PGC-1alpha binding surface via the helix 8–9 loop. The Journal of biological chemistry. 2008;283:20220–20230. doi: 10.1074/jbc.M801920200. [DOI] [PubMed] [Google Scholar]
- Hanai J, Cao P, Tanksale P, Imamura S, Koshimizu E, Zhao J, Kishi S, Yamashita M, Phillips PS, Sukhatme VP, et al. The muscle-specific ubiquitin ligase atrogin-1/MAFbx mediates statin-induced muscle toxicity. J Clin Invest. 2007;117:3940–3951. doi: 10.1172/JCI32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horard B, Vanacker JM. Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. Journal of molecular endocrinology. 2003;31:349–357. doi: 10.1677/jme.0.0310349. [DOI] [PubMed] [Google Scholar]
- Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SRD, Dowsett M. Aromatase Inhibitors For Breast Cancer Lessons From The Laboratory. Nature Reviews Cancer. 2003;3:821–831. doi: 10.1038/nrc1211. [DOI] [PubMed] [Google Scholar]
- Kallen J, Schlaeppi JM, Bitsch F, Filipuzzi I, Schilb A, Riou V, Graham A, Strauss A, Geiser M, Fournier B. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. The Journal of biological chemistry. 2004;279:49330–49337. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- Kim YG, Lou AC, Saghatelian A. A metabolomics strategy for detecting protein-metabolite interactions to identify natural nuclear receptor ligands. Mol Biosyst. 2011a;7:1046–1049. doi: 10.1039/c0mb00324g. [DOI] [PubMed] [Google Scholar]
- Kim YG, Lou AC, Saghatelian A. A metabolomics strategy for detecting protein-metabolite interactions to identify natural nuclear receptor ligands. Mol Biosyst. 2011b;7:1046–1049. doi: 10.1039/c0mb00324g. [DOI] [PubMed] [Google Scholar]
- Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, Morony S, Oliveirados-Santos AJ, Van G, Itie A, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- Kumar AS, Benz CC, Shim V, Minami CA, Moore DH, Esserman LJ. Estrogen receptor-negative breast cancer is less likely to arise among lipophilic statin users. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:1028–1033. doi: 10.1158/1055-9965.EPI-07-0726. [DOI] [PubMed] [Google Scholar]
- Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguere V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Molecular and cellular biology. 2003;23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JG, Thompson BM, McCrum EC, Russell DW. Extraction and analysis of sterols in biological matrices by high performance liquid chromatography electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:145–170. doi: 10.1016/S0076-6879(07)32006-5. [DOI] [PubMed] [Google Scholar]
- Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annu Rev Pathol. 2008;3:457–484. doi: 10.1146/annurev.pathmechdis.3.121806.151431. [DOI] [PubMed] [Google Scholar]
- Orozco P. Atherogenic lipid profile and elevated lipoprotein (a) are associated with lower bone mineral density in early postmenopausal overweight women. Eur J Epidemiol. 2004;19:1105–1112. doi: 10.1007/s10654-004-1706-8. [DOI] [PubMed] [Google Scholar]
- Patch RJ, Searle LL, Kim AJ, De D, Zhu X, Askari HB, O'Neill JC, Abad MC, Rentzeperis D, Liu J, et al. Identification of diaryl ether-based ligands for estrogen-related receptor alpha as potential antidiabetic agents. Journal of medicinal chemistry. 2011;54:788–808. doi: 10.1021/jm101063h. [DOI] [PubMed] [Google Scholar]
- Russell RG. Bisphosphonates: the first 40 years. Bone. 2011;49:2–19. doi: 10.1016/j.bone.2011.04.022. [DOI] [PubMed] [Google Scholar]
- Sathasivam S. Statin induced myotoxicity. Eur J Intern Med. 2012;23:317–324. doi: 10.1016/j.ejim.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Schaaf CP, Koster J, Katsonis P, Kratz L, Shchelochkov OA, Scaglia F, Kelley RI, Lichtarge O, Waterham HR, Shinawi M. Desmosterolosis-phenotypic and molecular characterization of a third case and review of the literature. American journal of medical genetics Part A. 2011;155A:1597–1604. doi: 10.1002/ajmg.a.34040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Molecular and cellular biology. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RA, McDonnell DP. Estrogen-related receptor alpha as a therapeutic target in cancer. Endocrine-related cancer. 2006;13(Suppl 1):S25–S32. doi: 10.1677/erc.1.01292. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miki Y, Moriya T, Shimada N, Ishida T, Hirakawa H, Ohuchi N, Sasano H. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer research. 2004;64:4670–4676. doi: 10.1158/0008-5472.CAN-04-0250. [DOI] [PubMed] [Google Scholar]
- Tagore R, Thomas HR, Homan EA, Munawar A, Saghatelian A. A global metabolite profiling approach to identify protein-metabolite interactions. J Am Chem Soc. 2008;130:14111–14113. doi: 10.1021/ja806463c. [DOI] [PubMed] [Google Scholar]
- Tarakida A, Iino K, Abe K, Taniguchi R, Higuchi T, Mizunuma H, Nakaji S. Hypercholesterolemia accelerates bone loss in postmenopausal women. Climacteric. 2011;14:105–111. doi: 10.3109/13697137.2010.507888. [DOI] [PubMed] [Google Scholar]
- Toledano JE, Partridge NC. Statins: not just for cholesterol? Trends in endocrinology and metabolism: TEM. 2000;11:255–256. doi: 10.1016/s1043-2760(00)00295-2. [DOI] [PubMed] [Google Scholar]
- Tomaszewski M, Stepien KM, Tomaszewska J, Czuczwar SJ. Statin-induced myopathies. Pharmacological reports : PR. 2011;63:859–866. doi: 10.1016/s1734-1140(11)70601-6. [DOI] [PubMed] [Google Scholar]
- Tremblay GB, Kunath T, Bergeron D, Lapointe L, Champigny C, Bader JA, Rossant J, Giguere V. Diethylstilbestrol regulates trophoblast stem cell differentiation as a ligand of orphan nuclear receptor ERR beta. Genes Dev. 2001;15:833–838. doi: 10.1101/gad.873401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayavekhin N, Saghatelian A. Discovery of a protein-metabolite interaction between unsaturated fatty acids and the nuclear receptor Nur77 using a metabolomics approach. J Am Chem Soc. 2011;133:17168–17171. doi: 10.1021/ja208199h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JS, Newman C, Eastell R. Heart drugs that affect bone. Trends in endocrinology and metabolism: TEM. 2012;23:163–168. doi: 10.1016/j.tem.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Wan Y. PPARgamma in bone homeostasis. Trends in endocrinology and metabolism: TEM. 2010;21:722–728. doi: 10.1016/j.tem.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Wei W, Wang X, Yang M, Smith LC, Dechow PC, Sonoda J, Evans RM, Wan Y. PGC1beta mediates PPARgamma activation of osteoclastogenesis and rosiglitazone-induced bone loss. Cell Metab. 2010;11:503–516. doi: 10.1016/j.cmet.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Zeve D, Suh JM, Wang X, Du Y, Zerwekh JE, Dechow PC, Graff JM, Wan Y. Biphasic and dosage-dependent regulation of osteoclastogenesis by beta-catenin. Mol Cell Biol. 2011;31:4706–4719. doi: 10.1128/MCB.05980-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Wang J, Wang Y, Kwok TT, Kong SK, Wong C. Estrogen-related receptor alpha (ERRalpha) inverse agonist XCT-790 induces cell death in chemotherapeutic resistant cancer cells. Chem Biol Interact. 2009;181:236–242. doi: 10.1016/j.cbi.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Xing W, Liu J, Cheng S, Vogel P, Mohan S, Brommage R. Targeted disruption of leucine-rich repeat kinase 1 but not leucine-rich repeat kinase 2 in mice causes severe osteopetrosis. J Bone Miner Res. 2013;28:1962–1974. doi: 10.1002/jbmr.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.