Heparan Sulfate Proteoglycan Is an Important Attachment Factor for Cell Entry of Akabane and Schmallenberg Viruses (original) (raw)
ABSTRACT
Akabane virus (AKAV) and Schmallenberg virus (SBV) are members of the genus Orthobunyavirus, which are transmitted by arthropod vectors with a broad cellular tropism in vitro as well as in vivo. Both AKAV and SBV cause arthrogryposis-hydranencephaly syndrome in ruminants. The main cellular receptor and attachment factor for entry of these orthobunyaviruses are unknown. Here, we found that AKAV and SBV infections were inhibited by the addition of heparin or enzymatic removal of cell surface heparan sulfates. To confirm this finding, we prepared heparan sulfate proteoglycan (HSPG)-knockout (KO) cells by using a clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system and measured the quantities of binding of these viruses to cell surfaces. We observed a substantial reduction in AKAV and SBV binding to cells, limiting the infections by these viruses. These data demonstrate that HSPGs are important cellular attachment factors for AKAV and SBV, at least in vitro, to promote virus replication in susceptible cells.
IMPORTANCE AKAV and SBV are the etiological agents of arthrogryposis-hydranencephaly syndrome in ruminants, which causes considerable economic losses in the livestock industry. Here, we identified heparan sulfate proteoglycan as a major cellular attachment factor for the entry of AKAV and SBV. Moreover, we found that heparin is a strong inhibitor of AKAV and SBV infections. Revealing the molecular mechanisms of virus-host interactions is critical in order to understand virus biology and develop novel live attenuated vaccines.
KEYWORDS: Akabane virus, heparan sulfate, Schmallenberg virus, cell entry
INTRODUCTION
Akabane virus (AKAV) and Schmallenberg virus (SBV) belong to the Simbu serogroup of the arthropod-borne Orthobunyavirus genus of the family Bunyaviridae. AKAV and SBV are phylogenetically closely related, and they also possess similar biological characteristics: (i) both cause abortion, stillbirth, premature birth, and congenital deformities in cattle, sheep, and goats; (ii) both primarily infect the central nervous system (CNS) of the fetus; and (iii) both are difficult to control, because they are transmitted by biting midges of the genus Culicoides (1). Both AKAV and SBV cause “abortion storms” that result in considerable economic losses to the livestock industry (2–4).
Despite these similarities, there are important features that distinguish these two viruses. AKAV is endemic throughout Australia, Southeast Asia, East Asia, the Middle East, and Africa, whereas SBV has emerged and has dispersed across a large area of Europe since 2011. AKAV comprises four genogroups (I to IV), whereas SBV comprises a single genotype. No differences in the pathogenicities of different SBV strains have been described. On the other hand, the OBE-1 strain of AKAV [AKAV(OBE-1)] (genogroup II) causes severe fetal malformation, whereas the Iriki strain [AKAV(Iriki)] (genogroup I) also causes fatal nonsuppurative encephalomyelitis in newborn cattle. Molecular determinants distinguishing the pathogenicities of these two different strains are unknown (5).
Orthobunyaviruses carry a tripartite, single-stranded, negative-sense RNA genome. The L segment encodes the L protein, a viral RNA-dependent RNA polymerase; the S segment encodes the N protein and the nonstructural protein NSs, both transcribed from an overlapping open reading frame; and the M segment encodes NSm and the two major viral envelope proteins, Gn and Gc (Gn/Gc), which form heterodimeric spikes on the virus particle. Gn/Gc are the proteins on the surface of the virion that bind to cell surface molecules in the initial step of orthobunyavirus infection (6, 7). Relatively few studies have investigated orthobunyavirus entry. La Crosse and Germiston neurotropic orthobunyavirus entry into the cell has been described to be promoted by DC-SIGN (8, 9). However, DC-SIGN is probably not the main attachment factor of ruminant orthobunyaviruses, because it is expressed on macrophages and dendritic cells, not in the CNS. Heparan sulfate proteoglycan (HSPG), one of major negatively charged transmembrane protein-linking glycosaminoglycans, is expressed by almost all cells, including neural cells. HSPG is involved in cell attachment of many viruses (e.g., herpes simplex virus [10, 11], adenovirus [12], respiratory syncytial virus [13, 14], human papillomavirus [15], foot-and-mouth disease virus [16], hepatitis B virus [17], hepatitis C virus [18], Ebola virus [19, 20], dengue virus [21], and human immunodeficiency virus [22]). In addition, HSPG is involved in cell attachment of phleboviruses in the family Bunyaviridae, including Rift Valley fever virus and Toscana virus (23–25). In a previous study, hemagglutination of AKAV was inhibited by the addition of heparin, a form of heparan sulfate (26). Therefore, it is possible that HSPGs are involved in AKAV and/or SBV infection, similarly to other viruses.
In this study, we examined the role of HSPGs in AKAV and SBV replication.
RESULTS
Heparin or heparinase treatment inhibits AKAV and SBV infections.
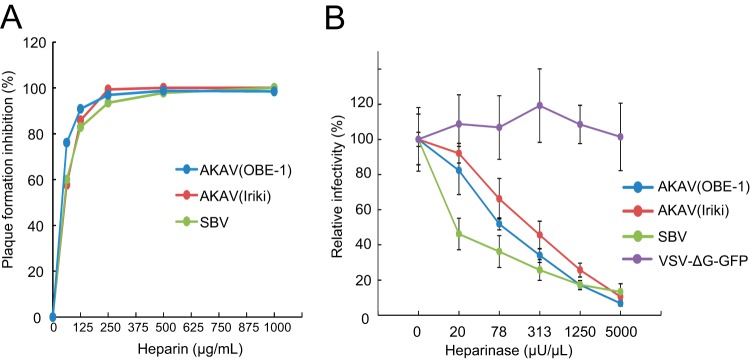
Heparin is a highly sulfated form of heparan sulfate (27) and is a known inhibitor of infection by various viruses. To test whether heparin inhibits AKAV or SBV replication, AKAV and SBV were incubated with different concentrations of heparin for 30 min, and the titers of the viruses neutralized by heparin were measured by plaque reduction assay in HmLu-1 cells (Fig. 1A), which support efficient replication of the Akabane and Schmallenberg viruses and abundantly express heparan sulfate. The number of plaques induced by either AKAV or SBV was reduced in a heparin concentration-dependent manner. These data suggest that heparin affects the replication cycles of both viruses.
FIG 1.
Effects of heparin and heparinase treatment on AKAV and SBV infections. (A) Effects of heparin on AKAV and SBV plaque reduction. Various concentrations of heparin were incubated for 30 min at room temperature with 100 PFU of AKAV(OBE-1), AKAV(Iriki), or SBV, and the ability of heparin to reduce plaque formation was assessed. (B) Effects of heparinase treatment on AKAV and SBV infectivity. HmLu-1 cells were treated with various concentrations of heparinase II, followed by AKAV or SBV infection. Cells were stained for AKAV or SBV antigen, and positive cells were counted under a fluorescence microscope. For VSV-ΔG-GFP-infected cells, GFP-positive cells were counted under a fluorescence microscope. Results are expressed in percentages relative to cells that were not treated with heparinase. The data are reported as the mean value with standard deviations for three independent experiments.
Next, we pretreated HmLu-1 cells with heparinase to remove HPSG from the cell surface and subsequently infected them with AKAV(OBE-1), AKAV(Iriki), and SBV. We also used VSV-ΔG-GFP, a vesicular stomatitis virus (VSV)-based vector expressing green fluorescent protein (GFP) that can complete only a single replication cycle within the cell (28), because this virus does not utilize HSPG during the early events of infection (23, 29). At 8 h postinfection (hpi), cells were fixed and stained with anti-AKAV or anti-SBV antibodies for the detection of AKAV- or SBV-infected cells, respectively. GFP-positive cells were instead counted to determine the number of VSV-ΔG-GFP-infected cells. As shown in Fig. 1B, VSV-ΔG-GFP was not susceptible to heparinase treatment, whereas the numbers of cells infected by AKAV and SBV were reduced in a heparinase concentration-dependent manner. SBV was more susceptible to heparinase treatment than the AKAVs (P < 0.05 at 20 and 78 μU/μl of heparinase). These data suggest that HSPG plays important roles in AKAV and SBV infections.
AKAV and SBV replication in HSPG-KO cells.
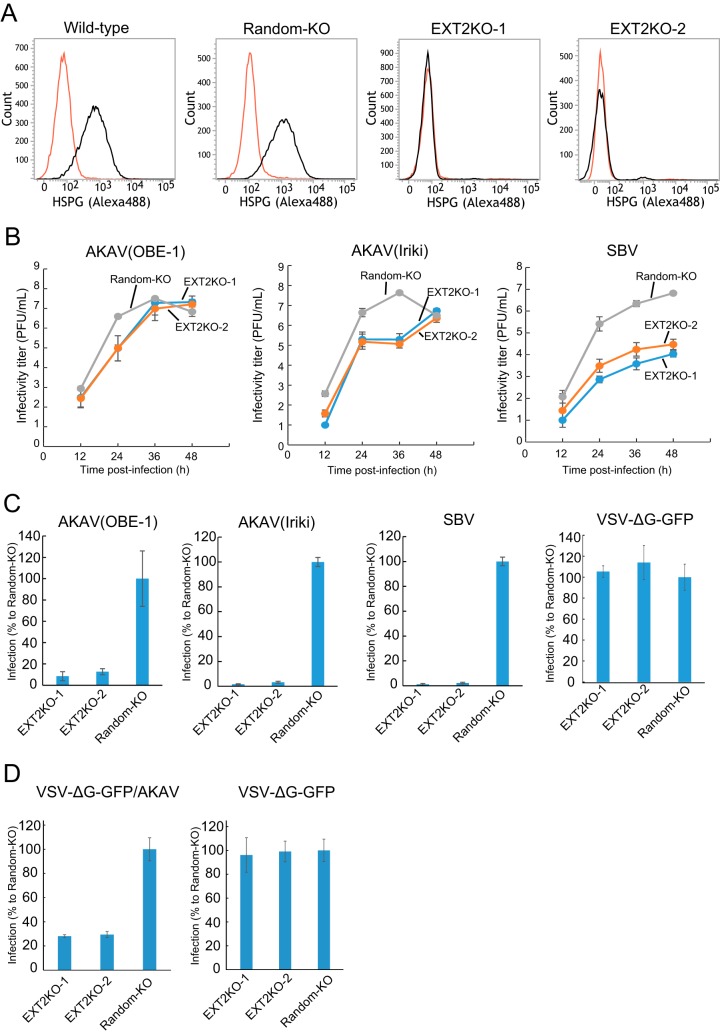
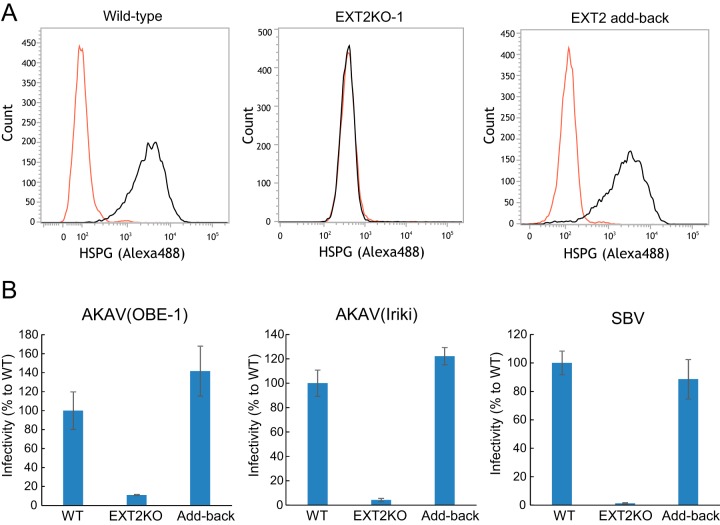
In order to further validate the data shown above, we established HSPG-knockout (HSPG-KO) cells using a clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system (30) disrupting the EXT2 gene, which encodes one of the HSPG-synthesizing enzymes (27). We designed two guide RNA (gRNAs) (EXT2-1 and EXT2-2) targeting different positions in the EXT2 gene and obtained three clones for each target (EXT2KO-1-1, -1-2, -1-3, -2-1, -2-2, and -2-3 cells). We also established control “random-KO” HmLu-1 cells by introducing a 20-nucleotide (nt) random target sequence in the gRNA with the CRISPR-Cas9 system and obtained three clones of the random-KO cells (random-KO-1, -2, and -3). Lack of HSPG expression in HSPG-KO cells, but not in random-KO or wild-type cells, was confirmed by flow cytometry analysis (Fig. 2A). In the HSPG-KO cells, titers of AKAV(OBE-1) and AKAV(Iriki) were about 100-fold lower than that in random-KO cells at 24 hpi but were at similar levels at later time points (Fig. 2B). SBV titers were instead between 10 and 1,000-fold lower in HSPG-KO cells than in random-KO cells throughout the course of the experiment (Fig. 2B). Next, we examined AKAV and SBV infectivity in the EXT2-KO cells and random-KO cells. EXT2-KO cells or random-KO cells were infected with AKAV(OBE-1), AKAV(Iriki), SBV, or VSV-ΔG-GFP (multiplicity of infection [MOI] of 0.1). At 8 hpi, AKAV antigen-positive cells, SBV antigen-positive cells, or GFP-positive cells were counted (Fig. 2C). Control VSV-ΔG-GFP-infected cell numbers were not significantly different between random-KO and HSPG-KO cells. Five to 10 times lower numbers of AKAV and SBV antigen-positive cells were detected in EXT2-KO cells than random-KO cells. To eliminate the possibility that the replication step of AKAV affected the results shown in Fig. 2C, we used a VSV-pseudotyped virus bearing AKAV glycoproteins (VSV-ΔG-GFP/AKAV). VSV-ΔG-GFP/AKAV or VSV-ΔG-GFP was inoculated into EXT2-KO cells or random-KO cells. At 8 hpi, GFP-positive cells were counted (Fig. 2D). As shown in Fig. 2C, control VSV-ΔG-GFP-infected cell numbers did not show a significant difference between random-KO and HSPG-KO cells. However, three-times-fewer VSV-ΔG-GFP-infected cells were detected in EXT2-KO cells than in random-KO cells (P < 0.01). These data indicated that HSPG was important for AKAV and SBV infections.
FIG 2.
AKAV and SBV growth kinetics and infectivity in HSPG-KO HmLu-1 cells. (A) Flow cytometric analysis of EXT2-KO HmLu-1 cells. CRISPR-Cas9-mediated EXT2-KO cell clones (EXT2-1 and EXT2-2) were labeled with anti-heparan sulfate mouse monoclonal antibody (10E4) (black) or with isotype control (red) and analyzed by flow cytometry (FACSVerse; BD Biosciences). Representative data (one out of three clones of random-KO, EXT2KO-1, and EXT2KO-2) are shown. (B) Growth kinetics of AKAV or SBV in HSPG-KO cells. AKAV(OBE-1), AKAV(Iriki), or SBV was inoculated onto three clones of random-KO, EXT2KO-1, and EXT2KO-2 cells at a multiplicity of infection of 0.01. Virus titers were determined by plaque assay in normal HmLu-1 cells. The data are reported as the mean titer for three clones of each KO cell (EXT2KO-1, EXT2KO-2, or random-KO) with standard deviations. (C) Infectivities of AKAV and SBV in HSPG-KO cells. Random-KO or HSPG-KO cells were infected with AKAV(OBE-1), AKAV(Iriki), SBV, or control VSV-ΔG-GFP. Cells were stained for AKAV or SBV antigen, and positive cells were counted under a fluorescence microscope. For VSV-ΔG-GFP-infected cells, GFP-positive cells were counted under a fluorescence microscope. Results are expressed as percentages relative to the number of positive random-KO cells. The data are reported as the mean value for three clones of each KO cell (EXT2KO-1, EXT2KO-2, or random-KO) with standard deviations. (D) Infectivities of VSV pseudotyped with AKAV Gn/Gc (VSV-ΔG-GFP/AKAV) in HSPG-KO cells. Random-KO or HSPG-KO cells were infected with VSV-ΔG-GFP/AKAV or control VSV-ΔG-GFP. GFP-positive cells were counted under a fluorescence microscope. Results are represented as percentages relative to the number of positive random-KO cells. The data are shown as the mean value for three clones of each KO cell (EXT2KO-1, EXT2KO-2, or random-KO) with standard deviations.
Quantification of AKAV and SBV bound to the HSPG-KO cell surface.
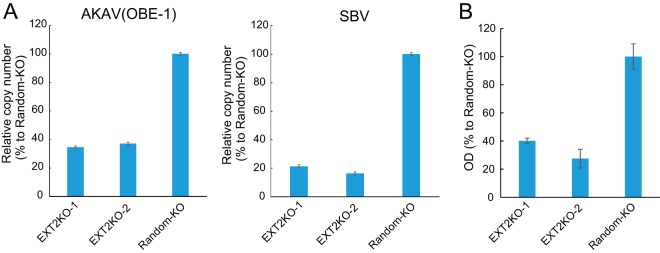
To determine whether HSPG is important for AKAV and SBV cell surface attachment, we quantified the amounts of cell surface-bound viruses. AKAV(OBE-1) and SBV were incubated with EXT2-KO-1, EXT2-KO-2, or random-KO cells for 1 h at 4°C, and virus-bound cells were collected. RNA was extracted from the cells, and AKAV or SBV RNA (S segment) was quantified by quantitative reverse transcription-PCR (qRT-PCR) (Fig. 3A). AKAV and SBV RNAs were significantly lower in HSPG-KO cells than in random-KO cells (P < 0.01). We also measured cell-bound AKAV by quantifying N protein using an N-detecting sandwich enzyme-linked immunosorbent assay (ELISA) (Fig. 3B). Cell surface-bound N proteins were significantly lower in EXT2-KO-1 and EXT2-KO-2 cells than in random-KO cells, confirming the qRT-PCR results. These data suggest that HSPG is an important molecule for AKAV and SBV cell surface attachment.
FIG 3.
AKAV and SBV binding assays in HSPG-KO cells. (A) Real-time RT-PCR for the quantification of cell surface-attached viruses. AKAV(OBE-1) or SBV was incubated with HSPG-KO cells at 4°C. After a washing step, total RNAs were extracted. AKAV or SBV S RNAs were quantified by one-step real-time RT-PCR. For relative quantification, standard curves of AKAV or SBV S RNA and GAPDH were prepared by serial dilution of a mixture of total RNA from uninfected HmLu-1 cells and RNA extracted from AKAV(OBE-1) or SBV-containing supernatants. Results are expressed as the percentages relative to the levels in random-KO cells. The results are representative of three different experiments. (B) Sandwich ELISA for the detection of N proteins of AKAV attached to cell surfaces. AKAV(OBE-1) was inoculated onto HSPG-KO or random-KO HmLu-1 cells and left for 1 h at 4°C. After a washing step, the cells were lysed, and the lysates were added to the anti-AKAV N monoclonal antibody (5E8)-coated wells of 96-well ELISA plates (Maxisorp, Nunc), followed by incubation with biotinylated anti-AKAV mouse polyclonal antibody. Subsequently, the wells were incubated with avidin-biotinylated horseradish peroxidase (HRP) complex (Vectastain ABC kit; Vector Laboratories). A 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution was used for detection, and optical density values were measured. Results are expressed as percentages relative to the number of positive random-KO cells. The results are representative of three independent experiments.
AKAV and SBV replication is restored in cells with EXT2 added back.
We next transduced the EXT2-KO cells with the EXT2 gene in order to confirm that AKAV and SBV replication could be restored by simply reintroducing the EXT2 gene back to EXT2-KO cells. We first confirmed that HSPG expression was restored in these cells by flow cytometry (Fig. 4A). Next, we examined AKAV and SBV infectivity in the EXT2 gene transduced EXT2-KO cells and wild-type cells. EXT2-KO cells or wild-type cells were infected with AKAV(OBE-1), AKAV(Iriki), or SBV. At 8 hpi, AKAV- and SBV-infected cells were detected (Fig. 4B). As expected, infectivity of AKAV(OBE-1), AKAV(Iriki), and SBV was restored in the cells with EXT2 added back.
FIG 4.
Rescue of AKAV and SBV infectivities by adding back the EXT2 gene to EXT2-KO cells. (A) Flow cytometric analysis of adding back the EXT2 gene in EXT2-KO-1 HmLu-1 cells. Cells were labeled with anti-heparan sulfate mouse monoclonal antibody (10E4) and analyzed by flow cytometry (FACSVerse; BD Biosciences). (B) Infectivities of AKAV and SBV in cells with EXT2 added back. The cells were infected with AKAV(OBE-1), AKAV(Iriki), or SBV. Cells were stained for AKAV or SBV antigen, and positive cells were counted under a fluorescence microscope. Results are expressed as percentages relative to the number of wild-type cells. The data are reported as the mean value with standard deviations for three independent experiments.
DISCUSSION
AKAV and SBV infect neurons (2, 4, 31) and a broad range of cells in the infected hosts. This suggests that AKAV and SBV use a cellular receptor(s) or attachment factor(s) that is expressed on a variety of cells. DC-SIGN was previously shown to promote La Crosse and Germiston neurotropic orthobunyavirus entry (8, 9). However, the distribution of DC-SIGN is not consistent with the tropisms of either AKAV or SBV, because DC-SIGN is expressed on a limited number of cell types (i.e., dendritic cells and macrophages). Here, we demonstrated that HSPG plays an important role in AKAV and SBV infections as an attachment factor. Cell surface HSPGs are expressed ubiquitously throughout the body (27), including on neuronal cells (32). Therefore, our data support the correlation between HSPG distribution and AKAV and SBV tropism. However, AKAV and SBV show a strong preference for neuronal cells in vivo, and there are likely other reasons for this. AKAV and SBV were still able to replicate in HSPG-KO cells, although at lower levels than those in HSPG-expressing cells. These data suggest the presence of another cellular factors(s) that defines the tissue tropisms of AKAV and SBV. Further studies are needed to identify the neuronal cell-specific receptors or reveal other defining steps after cell entry.
In AKAV(Iriki)-infected HSPG-KO cells, after the virus titer reached a plateau at 24 and 36 hpi, a second AKAV(Iriki) growth wave was observed at 48 hpi (Fig. 2B). During the AKAV(Iriki) replication in HSPG-KO cells, the virus may have acquired a mutation(s) in the receptor-binding site of the glycoproteins, possibly leading to adaptation to HSPG-KO cells. Thus, to investigate whether the AKAV(Iriki) recovered from HSPG-KO cells at a later time point of infection included mutations in the glycoproteins, we sequenced the entire M segment at 60 hpi but found no mutations. Hence, the mechanism of the second growth wave of AKAV(Iriki) still remains unknown. Continuous virus passage in HSPG-KO cells may result in mutations to adapt to another cellular attachment factor(s) or receptor(s).
Repetitive passage of some viruses in tissue culture induces one or two amino acid mutations in their glycoproteins, which increases their affinity toward HSPGs (33–38). This adaptation also induces viral attenuation in vivo (33–38). A previous study showed that cell culture-derived SBV showed slightly slowed replication in cattle; however, the involvement of HSPG underlying this is unknown (39). Rift Valley fever virus (23), dengue virus (40), and human T-lymphotropic virus 1 (HTLV-1) (41) do not acquire any mutations after passage in cell culture, which enhances their affinity toward HSPGs. This suggests that the species of the virus determines whether the virus adapts to cell culture and uses HSPGs for entry. AKAV(OBE-1), AKAV(Iriki), and SBV, used in the present study, were passaged several times in tissue culture before being cloned into reverse genetics plasmids. However, AKAV(Iriki) maintained its pathogenic potential against goat fetuses and mice (42, 43), implying that it did not undergo changes during passage in the cell culture. However, it is unclear whether serial passages in cell culture induced mutations in Gn/Gc proteins of AKAV and SBV to give them higher affinity to HSPG. Therefore, further studies are needed to compare the HSPG-binding affinities of the viral strains examined with those of our lab strains and isolates from clinical specimens which have original sequences.
In this study, we showed that the heparinase susceptibly of SBV was greater than that of AKAVs. In addition, SBV replication in HSPG-KO cells was limited compared to that of AKAVs. Moreover, AKAV(OBE-1) exhibited slightly lower HSPG dependency than AKAV(Iriki) did for virus infection and growth. One possible explanation for the difference in HSPG dependency is that AKAV(OBE-1), AKAV(Iriki), and SBV possess different sequences in their HSPG-binding domains and probably in Gn or Gc. Although HSPG-binding domains have been identified in other viral glycoproteins as well as in cellular proteins (44), we could not find known HSPG-binding motifs in AKAV and SBV glycoprotein sequences (data not shown). HSPG-binding domains are not simply defined by the secondary sequences of HS-binding proteins (44). The tertiary structure of these proteins and heparan sulfate interactions are also important for binding. Determining the three-dimensional (3D) structure of Gn/Gc proteins is likely required to define the binding site more precisely. Although the orthobunyavirus Gn/Gc 3D structure is not available currently, determining the 3D structure of Gn/Gc proteins is likely required to define the HSPG binding site.
Here, we clearly show that AKAV and SBV utilize HSPG for their initial cell surface attachment in gene-edited HSPG-KO cells. These findings further our understanding of the orthobunyavirus life cycle. Molecules inhibiting orthobunyavirus and HSPG interactions may be effective antivirals.
MATERIALS AND METHODS
Cells.
Baby hamster kidney cells stably expressing T7 RNA polymerase (BHK/T7-9 cells) (45) were kindly provided by Naoto Ito (Gifu University, Japan) and cultured in Dulbecco's modified Eagle's minimum medium (DMEM) supplemented with 5% fetal calf serum (FCS) and 10% tryptose phosphate broth at 37°C. Golden hamster lung (HmLu-1) cells were cultured at 37°C in DMEM supplemented with 5% FCS. Human embryonic kidney (HEK293T) cells were maintained in DMEM supplemented with 10% FCS.
Viruses.
AKAV strains OBE-1 and Iriki (42, 46) and SBV (31) were generated by reverse genetics. Briefly, to recover AKAV(Iriki), 1.2 μg of pT7riboSM2/IL, 0.6 μg of pT7riboSM2/IM, and 1.2 μg of pT7riboSM2/IS plasmids were mixed in 200 μl of Opti-MEM (GIBCO, Grand Island, NY, USA) with 9 μl of Trans-IT LT1 transfection reagent (Mirus Bio, Madison, WI, USA), incubated at room temperature for 15 min, and then added to BHK-T7 cells (45) grown in 6-well plates. To recover AKAV(OBE-1), 1.2 μg of pT7riboSM2/OL, 0.6 μg of pT7riboSM2/OM, and 1.2 μg of pT7riboSM2/OS plasmids were transfected into BHK-T7 cells as described for AKAV(Iriki) generation. To recover SBV, 1 μg each of pUCSBVST7, pUCSBVMT7, and pUCSBVLT7 plasmids and 2 μg of a plasmid expressing T7 polymerase under the control of the chicken β-actin promoter (pCAGGS-T7pol) were mixed in 300 μl of Opti-MEM (GIBCO) with 15 μl of Trans-IT 293 transfection reagent (Mirus), incubated at room temperature for 15 min, and then added to HEK293T cells grown in 6-well plates. At 3 days posttransfection, the culture supernatant of transfected cells was harvested and added to HmLu-1 cells. Viruses were propagated in HmLu-1 cells cultured in DMEM supplemented with 2% FCS. Nonspreading vesicular stomatitis virus (VSV-ΔG-GFP) pseudotyped with its own glycoprotein G was propagated following transfection of the VSV G protein-expressing plasmid (pCAGGS-VSVG) into HEK293T cells (28). VSV-pseudotyped virus with AKAV Gn and mutant Gc was also propagated in HEK293T cells following transfection of the mutant Gn/Gc/NSm of AKAV(OBE-1), which lacks a 10-amino-acid region in the C-terminal of the wild-type Gc. This mutant was made since VSV-pseudotyped virus with Gn and mutant Gc lacking the C termini of cytoplasmic tail glycoproteins of Crimian-Congo hemorrhagic fever (CCHF) virus, a member of Bunyaviridae, showed higher titer than VSV-pseudotyped virus with wild-type CCHF Gn/Gc proteins (47).
Production of PAbs to AKAV or SBV.
An anti-AKAV mouse polyclonal antibody (PAb) was prepared by two intraperitoneal injections of sucrose gradient-purified AKAV(OBE-1) into 6-week-old female ICR mice (Japan SLC, Hamamatsu, Japan) at 2-week intervals. An anti-SBV mouse PAb was prepared by two intraperitoneal injections of sucrose gradient-purified SBV in 6-week-old female ICR mice (Japan SLC) at 2-week intervals. Reactivities and specificities of both anti-SBV and anti-AKAV PAbs were confirmed by immunofluorescence assay, using AKAV(OBE-1)- or SBV-infected and mock-infected HmLu-1 cells.
Plaque assay.
A standard plaque assay was used to determine the infectivities of AKAV and SBV. After virus adsorption to HmLu-1 cells for 1 h at 37°C, the inocula were removed, and cells were overlaid with DMEM containing 0.6% agarose and 2% FCS. After incubation for 3 days, cells were stained with neutral red before counting PFU.
Plaque reduction assay.
One hundred microliters of serial 2-fold dilutions of heparin solution were prepared in minimal essential medium (MEM) containing 0.3% bovine serum albumin (BSA/MEM). An equal volume of the suspensions containing 100 PFU/100 μl of AKAV(OBE-1), AKAV(Iriki), or SBV was added to each dilution. After incubation for 30 min at room temperature, 200 μl of each virus-heparin mixture (containing 100 PFU of viruses) was titrated by the plaque assay.
Heparinase treatment.
Cells were seeded in 48-well plates at 24 h prior to infection. Medium was removed, and the cells were incubated with serial 4-fold dilutions of heparinase II (New England BioLabs) in BSA/MEM with 2 mM CaCl2 for 1 h at 37°C and then washed with BSA/MEM. The plates were immediately transferred on ice to suppress the synthesis and transport of HSPG. We then added the appropriate amounts of AKAV(OBE-1), AKAV(Iriki), or SBV to 100 μl of BSA/MEM and incubated the mixture for 1 h on ice. The supernatant was removed, and the cells were washed twice with BSA/MEM and with and DMEM with 5% FCS before incubation for 8 h at 37°C. The cells were then fixed with 4% paraformaldehyde for 15 min at room temperature. After removing paraformaldehyde, the cells were permeabilized with 0.1% Triton X-100 and incubated with an anti-AKAV N mouse monoclonal antibody (MAb) (5E8) (48) for AKAV-infected cells or an anti-SBV mouse PAb for SBV-infected cells, followed by incubation with Alexa Fluor 488-conjugated anti-mouse IgG antibody. The fluorescence-positive cells were counted as AKAV- or SBV-infected cells under fluorescence microscopy (Vert. A1; Carl Zeiss).
EXT2-KO cells.
EXT2-KO cells were established using the CRISPR-Cas9 system. EXT2 gene target sequences (EXT2-1 [CTATCCCCTGAAAAAGTACG] or EXT2-2 [CTACACGGATGACATCAGCC]) containing oligonucleotides were introduced into the guide RNA (gRNA) expression cassette of the plentiCRISPR vector (a gift from Feng Zhang; Addgene plasmid 52961) (30). A random target sequence (N20) containing oligonucleotides was also introduced into the gRNA expression cassette of the plentiCRISPR vector. One microgram of the plentiCRISPR plasmid containing each target gRNA sequence was transfected into HmLu-1 cells (1 × 105 cells) with TransIT-LT1 (Mirus). One day after transfection, the medium was replaced with 10 μg/ml of puromycin-containing medium for 5 days of selection. Surviving cells were passaged, diluted, and inoculated onto fresh dishes for colony formation. Each colony was picked, propagated, and genotyped. The genomic region surrounding the CRISPR-Cas9 target site for each gene in the cloned cell was PCR amplified with KOD-FX neo (Toyobo), and the PCR products were gel extracted and sequenced using a 3130 Genetic Analyzer (ABI). Primer sequences are available upon request. Clones with indels introduced at the targeted site were picked. Genotyped clones were detached with phosphate-buffered saline (PBS) containing EDTA and incubated with anti-heparan sulfate MAb (10E4) (USBio) (49), followed by incubation with Alexa Fluor 488-conjugated anti-mouse IgM antibody (AbCam). IgM clone MOPC 104E (Sigma, St. Louis, MO, USA) was used as an isotype control. Labeled cells were analyzed by flow cytometry (FACSVerse, BD Biosciences).
Real-time RT-PCR for the quantification of cell surface-attached viruses.
For virus absorption on cells, 1 × 107 PFU/ml of AKAV(OBE-1), AKAV(Iriki), or SBV was inoculated onto HSPG-KO or random-KO HmLu-1 cells and left for 1 h at 4°C. After 1 h of incubation, unbound viruses were washed three times with ice-cold BSA/MEM. Virus-bound cells were lysed, and the total RNA was extracted with Isogen (Nippon Gene). The extracted RNAs were assayed using RNA-direct SYBR green real-time PCR master mix (Toyobo), according to the manufacturer's instructions, in a Thermal Cycler Dice Real Time System (TaKaRa). One microgram of the extracted RNAs was amplified using the AKAV S RNA-specific primer set (forward, 5′-CCACAACCAAGTGTCGATCT-3′; reverse, 5′-AGATGCGGTGAAGCGTAAA-3′) or the SBV S RNA-specific primer set (forward, 5′-GGCCAAGATGGTCCTACATAAG-3′; reverse, 5′-TGTCTGGCACAGGATTTGAG-3′). The RNA was normalized to host GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA using a golden hamster GAPDH-specific primer set (forward, 5′-AAGGTCATCCCAGAGCTGAA-3′; reverse, 5′-CTGCTTCACCACCTTCTTGA-3′). For relative quantification, a standard curve of AKAV or SBV S RNA and GAPDH was prepared by serial dilution of the mixture of total RNA extracted from uninfected HmLu-1 cells and viral RNA extracted from AKAV(OBE-1)- or SBV-containing supernatants.
Sandwich ELISA for detection of N proteins of cell surface-attached AKAV.
For virus absorption on cells, 1 × 107 PFU of AKAV(OBE-1) was inoculated onto HSPG-KO or control HmLu-1 cells in 6-well plates and left for 1 h at 4°C. After 1 h of incubation, unbound viruses were washed three times with ice-cold BSA/MEM. Virus-bound cells were then lysed with lysis buffer containing 10 mM Tris-HCl (pH 7.4), 0.5% Triton X-100, 150 mM NaCl, and 1 mM EDTA for 10 min on ice. The lysates were collected and clarified by centrifugation (10,000 × g for 5 min at 4°C). Supernatants were then added to the anti-AKAV N MAb (5E8)-coated wells of 96-well ELISA plates (MaxiSorp; Nunc) and incubated for 30 min at room temperature. After washing with PBS–0.1% Tween 20 (PBS-T), biotinylated (biotin labeling kit-NH2; Dojindo) anti-AKAV mouse PAb was added to the wells and incubated for 30 min at room temperature. After washing with PBS-T, avidin-biotinylated horseradish peroxidase (HRP) complex (Vectastain ABC kit; Vector Laboratories) was added to the wells and incubated for 30 min at room temperature. A 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution was used to read the assay.
EXT2 gene reintroduction into HSPG-KO cells.
The golden hamster EXT2 open reading frame sequence (GenBank accession number XM_013118841) was amplified from a pool of cDNA, which was a product of reverse transcription of total RNA from HmLu-1 cells, using an _EXT2_-specific primer set. A pS lentivirus transfer vector under the control of the spleen focus-forming virus promoter was prepared by removing the Venus gene from the pSVenusfull vector (50) by BamHI and NotI digestion. Amplified EXT2 cDNA was cloned into the pS lentivirus vector and designated pS-EXT2. The lentivirus EXT2 expression vector was produced in HEK293T cells by cotransfection of the transfer vector pS-EXT2 and two lentivirus packaging plasmids, p8.9QV (51) and pCAGGS-VSVG. The lentivirus vector was concentrated by ultracentrifugation and inoculated onto EXT2-KO HmLu-1 cells. HSPG expression was confirmed by flow cytometry as described above.
Statistical analysis.
All samples were compared by Student's t test with two-tailed analysis to determine statistically significant differences.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Young Scientists (B) (grant number 15K19104), a Grant-in-Aid for Scientific Research (A) (grant number 26252048), and a Grant-in-Aid for Challenging Exploratory Research (grant number 26660227) from the Japan Society for the Promotion of Science (JSPS), Japan.
REFERENCES
- 1.Jennings M, Mellor PS. 1989. Culicoides: biological vectors of Akabane virus. Vet Microbiol 21:125–131. doi: 10.1016/0378-1135(89)90024-2. [DOI] [PubMed] [Google Scholar]
- 2.Kirkland PD. 2015. Akabane virus infection. Rev Sci Tech 34:403–410. doi: 10.20506/rst.34.2.2366. [DOI] [PubMed] [Google Scholar]
- 3.Wernike K, Elbers A, Beer M. 2015. Schmallenberg virus infection. Rev Sci Tech 34:363–373. doi: 10.20506/rst.34.2.2363. [DOI] [PubMed] [Google Scholar]
- 4.Wernike K, Conraths F, Zanella G, Granzow H, Gache K, Schirrmeier H, Valas S, Staubach C, Marianneau P, Kraatz F, Höreth-Böntgen D, Reimann I, Zientara S, Beer M. 2014. Schmallenberg virus-two years of experiences. Prev Vet Med 116:423–434. doi: 10.1016/j.prevetmed.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi T, Yanase T, Yamakawa M, Kato T, Yoshida K, Tsuda T. 2007. Genetic diversity and reassortments among Akabane virus field isolates. Virus Res 130:162–171. doi: 10.1016/j.virusres.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Albornoz A, Hoffmann AB, Lozach PY, Tischler ND. 24 May 2016. Early bunyavirus-host cell interactions. Viruses 8:143. doi: 10.3390/v8050143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott RM. 2014. Orthobunyaviruses: recent genetic and structural insights. Nat Rev Microbiol 12:673–685. doi: 10.1038/nrmicro3332. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann H, Li X, Zhang X, Liu W, Kühl A, Kaup F, Soldan SS, González-Scarano F, Weber F, He Y, Pöhlmann S. 2013. Severe fever with thrombocytopenia virus glycoproteins are targeted by neutralizing antibodies and can use DC-SIGN as a receptor for pH-dependent entry into human and animal cell lines. J Virol 87:4384–4394. doi: 10.1128/JVI.02628-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozach PY, Mancini R, Bitto D, Meier R, Oestereich L, Overby AK, Pettersson RF, Helenius A. 2010. Entry of bunyaviruses into mammalian cells. Cell Host Microbe 7:488–499. doi: 10.1016/j.chom.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WuDunn D, Spear PG. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol 63:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13–22. doi: 10.1016/S0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 12.Dechecchi MC, Tamanini A, Bonizzato A, Cabrini G. 2000. Heparan sulfate glycosaminoglycans are involved in adenovirus type 5 and 2-host cell interactions. Virology 268:382–390. doi: 10.1006/viro.1999.0171. [DOI] [PubMed] [Google Scholar]
- 13.Bourgeois C, Bour JB, Lidholt K, Gauthray C, Pothier P. 1998. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J Virol 72:7221–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldman SA, Audet S, Beeler JA. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol 74:6442–6447. doi: 10.1128/JVI.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giroglou T, Florin L, Schäfer F, Streeck RE, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson T, Ellard FM, Ghazaleh RA, Brookes SM, Blakemore WE, Corteyn AH, Stuart DI, Newman JW, King AM. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol 70:5282–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanlandschoot P, Van Houtte F, Serruys B, Leroux-Roels G. 2005. The arginine-rich carboxy-terminal domain of the hepatitis B virus core protein mediates attachment of nucleocapsids to cell-surface-expressed heparan sulfate. J Gen Virol 86:75–84. doi: 10.1099/vir.0.80580-0. [DOI] [PubMed] [Google Scholar]
- 18.Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem 278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 19.Salvador B, Sexton NR, Carrion R, Nunneley J, Patterson JL, Steffen I, Lu K, Muench MO, Lembo D, Simmons G. 2013. Filoviruses utilize glycosaminoglycans for their attachment to target cells. J Virol 87:3295–3304. doi: 10.1128/JVI.01621-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Hearn A, Wang M, Cheng H, Lear-Rooney CM, Koning K, Rumschlag-Booms E, Varhegyi E, Olinger G, Rong L. 2015. Role of EXT1 and Glycosaminoglycans in the early stage of filovirus entry. J Virol 89:5441–5449. doi: 10.1128/JVI.03689-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med 3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 22.Roderiquez G, Oravecz T, Yanagishita M, Bou-Habib DC, Mostowski H, Norcross MA. 1995. Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J Virol 69:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Boer SM, Kortekaas J, de Haan CA, Rottier PJ, Moormann RJ, Bosch BJ. 2012. Heparan sulfate facilitates Rift Valley fever virus entry into the cell. J Virol 86:13767–13771. doi: 10.1128/JVI.01364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riblett AM, Blomen VA, Jae LT, Altamura LA, Doms RW, Brummelkamp TR, Wojcechowskyj JA. 2016. A haploid genetic screen identifies heparan sulfate proteoglycans supporting Rift Valley fever virus infection. J Virol 90:1414–1423. doi: 10.1128/JVI.02055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pietrantoni A, Fortuna C, Remoli ME, Ciufolini MG, Superti F. 2015. Bovine lactoferrin inhibits Toscana virus infection by binding to heparan sulphate. Viruses 7:480–495. doi: 10.3390/v7020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jusa ER, Inaba Y, Ishibashi K, Noda M. 1995. Effect of heparin on hemagglutination by Akabane and Aino viruses belonging to the Simbu group of bunyaviruses. Vet Microbiol 45:251–258. doi: 10.1016/0378-1135(95)00040-H. [DOI] [PubMed] [Google Scholar]
- 27.Sarrazin S, Lamanna WC, Esko JD. 2011. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci U S A 94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shieh MT, WuDunn D, Montgomery RI, Esko JD, Spear PG. 1992. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol 116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varela M, Schnettler E, Caporale M, Murgia C, Barry G, McFarlane M, McGregor E, Piras IM, Shaw A, Lamm C, Janowicz A, Beer M, Glass M, Herder V, Hahn K, Baumgärtner W, Kohl A, Palmarini M. 2013. Schmallenberg virus pathogenesis, tropism and interaction with the innate immune system of the host. PLoS Pathog 9:e1003133. doi: 10.1371/journal.ppat.1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith PD, Coulson-Thomas VJ, Foscarin S, Kwok JC, Fawcett JW. 2015. “GAG-ing with the neuron”: The role of glycosaminoglycan patterning in the central nervous system. Exp Neurol 274:100–114. doi: 10.1016/j.expneurol.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Janowicz A, Caporale M, Shaw A, Gulletta S, Di Gialleonardo L, Ratinier M, Palmarini M. 2015. Multiple genome segments determine virulence of bluetongue virus serotype 8. J Virol 89:5238–5249. doi: 10.1128/JVI.00395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, Heinz FX. 2001. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J Virol 75:5627–5637. doi: 10.1128/JVI.75.12.5627-5637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernard KA, Klimstra WB, Johnston RE. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93–103. doi: 10.1006/viro.2000.0546. [DOI] [PubMed] [Google Scholar]
- 36.Byrnes AP, Griffin DE. 2000. Large-plaque mutants of Sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J Virol 74:644–651. doi: 10.1128/JVI.74.2.644-651.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olmsted RA, Baric RS, Sawyer BA, Johnston RE. 1984. Sindbis virus mutants selected for rapid growth in cell culture display attenuated virulence in animals. Science 225:424–427. doi: 10.1126/science.6204381. [DOI] [PubMed] [Google Scholar]
- 38.Gardner CL, Hritz J, Sun C, Vanlandingham DL, Song TY, Ghedin E, Higgs S, Klimstra WB, Ryman KD. 2014. Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: a model for rational arboviral vaccine design. PLoS Negl Trop Dis 8:e2719. doi: 10.1371/journal.pntd.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wernike K, Eschbaumer M, Breithaupt A, Hoffmann B, Beer M. 2012. Schmallenberg virus challenge models in cattle: infectious serum or culture-grown virus? Vet Res 43:84. doi: 10.1186/1297-9716-43-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Artpradit C, Robinson LN, Gavrilov BK, Rurak TT, Ruchirawat M, Sasisekharan R. 2013. Recognition of heparan sulfate by clinical strains of dengue virus serotype 1 using recombinant subviral particles. Virus Res 176:69–77. doi: 10.1016/j.virusres.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones KS, Petrow-Sadowski C, Bertolette DC, Huang Y, Ruscetti FW. 2005. Heparan sulfate proteoglycans mediate attachment and entry of human T-cell leukemia virus type 1 virions into CD4+ T cells. J Virol 79:12692–12702. doi: 10.1128/JVI.79.20.12692-12702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takenaka-Uema A, Bangphoomi N, Shioda C, Uchida K, Gen F, Kato K, Haga T, Murakami S, Akashi H, Hoimoto T. 2016. Characterization of a recombinant Akabane mutant virus with knockout of a nonstructural protein NSs in a pregnant goat model. Virol Sin 31:274–277. doi: 10.1007/s12250-015-3704-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogawa Y, Fukutomi T, Sugiura K, Kato K, Tohya Y, Akashi H. 2007. Comparison of Akabane virus isolated from sentinel cattle in Japan. Vet Microbiol 124:16–24. doi: 10.1016/j.vetmic.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 44.Xu D, Esko JD. 2014. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem 83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol Immunol 47:613–617. doi: 10.1111/j.1348-0421.2003.tb03424.x. [DOI] [PubMed] [Google Scholar]
- 46.Takenaka-Uema A, Murata Y, Gen F, Ishihara-Saeki Y, Watanabe K, Uchida K, Kato K, Murakami S, Haga T, Akashi H, Horimoto T. 2015. Generation of a recombinant Akabane virus expressing enhanced green fluorescent protein. J Virol 89:9477–9484. doi: 10.1128/JVI.00681-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suda Y, Fukushi S, Tani H, Murakami S, Saijo M, Horimoto T, Shimojima M. 2016. Analysis of the entry mechanism of Crimean-Congo hemorrhagic fever virus, using a vesicular stomatitis virus pseudotyping system. Arch Virol 161:1447–1454. doi: 10.1007/s00705-016-2803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akashi H, Inaba Y. 1997. Antigenic diversity of Akabane virus detected by monoclonal antibodies. Virus Res 47:187–196. doi: 10.1016/S0168-1702(96)01415-3. [DOI] [PubMed] [Google Scholar]
- 49.van den Born J, Gunnarsson K, Bakker MA, Kjellén L, Kusche-Gullberg M, Maccarana M, Berden JH, Lindahl U. 1995. Presence of N-unsubstituted glucosamine units in native heparan sulfate revealed by a monoclonal antibody. J Biol Chem 270:31303–31309. doi: 10.1074/jbc.270.52.31303. [DOI] [PubMed] [Google Scholar]
- 50.Shimojima M, Ströher U, Ebihara H, Feldmann H, Kawaoka Y. 2012. Identification of cell surface molecules involved in dystroglycan-independent Lassa virus cell entry. J Virol 86:2067–2078. doi: 10.1128/JVI.06451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimojima M, Ikeda Y, Kawaoka Y. 2007. The mechanism of Axl-mediated Ebola virus infection. J Infect Dis 196(Suppl 2):S259–S263. doi: 10.1086/520594. [DOI] [PubMed] [Google Scholar]