Transcription factor activities enhance markers of drug sensitivity in cancer (original) (raw)
. Author manuscript; available in PMC: 2019 May 17.
Abstract
Transcriptional dysregulation induced by aberrant Transcription factors (TFs) is a key feature of cancer, but its global influence on drug sensitivity has not been examined. Here we infer the transcriptional activity of 127 TFs through analysis of RNA-seq gene expression data newly generated for 448 cancer cell lines, combined with publicly available datasets to survey a total of 1,056 cancer cell lines and 9,250 primary tumors. Predicted TF activities are supported by their agreement with independent shRNA essentiality profiles and homozygous gene deletions, and recapitulate mutant-specific mechanisms of transcriptional dysregulation in cancer. By analysing cell line responses to 265 compounds, we uncovered numerous TFs whose activity interacts with anti-cancer drugs. Importantly, combining existing pharmacogenomic markers with TF activities often improves the stratification of cell lines in response to drug treatment. Our results, which can be queried freely at dorothea.opentargets.io, offer a broad foundation for discovering opportunities to refine personalised cancer therapies.
Keywords: Transcription Factor activity, drug response, cancer heterogeneity, genomic markers, cancer drivers
Introduction
Transcriptional dysregulation is required for tumor progression and drug resistance acquisition. Many cancer driver genes are transcription factors (TFs). Notable examples include TP53, the most commonly mutated tumor suppressor that controls cell growth arrest(1), and HIF1A, a key regulator of the adaptive response to hypoxia and angiogenesis(2). TFs are commonly dysregulated due to genomic alterations or aberrations in their regulatory proteins. For example, TP53 activity can be suppressed through amplification of its repressor MDM2(3) and HIF1A upregulation is often induced by loss-of-function mutations in VHL(4). Due to their role as downstream signalling effectors, aberrant activities of any pathway protein may dysregulate TF activities, altering the expression of its transcriptional targets or “regulon”. Different from driver alterations in kinase-mediated signalling cascades, where redundancy provides compensatory mechanisms, aberrant transcriptional regulators have been argued to be harder to circumvent by secondary genomic alterations(5). Consequently, TFs have been proposed as key nodal oncogenic drivers and their activity patterns used to characterise genomic aberrations in cancer(6,7) or their influence on a patient's prognosis(8).
Recently, the Genomics of Drug Sensitivity in Cancer (GDSC)(9,10), Cancer Therapeutics Response Portal (CTRP)(11) and Cancer Cell Line Encyclopedia (CCLE)(12) have generated large-scale public pharmacogenomic datasets spanning multiple molecular data types across hundreds of cancer cell lines. These datasets enabled the identification of genomic, transcriptomic and epigenomic markers of drug sensitivity(9,10,12) and have uncovered a complex network of genomic alterations interacting with sensitivity to hundreds of drugs. The challenge is now to dissect the underlying molecular mechanisms regulating drug response, for which novel and more systemic functional approaches are needed.
Here we used TF regulatory activities as sensors of pathway dysregulation. Assuming that the activity of a TF can be estimated from the mRNA levels of its direct target genes, defined from prior TF-gene regulatory data, we derived single-sample TF activity profiles across 9,250 primary tumors from the Cancer Genome Atlas (TCGA) and 1,056 cancer cell lines, employing newly generated RNA-seq data for 448 cancer models. We evaluated the prediction accuracy on independent genomics and gene-essentiality screens. Then, we mined for statistical interactions between somatic mutations and TF activities. To discriminate mutant-specific effects, we functionally re-annotated somatic mutations based on the affected protein feature (e.g. regulatory sites, protein interactions, truncation, etc.). Finally, we investigated TF activities alone or in combination with genomic markers as potential predictors of sensitivity to 265 compounds, performing a large-scale evaluation of TFs as markers of drug sensitivity in cancer. The collection of identified interactions is publicly available at http://dorothea.opentargets.io.
Materials and Methods
Cell lines and primary tumors data
RNA from 448 cell lines was sequenced in-house (Table S1A). Cell lines were sourced from collaborators or repositories and have been used for GDSC(9,10) and COSMIC cell line(13) projects. These have been cryopreserved in aliquots in liquid nitrogen for 7 years in the lab. A single cryovial were thawed for use and propagated for a maximum of 3 months before being discarded. All cell lines were mycoplasma negative. Cell line’s identity was compared, where possible, against those provided by the repositories (ATCC, Riken, JCRB and DSMZ) using a panel of 16 STRs (AmpFLSTR Identifiler KIT, ABI) and the corresponding genotype data is available at COSMIC database (http://cancer.sanger.ac.uk/cell_lines). RNA libraries were made with the Stranded mRNA library kit from KAPA Biosystems according to the manual using the Agilent Bravo platform. Libraries were sequenced on an Illumina HiSeq 2000. Raw and processed data were deposited on the European Genome-phenome Archive (EGAS00001000828) and ExpressionAtlas (E-MTAB-3983). For the other cell lines, RNA-seq fastq files were downloaded from CCLE(12) (PRJNA169425) and Klijn et al(14)(EGAS00001000610). To minimise technical biases, the 3 datasets were re-analysed using iRAP(15) to obtain raw counts. TCGA samples’ raw counts were downloaded from the Gene Expression Omnibus (GSE62944)(16). Raw counts were normalised and processed into counts per million reads (Supplementary Methods).
For cell lines, Whole Exome Sequencing (WES), Copy Number Alterations (CNA), methylation and drug response data were retrieved from the GDSC1000 web-portal(10), while gene essentiality scores were downloaded from the Project-Achilles web-portal(17). For primary tumors, WES, CNA and clinical data were retrieved from cBioportal (Supplementary Methods). Tables S1A-D list all samples and data types.
Consensus TF regulons data
We defined a set of high-confidence human TFs from the table S3 provided in Vaquerizas et al(18), by excluding unlikely TFs noted as “x”. Secondly, we retrieved TF-target regulatory interactions from public resources covering different TF-binding evidences, including TF binding site (TFBS) predictions, chromatin immunoprecipitation coupled with high-throughput data (ChIP-X), text-mining derived and manually curated TF-target interactions (Supplementary Methods). For each TF, we defined a consensus TF regulon (CTFR) selecting TF-target interactions reported in more than one source (Table S2). TF-target interactions are unsigned and unweighted.
Scoring basal TF activities
Given a matrix of normalised gene expression values per sample, the first step consisted in a gene-wise normalisation employing a kernel estimator of the cumulative density function (kcdf)(19). Next, the level of activity of a TF regulon in a sample was approximated as a function of the collective mRNA levels of its targets using aREA (analytic Rank-based Enrichment Analysis), a statistical method from the VIPER R package based on the mean ranks’ comparisons(6). aREA’s normalised enrichment scores (NES) were used as estimates of regulon relative activity. Positive and negative scores indicated, respectively, greater or weaker relative activity in a sample compared to the background population (cell lines or TCGA samples). The aREA algorithm was selected because it takes into account the effect (activation/repression) of the TF on each target, thus enabling comparisons with other types of regulatory networks. R code to compute TF activities is available at https://github.com/saezlab/DoRothEA
Comparison between normal and primary tumors
Differential TF activities contrasting normal and tumoral TCGA samples were computed using the limma R package. The matrix of relative activities per sample was used to fit a linear model for each TF (lmFit) and the eBayes test used to obtain the corresponding moderated t-statistics, nominal and adjusted p-values.
Association analysis with TF and driver mutations
We grouped somatic variants in driver genes according to their potential implications at the protein-level (Supplementary Methods). Next, we analysed the association between each group of mutations and the activity of a TF with an analysis-of-variance (ANOVA) test as in (10). To ensure comparable measurements among datasets and groups of mutations, we removed confounding effects by regressing-out effects associated with tissue lineage from the TF activity profiles. Next, for each mutation group/TF pair, the corrected TF activities were modeled as a function of the mutation status. The change in TF activity between mutants and wild-type samples was defined by Cohen’s d effect size and significance were estimated with type-II ANOVA using the car R package. P-values were adjusted for multiple testing corrections (Benjamini-Hochberg method) on a gene basis.
Association analysis with drug response
We used a linear model to correlate drug responses with TF activities. For each drug-TF pair, drug IC50s across all samples (YIC50) were modeled as a function of the dependent covariates (Xcovariates, including tissue-type in pan-cancer analyses, microsatellite instability status and screening medium), TF estimated activity (XTF) and noise (ψ):
YIC50=βcovariatesXcovariates+βTFXTF+ψ
The impact of the TF on drug response was defined by the regression coefficient (_β_TF) estimated with a multiple linear least squares regression. Significance of the regressors was estimated with a type-II ANOVA using the car R package. Finally, for each cancer-type, p-values were adjusted for multiple testing corrections using the Benjamini-Hochberg method.
In the pan-cancer analysis, the tissue-type was defined by the GDSC_description2 due to the presence of several cell lines without a matching TCGA label. In cancer-specific analyses, we grouped the samples according to the TCGA labels, for consistency with the GDSC1000 study(10). We additionally tested Ewing's sarcoma, leukemia, lymphoma, osteosarcoma and rhabdomyosarcoma tumor-types.
Association analysis between TF activities and known pharmacogenomic markers
Large-effect significant pharmacogenomic markers (p<0.001, FDR<20% and Glass∆s>1) were extracted from GDSC1000(10). For each pharmacogenomic marker (GM), we fit a null multiple regression model (Mnull) where the independent variable is the drug IC50 (YIC50) and the dependent variables are the cell line covariates (tumor type, MSI, and screening media; Xcovariates), the genomic marker (XGM) and a noise term (ψ):
Mnull:YIC50=βcovariatesXcovariates+βGMXGM+ψ
Next, for each TF in our panel, we built a nested regression model (MTF) containing the same variables of Mnull plus the activities of the TF (XTF) and their interaction with GM (XGM*TF):
MTF:YIC50=βcovariatesXcovariates+βGMXGM+βTFXTF+βGM∗TFXGM∗TF+ψ
Every MTF was compared to the corresponding null model Mnull using a Likelihood Ratio (LR) test. Resulting p-values were adjusted using the Benjamini-Hochberg method.
Results
TF activities estimation
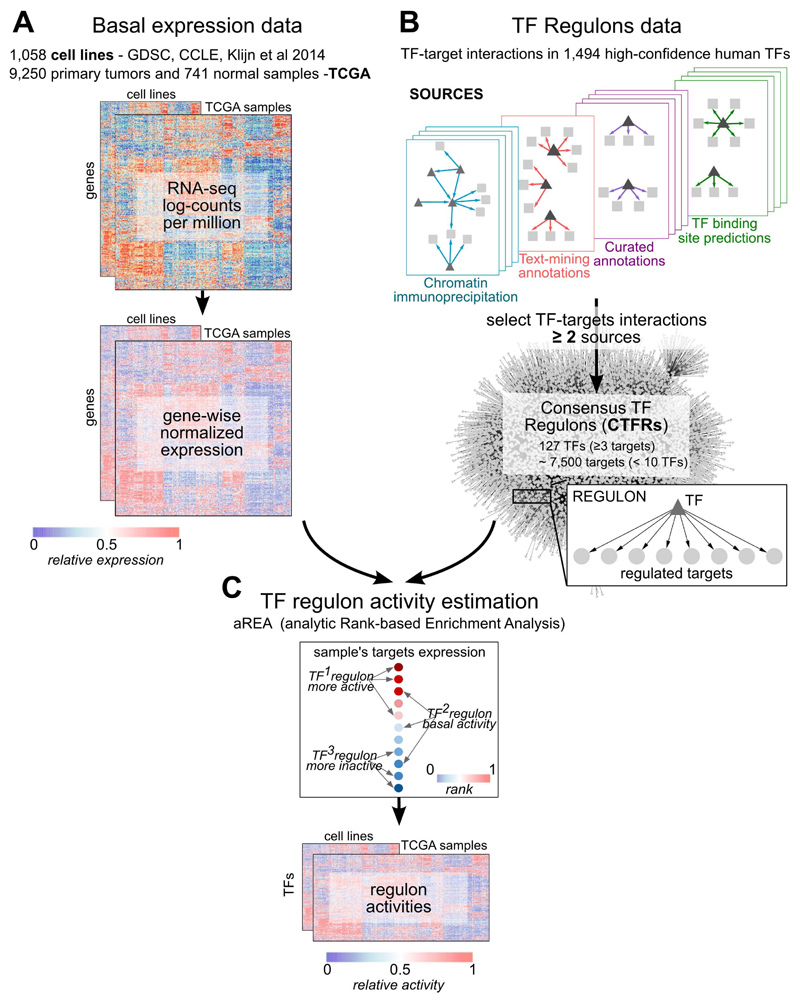
First, we assembled a collection of basal transcriptional profiles of immortalised human cancer cell lines and primary tumors (Figure 1A). For cancer cell lines, we newly derived RNA-seq data from 448 samples, that we complemented with RNA-seq profiles from 934 and 622 cell lines, respectively from the CCLE(12) and Klijn et al(14). This yielded a total of 1,362 unique cancer cell lines, of which 1,056 are in COSMIC(13) (Table S1A). To minimise technical biases, we derived raw counts using a common pipeline. For primary tumors, we downloaded RNA-seq raw counts for 9,250 TCGA primary tumors and 741 normal samples(16). Cell lines and TCGA samples were processed and normalised separately.
Figure 1. Estimation of Transcription Factor (TF) activities overview.
A) RNA-seq basal gene expression in cell lines data and TCGA samples (normal and tumors together) was processed separately to obtain normalised log-counts per million (CPM) that were then gene-wise normalised using a kernel estimation of the cumulative density function (kcdf). B) Consensus TFs regulons (CTFRs) were derived by selecting TF-target interactions observed in at least 2 sources among a collection of databases. In final CTFRs, targets under > 10 TFs and TFs with < 3 targets are removed. C) Estimation of single-sample TF activities from gene expression data and the CTFRs using the aREA algorithm from VIPER. Normal and tumor TCGA samples were analysed together.
To define the TF regulons (i.e. sets of genes whose transcription is regulated by a given TF), we collected 15,211 TF-targets interactions appearing in at least 2 publicly available resources (hereafter Consensus TF Regulons, CTFR; Figures 1B and S1A-B; Table S2). To ensure a minimum signal when we compute the TF activities, we removed targets regulated by more than 10 TFs and TFs with less than 3 targets in the expression matrix. The final CTFRs consisted of 7,445 targets for 127 TFs, with 111 targets per TF on average (Figure S1C). Pairwise overlap between regulons was low (average Jaccard Similarity Coefficient=0.0044, Figure S1D), indicating negligible levels of redundancy between CTFRs.
Next, we normalised the transcriptomic data gene-wisely to estimate relative levels of basal activity of each CTFR in each sample using the aREA algorithm(6). Cell lines and TCGA samples (tumor+normal) were analysed separately (Figure 1C; Tables S3A-B). Normalised enrichment scores (NES) (Figure S2A-D) were used as estimates of CTFR activity relative to the background population (hereafter simplified as ‘TF activities’). Subsampling analysis revealed that activity estimates were robust in populations with n≥20 (Figure S2E).
We evaluated the TF activity estimations using independent benchmark data derived from an essentiality screening(17), and CNA and WES data in cell lines (Supplementary Methods). Moreover, we investigated the inclusion of methylation data as a means to refine CTFRs on a cell line-basis excluding from the regulons those targets with hypermethylated promoters, not observing significant performance improvements (Supplementary File, Figures S3-S4). Finally, we compared the activities derived from the CTFRs against those derived from reverse-engineered regulons proposed in (6), observing slightly better performances for CTFRs (Supplementary File, Figures S3-S6). Hence, we selected CTFRs-based estimations (without including promoter methylation information) for our downstream analysis.
TF activities across primary tumors and cell lines
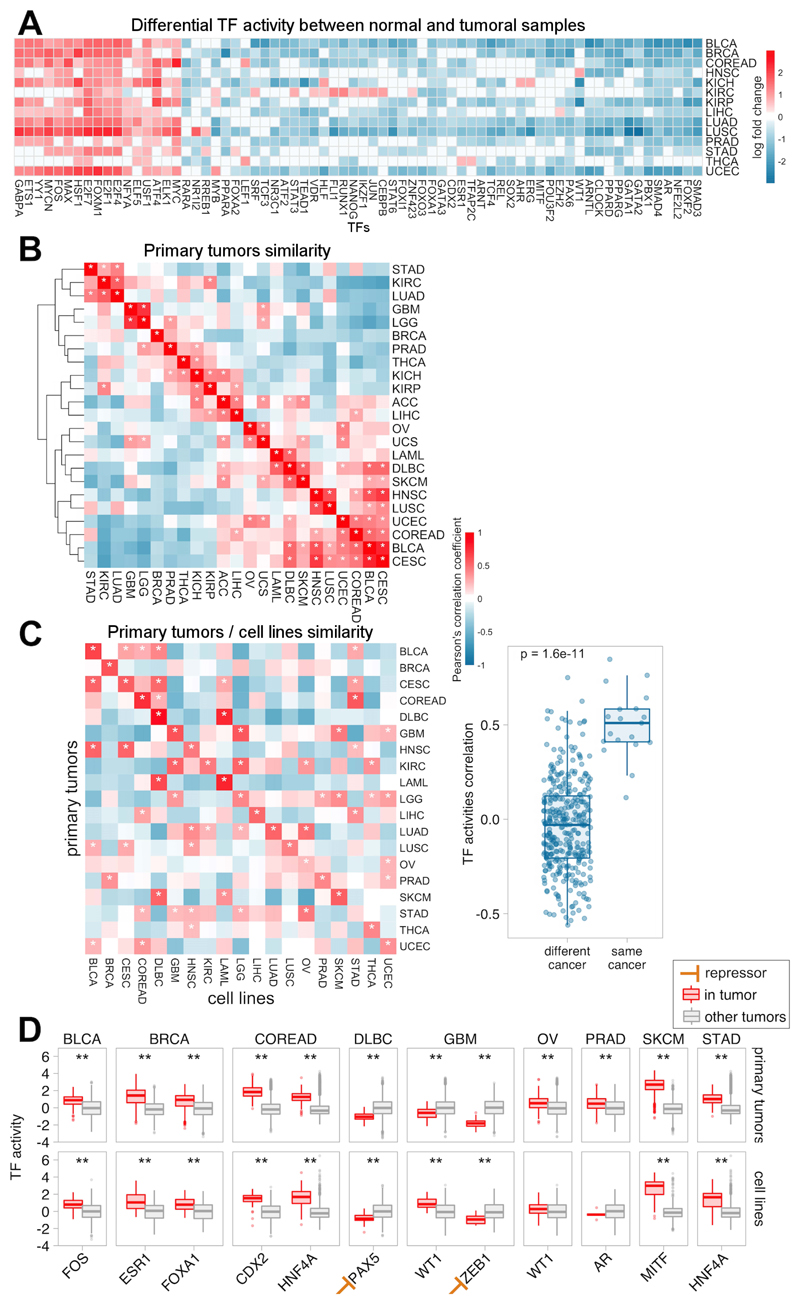
To obtain a global picture of TFs operating in primary tumors, we studied how TF activities distribute across TCGA samples. Differential activity analysis of normal versus tumor samples revealed groups of TFs consistently activated or repressed across the 14 tumor types with matched normal samples. While most TF regulons decrease their activity, a small subset undergoes a recurrent increase across tumor types (Figure 2A), including oncogenic regulators of cell cycle (MYC, MAX, E2F family members, FOS and FOXM1), tumor invasion and angiogenesis (ELK1 and ETS1)(20).
Figure 2. Transcription Factor (TF) activities across primary tumors and cancer cell lines.
A) Heatmap of the differential TF activity (log fold change) between normal and tumoral samples across 14 tumor types. Red and blue indicate lower- and higher-activity in tumors, respectively, whereas white indicates non-significant (adj. p>0.05) associations. Only TFs with significant in at least 50% of the tumors are plotted. B) Tumor type similarity: Correlation based hierarchical clustering of tumor type-level TF activities for 23 primary tumors. C) Comparison of TF activities between primary tumors and cell lines for 19 common tumor types. Each value in the heatmap represents the pearson correlation coefficients between tumor type-level TF activities. Asterisks indicate significant correlations (adj. p<0.05). D) Activity distributions for tissue-specific TFs. Each point represents the TF activity in a given patient or cell line.
Next, we compared the TF profiles between cancer types. First, we summarised sample-level activities into cancer-level activities. For each TF, we ranked the samples based on TF activity and quantified the enrichment of each cancer-type at the top of the ranks using the aREA algorithm (Figure S7A-B, Table S3C-D). Hierarchical clustering based on euclidean distance highlighted similar activity profiles for primary tumors from the same tissue lineage such as the diffuse gliomas GBM and LGG, hematopoietic and lymphoid LAML and DLBC or squamous-like tumors BLCA, CESC, HNSC and LUSC (Figure 2B). These clusters were also observed in the cell lines (Figure S7C). Correlation analysis revealed a significant agreement in the TF profiles between cell lines and primary tumors from the same tissue lineage (average Pearson correlation 0.5 and -0.035 within and between different tumor types respectively, Figure 2C).
Closer examination of well-established tissue-specific TFs (retrieved from the Human Cancer Protein Atlas v15(21)) showed that our approach captures 11 out of 12 TFs operating preferentially in specific tissues in primary tumors (Figure 2D) such ESR1 and FOXA1 in BRCA or MITF in SKCM. Note that for ZEB1, a transcriptional repressor involved in epithelial-to-mesenchymal transition (EMT)(22), higher protein activities correspond to a downregulation of the regulon. Importantly, these tendencies are maintained in the cell lines with the exception of AR, where 4 out of 6 prostate cell lines display AR-independent proliferation (Table S1C). Taken together, these results show that our approach captures expected activity patterns of known cancer-specific TFs.
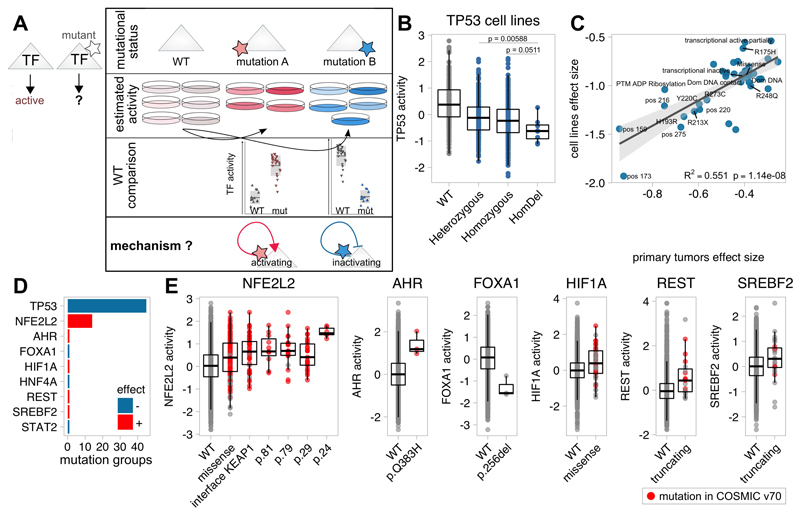
TF activities dissect mutant-specific aberrations
Previous studies demonstrated that different mutations in the same protein could cause a continuum of effects, ranging from neutrality to a significant functional impact(23). We thus set-out to characterise the effect of mutations occurring in TFs on their own activity. As proof of concept, we focused on TP53 due to its high mutation frequency and heterogeneity. We curated TP53 mutations according to specific mutations, hotspots, protein consequence, zygosity (in cell lines), affected domain, PTM or structural property and previously proposed mutation stratifiers(24) (Table S4A). Subsequently, we compared predicted TP53 activities of each TP53 mutation group with wild-type samples (Figure 3A). To avoid confounding effects due to the use of different samples and tumor types, we regressed-out the tissue lineage from the TF activity profiles through linear modelling. Our results indicated that all TP53 mutation groups significantly affecting TP53 transcriptional activity decreased it (Figure S8; Table S4B). Overall, homozygous mutations and deletions had a stronger effect size than heterozygous mutations (Figure 3B). Focusing on the most frequent mutational hotspots revealed that R231X and R273C reached larger effect sizes than R248Q and R175H substitutions in both primary tumors and cell lines. However, direct pairwise comparison between mutants did not yield significant results alone. Importantly, these significant changes in activity were correlated between primary tumors and cell lines (R2=0.551, p=1.14×10-8, Figure 3C). This suggests that transcriptional activity prediction may better capture effect on TP53 activity than mutation alone. This is supported by comparison of activity predictions with experimentally defined TP53 mutant yeast transactivation class from the IARC TP53 Database(25)where each possible TP53 missense mutation is assigned to a transactivation class -functional, partially or non-functional- according to its effects on the transcription of 8 TP53-responsive promoters in yeast). Comparison between non-functional and the other missense mutants showed a significant agreement with our predictions in cell lines (one-tailed t-tests, p=0.00535) and, although marginally significant, in primary tumors (p=0.0418).
Figure 3. Functional characterisation of mutant Transcription Factor (TF) on transcriptional activities.
A) Functional evaluation of TF mutations on their own activity. B) Boxplot depicting TF activities according TP53 mutation zygosity in cell lines. C) Comparison of the predicted effect size of significant TP53 mutations between primary tumors and cell lines. D) Systematic characterisation of mutant TFs in primary tumors. Each bar represents the number of significant mutant groups in the TF impacting its activity. Red and blue indicate positive and negative effects, respectively. E) Boxplots comparing TF activities across different variants for NFE2L2, AHR, FOXA1, REST and SREBF2. Red dots indicate that the mutation is reported in COSMIC v70.
Motivated by these results, we investigated systematically the effect of mutations in TFs on their activity. To distinguish mutant-specific effects, these were studied individually. Importantly, to consider non-recurrent yet potentially functional driver mutations, we also grouped mutations that, although introducing different changes in different residues, could affect protein function in a similar way (e.g. same structural region, interaction or post-translational modification site). We recovered 1,200 mutation groups in 122 TFs from primary tumors (n≥3). Pancancer analysis in primary tumors identified 9 TFs that, when mutated, exhibit a significant change in activity(FDR<5%; Figure 3D, Table S4C). In general, mutations in TFs with known oncogenic roles, such as NFE2L2, HIF1A and AHR were associated with increased regulatory activity, pointing to gain-of-function mutations. In contrast, mutations in the proposed tumor suppressors STAT2 and FOXA1 are associated with decreased activity. Also, truncating mutations in the transcriptional repressor REST resulted in increased regulon expression (Figure 3E). Analysing cell lines showed similar trends for REST truncating mutations (p=0.00367, FDR=0.0367) and NFE2L2 missense mutation in D29 (p=0.009, FDR=0.099).
Closer examination of results revealed again differences in the effect of mutation types on protein activity. In NFE2L2, a cytoprotective oncogene, missense mutations affecting p.W24/p.D29 residues at the surface or at the KEAP1-interface (positions 77-82) are associated with higher NFE2L2 activity, with NFE2L2W24R/C mutations causing the strongest increase(Figure 3E). Mutations at the KEAP1-binding site were already proposed to be positively selected to abolish NFE2L2 degradation(26).
Associations with known driver mutations
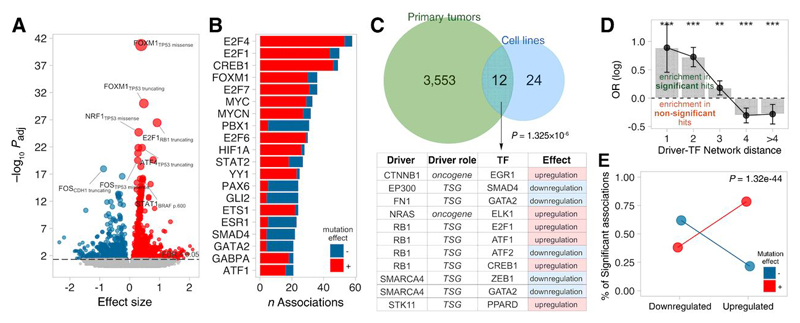
Next, we evaluated how mutations in any cancer driver genes, proposed in(27,28), could impact TF activities. We grouped mutations in driver genes following the same strategy described for TFs. This yielded 1,774 mutation groups (n ≥ 5) in 171 driver genes.
Systematic comparisons of TF activities in mutant against wild-type primary tumors yielded 3,565 driver mutation groups-TF associations involving 97 driver genes and 75 TFs (FDR<5%, Figure 4A-B; Table S5A). The same analysis in cell lines allowed us to study only 533 mutation groups, and rendered fewer associations (probably due to lower sample number) that involved 36 interactions between 17 drivers and 25 TFs (FDR<5%, Table S5B). Importantly, 12 hits were shared between primary tumors and cell lines with concordant effect (Fisher’s exact test (FET), odds-ratio=7.89 p<1.32×10-6, Figure 4C). Some of these associations represent proposed mechanisms of TF regulation, such as the repression of E2F1 by RB1, perhaps the best-described inhibitor of TF function(29), or ELK1 regulation by ERK-MAPK pathway(20,30).
Figure 4. Functional characterisation of driver mutations on Transcription Factor (TF) activities.
A) Volcano plot with effect size (x) and adjusted p-value (y) of all tested pancancer associations. B) Number of significant associations per TF in primary tumors and cell lines, coloured according to the sign of the association: red and blue correspond to significantly higher or lower TF activities in mutants compared to wild-type, respectively. C) Significant (FDR<5%) the TF-driver associations from primary tumors and cell lines and the overlap. Shared driver-TF pairs are indicated in the table. D) Log odds-ratio of finding a significant interaction by network distance (minimum number of directed intermediates between the driver and the corresponding TF). ** indicates p<0.05; ***p<0.001 (FET). E) Enrichment in positive/negative driver-TF associations (red/blue, respectively) to involve oncogenic/tumor-suppressor TFs, respectively.
To assess whether the detected associations represent plausible driver-TF regulatory events, we extracted directed edges from literature-curated signalling networks from OmniPath(31) and quantified shortest path lengths between every driver-gene/TF pair. Enrichment analysis confirmed that significant driver-gene/TF associations tend to be closer than non-significant associations (Figure 4D). Next, we investigated whether the predicted effect of driver mutations on TF activities (association sign) agrees with the TF’s role in cancer. We classified TFs into 3 groups according to their role in cancer: (i) up-regulated in cancer, if the TF displays significant greater activity in tumor than in normal samples or is a known oncogene(27,28); (ii) down-regulated, if the TF function is repressed in tumor samples or is a tumor suppressor; or (iii) neutral. Enrichment analysis revealed that positive driver/TF interactions (i.e. potential TF-activating events) tend to involve cancer upregulated TFs, in contrast, negative interactions are more prone to involve cancer downregulated TFs (Figure 4E). Taken together, our results suggest that the identified associations point to potential mechanisms of driver-mediated transcriptional dysregulation in cancer.
Drug sensitivity interactions in 984 cancer cell lines
We next investigated the potential of TF activities as markers of response to 265 compounds across 984 cancer cell lines(10). Association between TF-drug pairs was tested with a linear regression approach accounting for potentially confounding factors (tissue lineage, microsatellite instability and cell lines growth media).
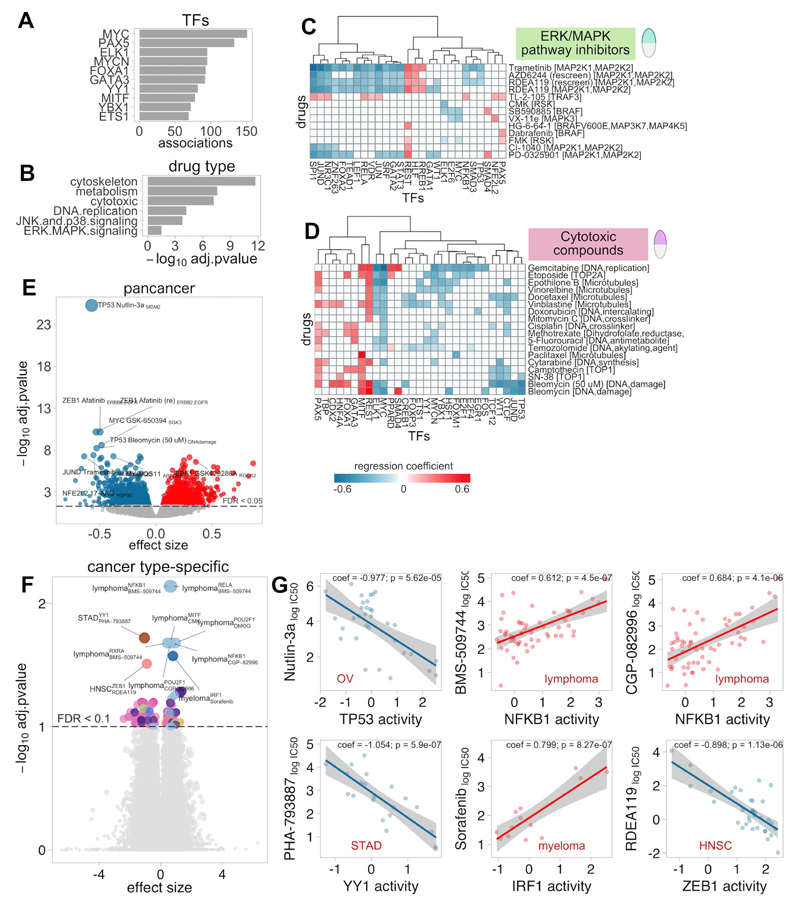
A pancancer analysis identified 3,300 significant TF-drug associations (p<0.001, FDR<5%), with 251 drugs (95%) and 123 TFs (97%) implicated in at least one interaction (Table S6A). Most drugs were associated with multiple TFs, which, considering the relatively low overlap between regulons (Figure S1D) may correspond to functional cooperation in transcriptional regulators rather than target redundancy. In fact, interacting TFs display similar activity profiles (Figure S9). Most TF-drug associations involved relevant oncogenic TFs such as MYC, PAX5, MYCN, FOXA1 and GATA3 (Figure 5A; Table S6B-C). Significant associations were enriched for cytotoxic drugs and compounds targeting cytoskeleton, metabolism, DNA replication, JNK-p38 and ERK-MAPK signalling (FET p<0.001, Figure 5B; Table S6D).
Figure 5. Associations between Transcription Factor (TF) activities and drug sensitivity.
A) Frequency of TFs in significant pancancer TF-drug associations. B) Enrichment p-values for drug classes that are overrepresented among significant pancancer associations. C and D) Heatmaps of significant associations with with drugs targeting ERK-MAPK pathway (C) and cytotoxic drugs (D). E) Volcano plot with effect size (x) and adjusted p-value (y) of all tested pancancer TF-drug associations. Red and blue indicate positive (resistance) and negative (sensitivity) effects, respectively. F) Volcano plot with effect size (x) and adjusted p-value (y) of all tested cancer-specific TF-drug associations. G) Examples of cancer-specific TF-drug associations. Red and blue indicate positive (resistance) and negative (sensitivity) effects, respectively.
Some of the investigated TFs are recurrently mutated in cancer and have already been proposed as genomic markers of drug sensitivity. To validate if TF activities are able to recapitulate the same drug-gene associations observed at the genomic-level, we compared our findings with the pharmacogenomic interactions (FDR<25% and p<0.001) previously identified for these cell lines(10). Our approach identified 12 out of the 21 significant pharmacogenomic interactions involving a TF in our panel (FET p=8.39×10-4, odds-ratio=4.45), including TP53 mutations interacting with response to Nutlin-3a; MYC with Vismodegib and PAX5 with Bleomycin, among others. The same drug association analysis on TF activities derived from reverse-engineered regulons(6) instead of CTFRs (on the overlapping TFs) rendered fewer hits than CTFRs, none in the pharmacogenomic marker list (Figure S10).
Next, we investigated whether TF activity predicts sensitivity to direct intervention of their upstream regulators with targeted drugs. We extracted from OmniPath the interactions involving the proteins targeted by the drugs. Enrichment analysis confirmed that significant hits were more likely to involve TFs directly connected to the targets of the associated drug (FET p=0.0155, odds-ratio=1.2), suggesting that predicted activities may be indeed indicative of upstream pathway activation and therefore useful sensitivity markers for drugs targeting their components. For example, sensitivity to drugs targeting the ERK-MAPK pathway (Figure 5C) was associated with increased activities in several MEK targeted TFs including SPI1, JUN, JUND and STAT3(32,33), whereas vulnerability to the two tested RSK-inhibitors correlates with ELK1, another known downstream MAPK target(20,30).
Also, our analyses also identified TFs showing simultaneous sensitivity interactions to drugs targeting common processes. For example, sensitivity to cytotoxic compounds was associated with TFs classically upregulated in actively proliferating cells such as MYC, while activity of tissue-specific TFs (such as MITF, REST or HNF4A) was associated with resistance to these drugs (Figure 5D).
The strongest detected association involved TP53 and Nutlin-3a (regression coefficient (coeff)=-0.57, p=x1.58×10-30, Figure 5E). Nutlin-3a is an MDM2-inhibitor that blocks MDM2-mediated TP53 degradation. Our results agree with pharmacogenomic studies in that samples with lower TP53 activities show lower sensitivity to MDM2-inhibition(9,10). Another strong interaction was ZEB1 upregulation, an EMT marker, associated with resistance to EGFR inhibitor Afatinib (coeff=-0.53, p=5.19×10-15) and Gefitinib (coeff=-0.24, p=5.9×10-7). This is in agreement with a recent study in NSCLC describing ZEB1-mediated acquired resistance to EGFR-inhibitors(34).
Cancer-specific analysis revealed fewer associations compared to the pancancer analysis, probably due to reduced sample size (Figure 5F; Table S6E). Still, we recovered 125 TF-drug associations (p<0.001, FDR<10%), most in lymphoma, the largest subpopulation. Some hits involved drugs with no associated genomic markers (Figure 5G). Among the top hits we found that NFKB1 activity was associated with sensitivity to ITK inhibitor BMS-509744 in Lymphoma cells (coeff=0.612, p=4.5×10-7); in STAD, sensitivity to PHA-793887, a pan-CDK inhibitor, was associated with YY1, recently proposed to contribute to gastric oncogenesis(35) (coeff=-1.05, p=5.9×10-7); in Myeloma, resistance to the tyrosine kinase inhibitor Sorafenib was associated with the activity of IRF1, a proposed tumor suppressor for Acute Myeloid Leukemia(36) (coeff=0.8, p=8.27×10-7); finally, sensitivity to the MEK inhibitor RDEA119 in HNSC associated with ZEB1 activity (coeff=-0.898, p=1.13×10-6) a key EMT effector in HNSC development(37).
TF activities enhance the predictive ability of genomic markers
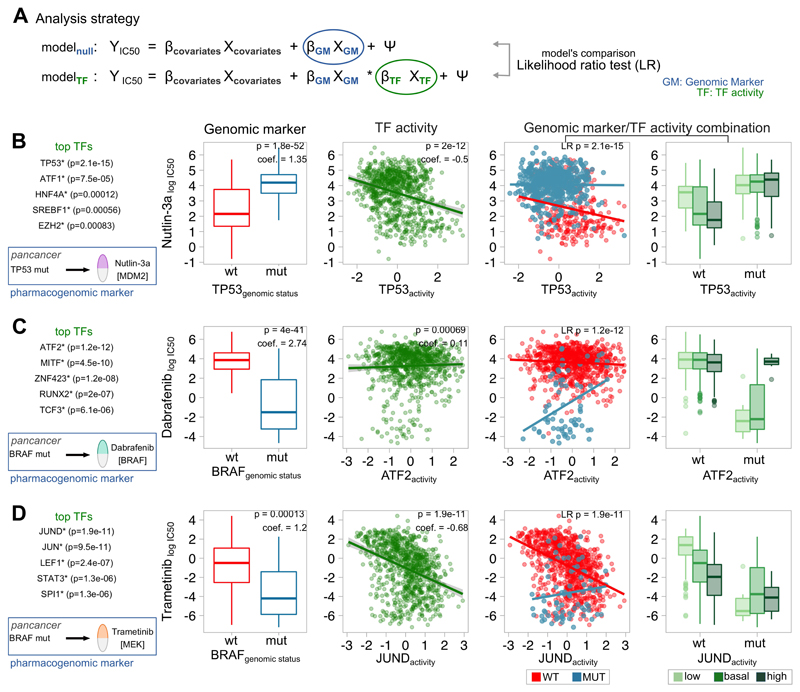
We showed before that the strongest TF-drug association detected involved the well-known interaction between TP53 and Nutlin-3a. According to previous studies, samples with TP53 mutations are Nutlin3a-resistant(9,10), while our results suggest that samples with higher TP53 activities are more sensitive. We reasoned that protein activities might complement mutation-based markers to further improve the stratification of sensitive and nonsensitive cell lines. To test this hypothesis, we used a Likelihood Ratio test (LR) to compare pharmacogenomic models with and without including TF activities (Figure 6A). We confirmed that TP53 activity was able to further identify sensitive cell lines among wild--type samples (Figure 6B, p=2.1×10-15). None other TF outperformed TP53. This observation was reproduced in 3 out of 5 individual tumor types (OV, GBM and LAML; p<0.05) where TP53 mutations are markers of Nutlin-3a response.
Figure 6. Modelling the combined effect on drug sensitivity of known pharmacogenomic markers and Transcription Factor (TF) activities.
A) Analysis strategy: 2 pharmacogenomic regression models are built, one without any TF information (null model) and another including the activity of a TF (test model). Both models are compared using a Likelihood Ratio test. B) Improvement of the association TP53MUTNutlin-3a by including TP53 TF activity. C) Improvement of the association BRAFMUT-Dabrafenib by including ATF2 TF activity. D) Improvement of the association BRAFMUT-Trametinib by including JUND TF activity. Left box represents the top TFs improving the null pharmacogenomic model. Indicated p-values are nominal, with FDR<0.05. First boxplot represents the IC50 (y) in mutant (blue) and WT (red) samples (x). The second scatterplot represents the IC50 (y) and the predicted TF activity (x). The third scatterplot represents the interaction between the IC50 (y) and the predicted TF activity (x) in mutant (blue) and WT (red) samples. The fourth boxplot represents the IC50 (y) in mutant and WT samples (x) coloured according to the TF activity (low: activity<-1; basal: -1<activity<1; high: activity>1).
Motivated by this finding we ran a systematic analysis to search for TFs able to refine known pharmacogenomic interactions. Overall, 95 out of 160 (59.4%) tested strong-effect pharmacogenomic interactions identified in(10) are improved by at least one TF (FDR<5%, LR test; Table S7). The second most significant hit after TP53-Nutlin3a involved the interaction between BRAF mutations and the FDA-approved BRAF inhibitor Dabrafenib. Specifically, in BRAF mutant samples, resistance to Dabrafenib interacts with ATF2 and MITF activity (Figure 6C, p=1.2×10-12 and p=4.68×10-10). Resistance in BRAF mutants to Dabrafenib was still observable in SKCM samples with higher ATF2-targets expression (p=0.00123). The importance of ATF2 in melanoma is supported by several lines of evidence; ATF2 is required for melanoma tumor development(38) nuclear ATF2 (transcriptionally active) is associated with poor prognosis and genotoxic stress-resistance(39). Moreover, PKCε, the kinase mediating ATF2 transcriptional activity, is among the top 10 kinases associated with BRAF-inhibition resistance, which supports the relationship between ATF2 and Dabrafenib resistance(40). In fact, ATF2 essentiality scores from Achilles project(41) correlate with the predicted activity for ATF2 in BRAFV600E mutant samples at the pancancer-level (Pearson correlation, R=-0.615, p=0.0332; Figure S11A) but not in BRAFwt (R=0.082, p=0.347; Figure S11B).
Interestingly, the most significant improvements in predictions were observed between drugs targeting ERK-MAPK signalling (FET p=5.28×10-6) and the driver genes BRAF, KRAS or HRAS. For example, in BRAF wild-type samples, sensitivity to MEK inhibitors improved including JUND in the model, among others (p=1.86×10-11, p=3.12×10-11 and p=1.77×10-8; Trametinib, RDEA119 and AZD6244, respectively; Figure 6D). JUND is a downstream substrate in ERK-MAPK signalling(32). Our previous analysis already suggested JUND activity be predictive of MEK-inhibition sensitivity alone. Here we show how JUND also improves response prediction to MEK inhibitor AZD6244 within HRAS mutant pancancer samples (p=1.21×10-7). Taken together, our results suggest that JUND regulon expression may be used as a sensor of ERK-MAPK pathway activity and vulnerability to MEK-inhibition.
Finally, other potential interactions affecting well-established pharmacogenomic markers are: the interaction of JUND with cell cycle CDK4/CDK6 inhibitor in RB1 mutants (p=1.9×10-6), which modulate cyclins(42); sensitivity to AKT inhibitor GSK690693 interaction with several TFs in OV PIK3CA mutants, where the stronger hits involve EGR1 and CREB1 (p=5.1×10-6 and p=7.6×10-3), downstream effectors of PI3K-Akt pathway(43); and, in HER2+ BRCA samples, sensitivity interaction between ELF1 activity and ERBB2 inhibitors Lapatinib and CP724714 (p=3.9×10-6 and p=8.9×10-5), a candidate regulator of ERBB2 expression(44).
Discussion
TFs activities derived from gene expression data have attracted much attention in cancer research during the last years. Recent studies have used different strategies to derive TF activity profiles across different cancers, evaluated their potential as prognostic markers(8), and applied them to characterise the impact of cancer somatic alterations(6,7). Although based on different definitions of TF regulons, the common outcome is that estimating regulatory activities from the mRNA levels of the targeted genes can reveal known and novel mechanisms involved in tumor development. However, the potential of TF activities as markers to guide personalised treatments, alone or in combination with established genomic markers, has not yet been explored.
Here, we applied a pipeline to derive signatures of TF activity from new and existing RNA-seq data in 1,056 cancer cell lines and 9,250 primary tumors. Our approach combines consensus TF regulons (CTFRs) and gene-wise normalised expression data with unsupervised single-sample enrichment algorithms. This circumvents the need for a prior classification of samples into subtypes, of particular benefit when working with heterogeneous group of cancer samples, and does not require of unperturbed reference samples (often not available). Moreover, comparable TF activity signatures can be obtained for new samples by normalising the expression values of each gene against our reference panel of samples.
TF activity profiles enabled us to (i) functionally characterise different TF mutations; (ii) link genomic aberrations in cancer driver genes with TF dysregulation; (iii) suggest new mechanisms for response to specific compounds in cancer models; and (iv) propose new markers of drug response, alone or in combination with genomic markers. Although we expect some interactions to reflect the cooperative behaviour between TFs controlling common processes rather than causal associations, these recapitulated known pharmacogenomic relationships and were enriched for TF-drug pairs close in the signalling network. Thus, we envision that the identified associations provide reliable evidence to refine existing hypotheses or formulate new ones to understand therapeutic outcomes. Particularly, our study shows that predictions of therapeutic response can be improved if, in addition to the mutational status of a marker gene, the regulatory activity of the coded protein is also considered. This can be achieved directly when the marker gene codes for a TF (as exemplified by TP53-Nutlin3a response), or indirectly when the protein targeted by the drug regulates a TF (as the case of JUND in MEK inhibitors).
The critical factor in the quantification of TF activities is the definition of the targets putatively regulated. Here, we chose to use a curated compendium of regulatory interactions (Consensus TF Regulons; CTFR) derived from different TF-DNA binding evidences such as in vivo ChIP-Seq experiments, in silico TFBS predictions and manual curations. The major limitations of our approach are: (i) CTFRs are restricted by prior knowledge, which may render incomplete regulons; (ii) the assumption that a TF either induces or represses its targets (but TFs may have both roles) and (iii) the cell-type dependencies of some TF-target interactions. Taken together, these limitations may cause inaccurate activity estimations for TFs with dual activator/repressor role or for TFs whose targets vary across cell types(45). Under these considerations, approaches inferring condition-specific regulons from transcriptomic associations have become popular(46). The underlying principle is that TF circuits can be inferred by correlating mRNA levels of the TFs with all other genes(47). However, our comparison revealed that activities derived from CTFRs perform slightly better than those from inferred regulons. Potential explanations may be that: (i) inference methods assume that mRNA levels are good activity indicators of the coded proteins, which may fail for TFs whose activity depends on post-translational regulation (such phosphorylation) or indeed their stoichiometric assembly as heteromeric complexes(48); (ii) these methods are susceptible of being confounded by indirect associations or co-expression with other TFs(49); (iii) regulons inferred from primary tumors may not capture regulatory events occurring in cell lines; and (iv) the pervasiveness of somatic mutations changing the function of TFs. Pertinent examples are loss-of-function TP53 missense mutants which, while abundantly present at mRNA and protein level, are unable to regulate the expression of its canonical targets. Finally, the inference of such condition-specific networks requires a prior classification of samples, which may not be trivial for heterogeneous cancer cell line panels. An alternative could combine CTFRs with network inference approaches(50).
Nonetheless, our TF predictions based on CTFRs agree with independent essentiality screenings and genomic data, and mimic changes in transactivation potential observed in mutagenesis studies. Importantly, CTFRs are able to reproduce known pharmacogenomic interactions while inferred regulons fail to do so. However, it is worth mentioning that our strategy to retrieve CTFRs may favour well-studied TFs, whose targets are thoroughly characterised, thus resulting in biased performances. Further refinement of the approaches to define TF regulons activity in cancer should enable to find further pharmacogenomic interactions, novel markers and therapeutic opportunities.
Briefly, our results demonstrate that TF activity profiles derived from CTFRs can be used to characterise genomic alterations and drug response in cancer patients, proposing these as promising complementary therapeutic markers. The proposed approach may have strong implications in the refinement of personalised treatment methodologies. We envision that with the increase in the coverage and quality of the CTFRs, the proposed strategy will become instrumental to interpret transcriptional dysregulation in cancer and elucidate its clinical implications.
Supplementary Material
Supplementary Data
Acknowledgements
This work was supported by Open Targets (grant number OTAR016). Research in MJG laboratory is funded by the Wellcome Trust (102696) and Open Targets (OTAR014). We thank the Gene Expression Atlas team for the help with the RNA-sequencing processing, especially Laura Huerta for the curation of sample annotations and Robert Petryszak and Alvis Brazma for the general support. We thank Nils Blüthgen for the help in the curation of JASPAR data. We thank Ultan McDermott, Simon Cook, Stacey Price, Jayeta Saxena and Hayley Francies for feedback on targeted therapies in melanoma and colorectal cancer cells. We thank Pedro Beltrao, Ivan Costa, Luis Tobalina and Denes Turei for insightful discussions and providing valuable feedback on the manuscript; and Euan Stronach, Paul Fisher and Glyn Bradley for input in design and analysis. We thank Roberto Battisti for designing and implementing a first prototype version of the DoRothEA online tool.
Footnotes
Disclosure of Potential Conflicts of Interest
All the authors declare that they have no competing interests.
References
- 1.Levine AJ. p53, the Cellular Gatekeeper for Growth and Division. Cell. 1997;88:323–31. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med. 2001;7:345–50. doi: 10.1016/s1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- 3.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–3. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 4.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 5.Gonda TJ, Ramsay RG. Nat Rev Cancer. Vol. 15. Nature Research; 2015. Directly targeting transcriptional dysregulation in cancer; pp. 686–94. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016;48:838–47. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osmanbeyoglu HU, Toska E, Chan C, Baselga J, Leslie CS. Pancancer modelling predicts the context-specific impact of somatic mutations on transcriptional programs. Nat Commun. 2017;8 doi: 10.1038/ncomms14249. 14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falco MM, Bleda M, Carbonell-Caballero J, Dopazo J. The pan-cancer pathological regulatory landscape. Sci Rep. 2016;6 doi: 10.1038/srep39709. 39709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12:109–16. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–83. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, et al. Nat Biotechnol. Vol. 33. Nature Research; 2014. A comprehensive transcriptional portrait of human cancer cell lines; pp. 306–12. [DOI] [PubMed] [Google Scholar]
- 15.Fonseca NA, Petryszak R, Marioni J, Brazma A. iRAP - an integrated RNA-seq Analysis Pipeline. [cited 2017 Feb 27];bioRxiv. 2014 :005991. [Internet]. Available from: http://biorxiv.org/content/early/2014/06/06/005991. [Google Scholar]
- 16.Rahman M, Jackson LK, Johnson WE, Li DY, Bild AH, Piccolo SR. Alternative preprocessing of RNA-Sequencing data in The Cancer Genome Atlas leads to improved analysis results. Bioinformatics. 2015;31:3666–72. doi: 10.1093/bioinformatics/btv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1 doi: 10.1038/sdata.2014.35. 140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 19.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller JM, Krauss B, Kaltschmidt C, Baeuerle PA, Rupec RA. Hypoxia induces c-fos transcription via a mitogen-activated protein kinase-dependent pathway. J Biol Chem. 1997;272:23435–9. doi: 10.1074/jbc.272.37.23435. [DOI] [PubMed] [Google Scholar]
- 21.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 22.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–40. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Ory K, Legros Y, Auguin C, Soussi T. Analysis of the most representative tumour-derived p53 mutants reveals that changes in protein conformation are not correlated with loss of transactivation or inhibition of cell proliferation. EMBO J. 1994;13:3496–504. doi: 10.1002/j.1460-2075.1994.tb06656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–11. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37:865–76. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 26.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–73. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10:1081–2. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–62. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R, Ernst WH, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12:5097–104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Türei D, Korcsmáros T, Saez-Rodriguez J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat Methods. 2016;13:966–7. doi: 10.1038/nmeth.4077. [DOI] [PubMed] [Google Scholar]
- 32.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–73. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 33.Ceresa BP, Horvath CM, Pessin JE. Signal transducer and activator of transcription-3 serine phosphorylation by insulin is mediated by a Ras/Raf/MEK-dependent pathway. Endocrinology. 1997;138:4131–7. doi: 10.1210/endo.138.10.5266. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida T, Song L, Bai Y, Kinose F, Li J, Ohaegbulam KC, et al. ZEB1 Mediates Acquired Resistance to the Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors in Non-Small Cell Lung Cancer. PLoS One. 2016;11:e0147344. doi: 10.1371/journal.pone.0147344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang W, Tong JHM, Chan AWH, Zhao J, Dong Y, Wang S, et al. Yin Yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. J Transl Med. 2014;12:80. doi: 10.1186/1479-5876-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green WB, Slovak ML, Chen IM, Pallavicini M, Hecht JL, Willman CL. Lack of IRF-1 expression in acute promyelocytic leukemia and in a subset of acute myeloid leukemias with del(5)(q31) Leukemia. 1999;13:1960–71. doi: 10.1038/sj.leu.2401596. [DOI] [PubMed] [Google Scholar]
- 37.Duque-Afonso J, Wei MC, Lin C-H, Feng J, Buechele C, Wong SH-K, et al. Blood. Vol. 126. American Society of Hematology; 2015. Oncogenic Role for the Lck/ZAP70/PLCG2 Signaling Pathway in Pre-B-ALL Pathogenesis; pp. 810–810. [Google Scholar]
- 38.Berger AJ, Kluger HM, Li N, Kielhorn E, Halaban R, Ronai Z, et al. Subcellular localization of activating transcription factor 2 in melanoma specimens predicts patient survival. Cancer Res. 2003;63:8103–7. [PubMed] [Google Scholar]
- 39.Lau E, Kluger H, Varsano T, Lee K, Scheffler I, Rimm DL, et al. PKCε promotes oncogenic functions of ATF2 in the nucleus while blocking its apoptotic function at mitochondria. Cell. 2012;148:543–55. doi: 10.1016/j.cell.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma V, Young L, Cavadas M, Owen K, Reproducibility Project: Cancer Biology Registered Report: COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Elife. 2016;5 doi: 10.7554/eLife.11414. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao DD, Tsherniak A, Gopal S, Weir BA, Tamayo P, Stransky N, et al. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res. 2013;23:665–78. doi: 10.1101/gr.143586.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanden Bush TJ, Bishop GA. CDK-mediated regulation of cell functions via c-Jun phosphorylation and AP-1 activation. PLoS One. 2011;6:e19468. doi: 10.1371/journal.pone.0019468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarkson AN, Parker K, Nilsson M, Walker FR, Gowing EK. Combined ampakine and BDNF treatments enhance poststroke functional recovery in aged mice via AKT-CREB signaling. J Cereb Blood Flow Metab. 2015;35:1272–9. doi: 10.1038/jcbfm.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott GK, Chang CH, Erny KM, Xu F, Fredericks WJ, Rauscher FJ, 3rd, et al. Ets regulation of the erbB2 promoter. Oncogene. 2000;19:6490–502. doi: 10.1038/sj.onc.1204041. [DOI] [PubMed] [Google Scholar]
- 45.Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, Rohs R. Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci. 2014;39:381–99. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marbach D, Costello JC, Küffner R, Vega NM, Prill RJ, Camacho DM, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tegnér J, Yeung MKS, Hasty J, Collins JJ. Reverse engineering gene networks: Integrating genetic perturbations with dynamical modeling. Proceedings of the National Academy of Sciences. 2003;100:5944–9. doi: 10.1073/pnas.0933416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margolin AA, Califano A. Theory and limitations of genetic network inference from microarray data. Ann N Y Acad Sci. 2007;1115:51–72. doi: 10.1196/annals.1407.019. [DOI] [PubMed] [Google Scholar]
- 49.Marbach D, Prill RJ, Schaffter T, Mattiussi C, Floreano D, Stolovitzky G. Revealing strengths and weaknesses of methods for gene network inference. Proc Natl Acad Sci U S A. 2010;107:6286–91. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ernst J, Beg QK, Kay KA, Balázsi G, Oltvai ZN, Bar-Joseph Z. PLoS Comput Biol. Vol. 4. Public Library of Science; 2008. A Semi-Supervised Method for Predicting Transcription Factor–Gene Interactions in Escherichia coli; p. e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data