VP1 Amino Acid Residue 145 of Enterovirus 71 Is a Key Residue for Its Receptor Attachment and Resistance to Neutralizing Antibody during Cynomolgus Monkey Infection (original) (raw)
The contribution of VP1-145 variants in humans is not fully understood. In some studies, VP1-145G/Q viruses were isolated more frequently from severely affected patients than from mildly affected patients, suggesting that VP1-145G/Q viruses are more virulent. In the accompanying paper (K. Kobayashi, Y. Sudaka, A. Takashino, A. Imura, K. Fujii, and S. Koike, J Virol 92:e00681-18, 2018, https://doi.org/10.1128/JVI.00681-18), we showed that VP1-145E viruses are more virulent than VP1-145G viruses in human SCARB2 transgenic mice. Heparan sulfate acts as a decoy to specifically trap the VP1-145G viruses and leads to abortive infection. Here, we demonstrated that VP1-145G was attenuated in cynomolgus monkeys, suggesting that this hypothesis is also true in a nonhuman primate model. VP1-145E viruses, but not VP1-145G viruses, were highly resistant to neutralizing antibodies. We propose the difference in resistance against neutralizing antibodies as another mechanism of EV71 virulence. In summary, VP1-145 contributes to virulence determination by controlling attachment receptor usage and antibody sensitivity.
KEYWORDS: animal models, enterovirus, neutralizing antibodies, pathogenesis
ABSTRACT
Enterovirus 71 (EV71) is a causative agent of hand, foot, and mouth disease and sometimes causes severe or fatal neurological complications. The amino acid at VP1-145 determines the virological characteristics of EV71. Viruses with glutamic acid (E) at VP1-145 (VP1-145E) are virulent in neonatal mice and transgenic mice expressing human scavenger receptor B2, whereas those with glutamine (Q) or glycine (G) are not. However, the contribution of this variation to pathogenesis in humans is not fully understood. We compared the virulence of VP1-145E and VP1-145G viruses of Isehara and C7/Osaka backgrounds in cynomolgus monkeys. VP1-145E, but not VP1-145G, viruses induced neurological symptoms. VP1-145E viruses were frequently detected in the tissues of infected monkeys. VP1-145G viruses were detected less frequently and disappeared quickly. Instead, mutants that had a G-to-E mutation at VP1-145 emerged, suggesting that VP1-145E viruses have a replication advantage in the monkeys. This is consistent with our hypothesis proposed in the accompanying paper (K. Kobayashi, Y. Sudaka, A. Takashino, A. Imura, K. Fujii, and S. Koike, J Virol 92:e00681-18, 2018, https://doi.org/10.1128/JVI.00681-18) that the VP1-145G virus is attenuated due to its adsorption by heparan sulfate. Monkeys infected with both viruses produced neutralizing antibodies before the onset of the disease. Interestingly, VP1-145E viruses were more resistant to neutralizing antibodies than VP1-145G viruses in vitro. A small amount of neutralizing antibody raised in the early phase of infection may not be sufficient to block the dissemination of VP1-145E viruses. The different resistance of the VP1-145 variants to neutralizing antibodies may be one of the reasons for the difference in virulence.
IMPORTANCE The contribution of VP1-145 variants in humans is not fully understood. In some studies, VP1-145G/Q viruses were isolated more frequently from severely affected patients than from mildly affected patients, suggesting that VP1-145G/Q viruses are more virulent. In the accompanying paper (K. Kobayashi, Y. Sudaka, A. Takashino, A. Imura, K. Fujii, and S. Koike, J Virol 92:e00681-18, 2018, https://doi.org/10.1128/JVI.00681-18), we showed that VP1-145E viruses are more virulent than VP1-145G viruses in human SCARB2 transgenic mice. Heparan sulfate acts as a decoy to specifically trap the VP1-145G viruses and leads to abortive infection. Here, we demonstrated that VP1-145G was attenuated in cynomolgus monkeys, suggesting that this hypothesis is also true in a nonhuman primate model. VP1-145E viruses, but not VP1-145G viruses, were highly resistant to neutralizing antibodies. We propose the difference in resistance against neutralizing antibodies as another mechanism of EV71 virulence. In summary, VP1-145 contributes to virulence determination by controlling attachment receptor usage and antibody sensitivity.
INTRODUCTION
Enterovirus 71 (EV71) is a member of the genus Enterovirus in the family Picornaviridae and is classified as a human enterovirus species A (HEV-A) (1). EV71 and other members of HEV-A are major causative agents of hand, foot, and mouth disease (HFMD) (1). They usually cause mild or subclinical infection. However, in some patients, EV71 can cause severe neurological symptoms, including meningitis, brainstem encephalitis, poliomyelitis-like paralysis, pulmonary edema, and death (2). However, virulence determinants of EV71 have not been identified.
EV71 is a nonenveloped, positive-stranded RNA virus with a genome approximately 7,400 bases long (1). The EV71 RNA genome is enclosed within an icosahedral capsid comprising 60 copies of each of four viral structural proteins, VP1 to VP4 (3, 4). Human scavenger receptor B2 (hSCARB2) is the essential receptor for establishment of infection of all EV71 strains (5). hSCARB2 supports attachment of the virion at the cell surface, internalization, and initiation of virion uncoating (6, 7). hSCARB2 is speculated to bind to the canyon of the virion like other picornavirus receptors that can initiate uncoating, but this has not been experimentally proven. EV71 also uses molecules that support attachment of the virion to the cell surface but not uncoating (6). These molecules, including P-selectin glycoprotein ligand (PSGL-1) (8), heparan sulfate (HS) (9), annexin II (10), sialic acid (11), nucleolin (12), and vimentin (13), are called attachment receptors (14). HS, a highly sulfated glycosaminoglycan (GAG), is expressed as HS proteoglycan on cell surfaces in its membrane-bound form and on the extracellular matrix in its secreted form (15). Amino acid residue 145 of the capsid protein VP1 (VP1-145) is located within the DE loop near the center of the 5-fold mesa of EV71 virions (3, 4) and is variable among EV71 isolates. The viruses that have glycine (G), glutamine (Q), or glutamic acid (E) at this position, which we refer to as VP1-145G, VP1-145Q, and VP1-145E, respectively, have been isolated. VP1-145E is most common (81%), whereas VP1-145G (9%) and VP1-145Q (9%) are less common (16). This amino acid variation also influences the attachment receptor usage and the virulence of the virus. The region surrounding VP1-145 is rich in positively charged amino acids, and VP1-145G and VP1-145Q viruses bind HS (17) and PSGL-1 (16) via electrostatic interactions, whereas the negative charge of the E residue in VP1-145E impairs these interactions. The contribution of this variation to pathogenicity is not fully understood. Change of VP1-145G/Q to VP1-145E is responsible for murine adaptations and/or virulence because VP1-145E strains of EV71 are more virulent than VP1-145G/Q strains in different neonatal mouse models (18–22). It is not clear whether this mutation is merely a murine adaptation determinant or a virulence determinant in general. To elucidate PSGL-1-dependent viral replication in vivo and pathogenesis, Kataoka et al. produced viruses that can and cannot bind PSGL-1, namely, PSGL-1-binding (PB) (VP1-98E145G) and PSGL-1-nonbinding (non-PB) (VP1-98K145E) strains, respectively, and used them to infect cynomolgus monkeys, a nonhuman primate model (23). The results suggested that non-PB strains are mainly responsible for the development of viremia and neuropathogenesis. In humans, VP1-145G or VP1-145Q was isolated more frequently from severely affected patients than from mildly affected patients, although the majority of viruses isolated from both patient types have E at position 145 (24–27). In the accompanying paper (28), we showed that VP1-145E is more virulent than VP1-145G in an hSCARB2 transgenic (tg) mouse model. We found that VP1-145G viruses use HS as an attachment receptor but that this binding capacity results in adsorption of the virus in nonsusceptible tissues. Therefore, HS-binding capacity did not contribute to viral replication in vivo. We proposed a mechanism of attenuation whereby VP1-145G virus is selectively adsorbed by HS and undergoes abortive infection.
To investigate how VP1-145 residues contribute to pathogenesis of EV71 in a nonhuman primate model closely related to humans (29–34), we infected cynomolgus monkeys with VP1-145 variants. We showed that VP1-145E viruses are more virulent than corresponding VP1-145G viruses. VP1-145G viruses did not spread efficiently, consistent with our hypothesis that HS-binding strains are trapped by HS. Furthermore, VP1-145E viruses were more resistant to neutralizing antibodies than VP1-145G viruses. We propose that the resistance to neutralizing antibodies is another determinant of EV71 fitness in vivo and virulence.
RESULTS
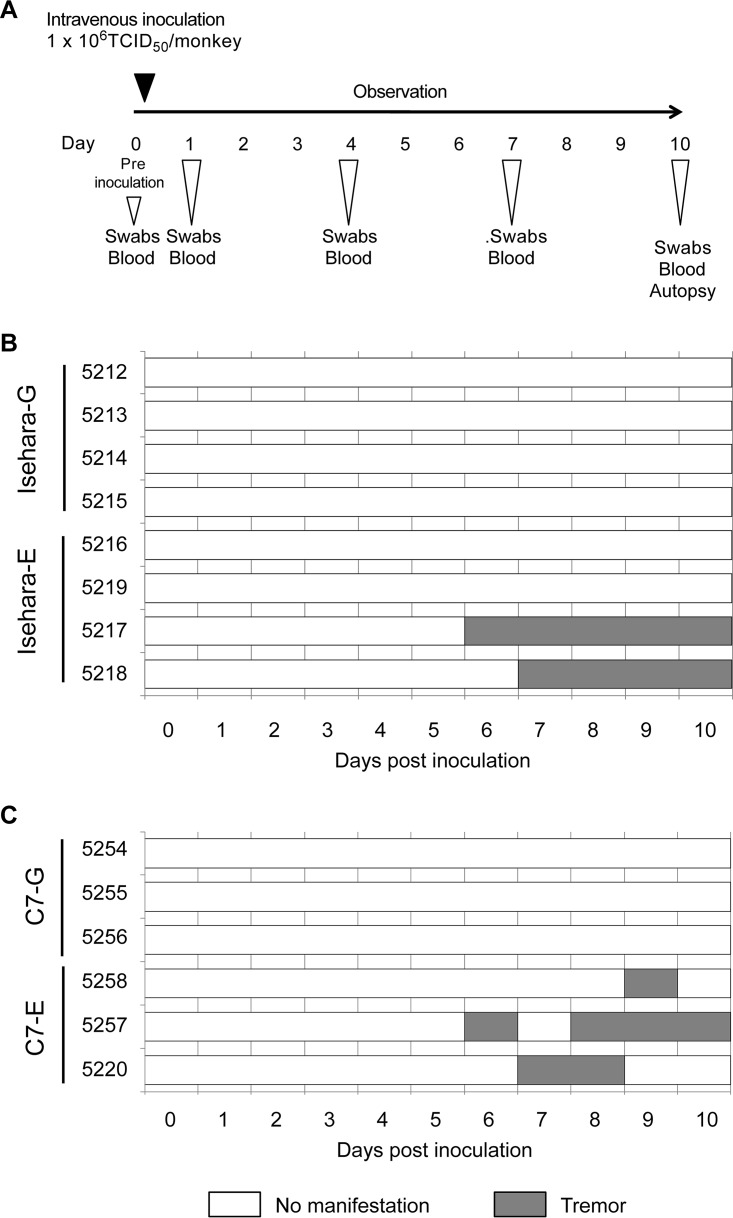
Clinical symptoms in cynomolgus monkeys inoculated with VP1-145 viruses.
We generated recombinant viruses, based on the Isehara, C7/Osaka, and SK-EV006 strains, that have G or E at VP1-145 and determined the virulence of these viruses in an hSCARB2 tg mouse model (28). VP1-145E resulted in greater virulence than VP1-145G in all virus strains. To confirm whether VP1-145E is more virulent than VP1-145G in a nonhuman primate model, we inoculated cynomolgus monkeys with VP1-145 mutants of the Isehara and C7/Osaka strains. Four monkeys each were intravenously inoculated with 106 50% tissue culture infective doses (TCID50) of Isehara-G or Isehara-E. Three monkeys each were also intravenously inoculated with 106 TCID50 of C7-G or C7-E. After inoculation, clinical manifestations of the monkeys were observed daily for 10 days (Fig. 1A). During the observation period, no monkeys in either group demonstrated very severe clinical manifestations (Fig. 1B and C). Two monkeys inoculated with Isehara-E (monkeys 5217 and 5218) exhibited tremors beginning 6 to 7 days postinoculation (dpi) (Fig. 1B), as did two monkeys inoculated with C7-E (5257 and 5220) (Fig. 1C). One monkey inoculated with C7-E (5258) exhibited tremors at 9 dpi (Fig. 1C). No monkeys inoculated with Isehara-G or C7-G had apparent neurological manifestations. Neither HFMD-like symptoms nor pulmonary edema was observed in any group. Thus, these results indicated that VP1-145E viruses induce neurological symptoms more frequently than VP1-145G viruses.
FIG 1.
Experimental infection of cynomolgus monkeys with VP1-145 mutants and time course of clinical observations. (A) Experimental schedule. Monkeys were intravenously inoculated with 1 ml of virus solution containing 106 TCID50 of VP1-145G or VP1-145E virus at day 0, and the clinical manifestations were observed daily for 10 days. Under anesthesia, clinical samples were collected on indicated days, and postmortem tissue samples were collected at 10 dpi. (B) Monkeys 5212, 5213, 5214, and 5215 were inoculated with Isehara-G and monkeys 5216, 5217, 5218, and 5219 with Isehara-E. (C) Monkeys 5254, 5255, and 5256 were inoculated with C7-G and monkeys 5257, 5258, and 5220 with C7-E.
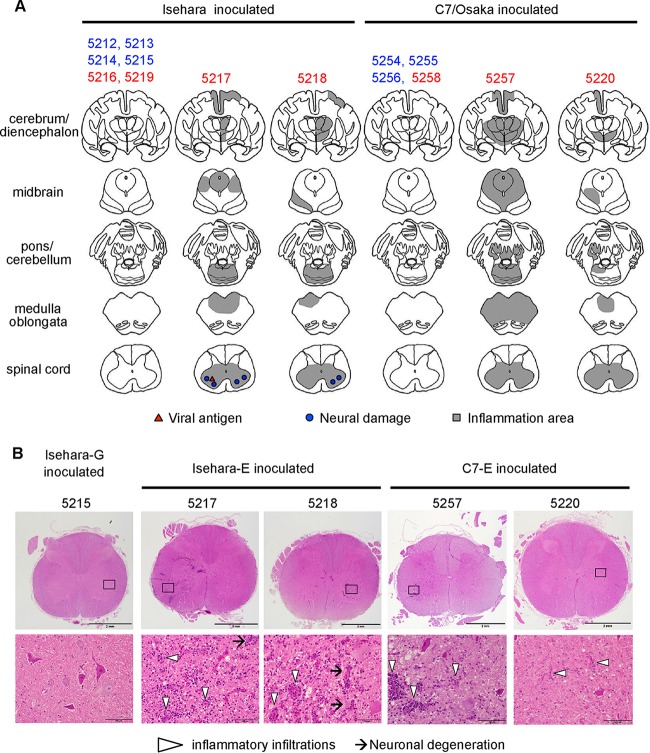
Histological analysis of CNS tissues and neurovirulence of VP1-145 viruses.
The monkeys were sacrificed at 10 dpi, and central nervous system (CNS) tissue samples were histopathologically examined (Fig. 2). In the Isehara-E-inoculated group, extensive inflammation was found in many areas of the CNS, including the spinal cord and gray matter of the cerebrum in the two monkeys that exhibited neurological symptoms (monkeys 5217 and 5218). There were mild neurodegenerations in the anterior horn of the spinal cord (Fig. 2B). Viral antigen was detected in the anterior horn of the spinal cord of monkey 5217 (Fig. 2A). Pathological changes were not found in the CNS tissues of the two monkeys (5216 and 5219) with no neurological symptoms (Fig. 2A). In the Isehara-G-inoculated group, no monkeys had any apparent CNS lesions (Fig. 2A, monkeys 5212, 5213, 5214, and 5215). Similarly, in the C7-E-inoculated group, inflammations were found in the CNS tissues of the two monkeys with tremors (5257 and 5220) (Fig. 2A). Although monkey 5258, inoculated with C7-E, exhibited mild neurological symptoms at 8 dpi, inflammation and neuronal degeneration were not found in the CNS tissues (Fig. 2A). The monkeys inoculated with C7-G had no apparent CNS lesions (Fig. 2A, monkeys 5254, 5255, and 5256). In conclusion, histopathological analysis of CNS tissues at 10 dpi revealed that Isehara-E and C7-E induced inflammation and neuronal damage, whereas Isehara-G and C7-G did not.
FIG 2.
Histopathological comparison of CNS tissues in EV71-inoculated monkeys. (A) Distribution of EV71-induced lesions. Postmortem CNS tissues at 10 dpi from monkeys inoculated with Isehara or C7/Osaka are shown. Filled triangles and circles indicate viral antigen-positive and neuronal damage, respectively. The gray areas show inflammation in the parenchyma and the meninges of the CNS. (B) Histopathological findings in spinal cord. Typical histopathological changes in the spinal cords of VP1-145E-inoculated groups are shown. Upper panels, histology of the lumbar part of the spinal cord of a monkey inoculated with Isehara-G (monkey 5215) and monkeys inoculated with Isehara-E (5217 and 5218) or C7-E (5257 and 5220), stained using H&E. Lower panels, higher magnifications of the boxes in the anterior horns in the upper panels. Prominent inflammatory infiltrations are indicated by arrowheads. Neuronal degenerations were seen in the anterior horns of the spinal cords within the lesion (arrows).
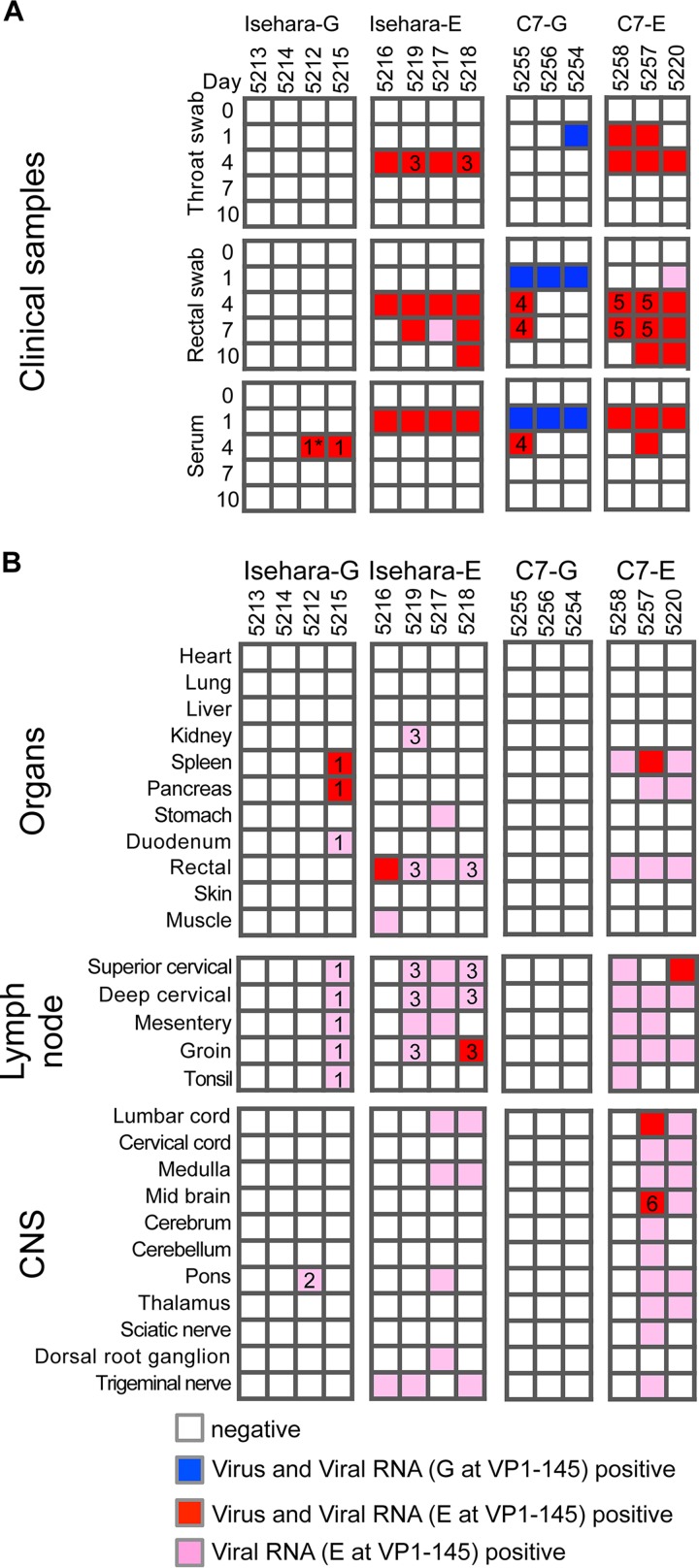
Virus isolation and viral RNA detection from clinical and tissue samples.
To monitor viral spread following intravenous inoculation, clinical samples (throat swabs, rectal swabs, and sera) were collected at 0 (preinoculation), 1, 4, 7, and 10 dpi, and autopsy samples were obtained at 10 dpi (Fig. 1A). As viral titers in clinical and autopsy samples were low (23), we isolated viruses using RD-SCARB2 cells (28), which are approximately 100-fold more susceptible to EV71 infection than RD-A cells. Almost all cytopathic effects (CPEs) appeared during the second blind passage. We also performed direct detection of viral RNA from clinical and autopsy samples by reverse transcription-PCR (RT-PCR). The sensitivity of RT-PCR is higher than that of virus isolation using RD-SCARB2 cells. Viral RNA was detected in all samples positive for viral isolation (Fig. 3). The samples positive for virus isolation and/or viral RNA detection are indicated by colored boxes in Fig. 3.
FIG 3.
Schematic illustration of EV71 detection. Virus isolation and viral RNA detection from clinical samples and autopsy samples are summarized. The VP1 region of the viral RNA was amplified, and the sequence was determined. The samples positive for virus isolation and viral RNA with G at VP1-145 are indicated by blue. The samples positive for virus isolation and viral RNA, or viral RNA alone, with E are indicated by red and pink, respectively. The numbers in the boxes indicate the samples containing mutations in the VP1 region, which are summarized in Table 1.
Virus and viral RNA were detected in throat swabs at 1 and/or 4 dpi in the monkeys inoculated with Isehara-E, C7-E, and C7-G. Virus and/or viral RNA was detected in rectal swabs at 1 dpi and later in the monkeys inoculated with Isehara-E, C7-E, or C7-G. Virus and/or viral RNA was not detected in throat or rectal swabs of monkeys inoculated with Isehara-G. Transient viremia was detected at 1 and/or 4 dpi in monkeys infected with all viruses (Fig. 3A).
We detected virus and/or viral RNA in various autopsy samples more frequently from monkeys inoculated with Isehara-E and C7-E than from monkeys inoculated with Isehara-G and C7-G, respectively (Fig. 3B). Notably, among VP1-145E-inoculated monkeys, the monkeys with clinical signs and histopathological changes (5217, 5218, 5257, and 5220) retained detectable amounts of viral RNA in their CNS, whereas the other monkeys did not. These results indicate that VP1-145E viruses are more virulent than VP1-145G viruses as judged by clinical manifestation, histopathological changes, and virus recovery.
In vivo genetic stability of VP1-145 viruses.
To elucidate the genetic stability of the viruses, we determined the VP1 sequences of all the samples in which viral RNA was detected (Fig. 3). We found some mutations in the recovered viral sequences, and the samples with mutations are indicated with the numbers in the boxes in Fig. 3. Interestingly, a G-to-E mutation was found at VP1-145 (Fig. 3 and Table 1, viruses 1 and 2) in all 11 samples from monkeys inoculated with Isehara-G. In addition to this mutation, VP1-77 Glu-to-Gly, VP1-150 Leu-to-Ser, and VP1-243 Ser-to-Pro mutations were found in the pons of monkey 5212 (Fig. 3 and Table 1, virus 2). In all samples recovered from monkeys that had been inoculated with Isehara-E, VP1-145 did not change. In samples recovered from monkeys 5218 and 5219, we found a VP1-19 Ala-to-Ser mutation (Fig. 3 and Table 1, virus 3).
TABLE 1.
Mutations in VP1 regions of recovered viral genomes from EV71-infected monkeys
| No. in Fig. 3 | Inoculated virus | Nucleotide mutation | Amino acid mutation |
|---|---|---|---|
| 1 | Isehara-G | 2873 G → A | VP1-145 G → E |
| 2 | Isehara-G | 2671 A → G | VP1-77 E → G |
| 2873 G → A | VP1-145 G → E | ||
| 2890 T → C | VP1-150 L → S | ||
| 3168 T → C | VP1-243 S → P | ||
| 3 | Isehara-E | 2496 G → T | VP1-19 A → S |
| 3143 G → A | VP1-234 S → S | ||
| 4 | C7-G | 2876 G → A | VP1-145 G → E |
| 5 | C7-E | 2885 C → C and T (mixed) | VP1-148 P → P and L (mixed) |
| 6 | C7-E | 3139 T → T and C (mixed) | VP1-233 F → F and L (mixed) |
In samples inoculated with C7-G, viral RNA in the throat swab, rectal swab, and serum samples at 1 dpi did not contain any mutations (Fig. 3A). However, viral RNA in the rectal swab and serum of monkey 5255 at 4 to 7 dpi had a G-to-E substitution at VP1-145 (Fig. 3A and Table 1, virus 4). VP1-145 of all samples recovered from C7-E-inoculated monkeys was unchanged. We detected partial substitution of VP1-148 Pro to Leu and VP1-233 Phe to Leu in monkeys 5257 and 5258 (Fig. 3 and Table 1, viruses 5 and 6).
Importantly, E at VP1-145 did not change in VP1-145E-inoculated groups but VP1-145E variants emerged in the monkeys inoculated with VP1-145G viruses after 1 dpi. These results indicated that VP1-145E is more stable than VP1-145G in cynomolgus monkey and that a strong selection pressure for G-to-E mutation at VP1-145 operates in vivo.
Induction of circulating cytokines.
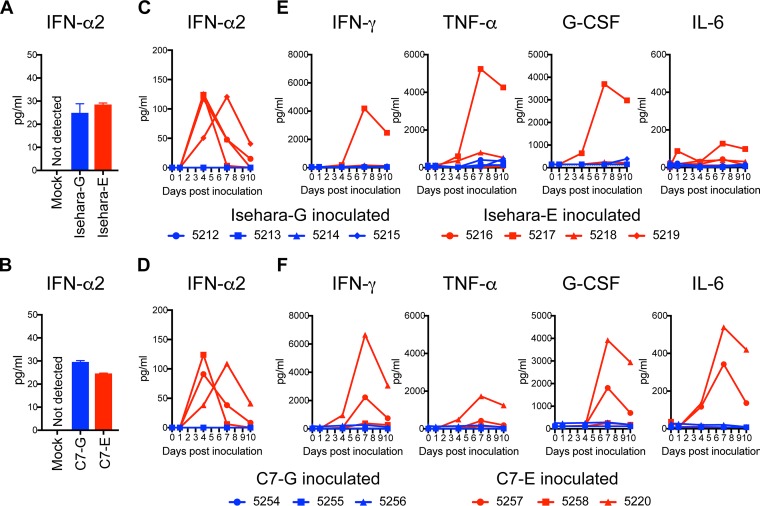
Innate immune responses, including type I interferon (IFN) and proinflammatory cytokines, play important roles in host defense against viral infection. Before comparison of innate immune responses in the monkey, we verified type I IFN production in cultured monkey cells. We infected COS7 cells with VP1-145 variants and measured amounts of IFN-α2 in culture supernatants by enzyme-linked immunosorbent assay (ELISA). Under this condition, IFN-α2 was not detected in the supernatant of mock-infected COS7 cells (Fig. 4A and B). Similar levels of IFN-α2 were detected in culture supernatants of both VP1-145G- and VP1-145E-infected cells (Fig. 4A and B), suggesting that both VP1-145G and VP1-145E have IFN-inducing activity in monkey cells in vitro. We measured serum IFN-α2 levels and cytokines using ELISA following inoculations with VP1-145G and VP1-145E viruses. A significant increase in serum IFN-α2 level was observed at 4 or 7 dpi in all monkeys inoculated with Isehara-E or C7-E (Fig. 4C and D). In contrast, an increase in serum levels of IFN-α2 was not evident in monkeys inoculated with Isehara-G or C7-G (Fig. 4C and D). These results indicated that innate immune systems were induced from 4 dpi in VP1-145E-inoculated groups but not in VP1-145G-inoculated groups under these experimental conditions. We conclude that infection with VP1-145E viruses progresses more extensively than that with VP1-145G viruses.
FIG 4.
Serum cytokine levels in the inoculated monkeys. (A and B) Production of IFN-α2 in culture supernatant of COS7 cells infected with VP1-145 variants of the Isehara (A) or C7/Osaka (B) strain was analyzed using ELISA. The means ± standard error for the two experiments are shown. (C and D) Serum levels of IFN-α2 collected from the monkeys inoculated with the Isehara (C) or C7/Osaka (D) strain were analyzed using ELISA. (E and F) Serum levels of the cytokines IFN-γ, TNF-α, G-CSF, and IL-6 in the monkeys inoculated with the Isehara (E) or C7/Osaka (F) strain were analyzed using Luminex 200. Serum cytokine levels, collected at 0 (preinoculation) and 1, 4, 7, and 10 dpi, from the VP1-145G virus-inoculated group are indicated in blue, and those from the VP1-145E virus-inoculated group are indicated in red.
We also compared the serum levels of IFN-γ, tumor necrosis factor alpha (TNF-α), granulocyte colony-stimulating factor (G-CSF), and interleukin-6 (IL-6) during EV71 infection using a multiplex cytokine assay system (Fig. 4E and F). Significant increases in serum levels of IFN-γ, TNF-α, and G-CSF were observed at 7 to 10 dpi in monkey 5217 in the Isehara-E-inoculated group and in monkeys 5257 and 5220 in the C7-E-inoculated group, all of which showed tremors and pathological changes (Fig. 4C and F). In contrast, no increase in serum levels of the cytokines was observed in monkeys inoculated with Isehara-G or C7-G viruses (Fig. 4E and F).
In summary, the induction of innate immune responses after infection with VP1-145E and VP1-145G viruses was different. VP1-145E viruses can be detected more frequently and in more tissues than VP1-145G viruses, suggesting that they disseminate more widely. The strong cytokine secretions may be induced in response to progressive infection by VP1-145E viruses.
Distribution of HS in cynomolgus monkeys.
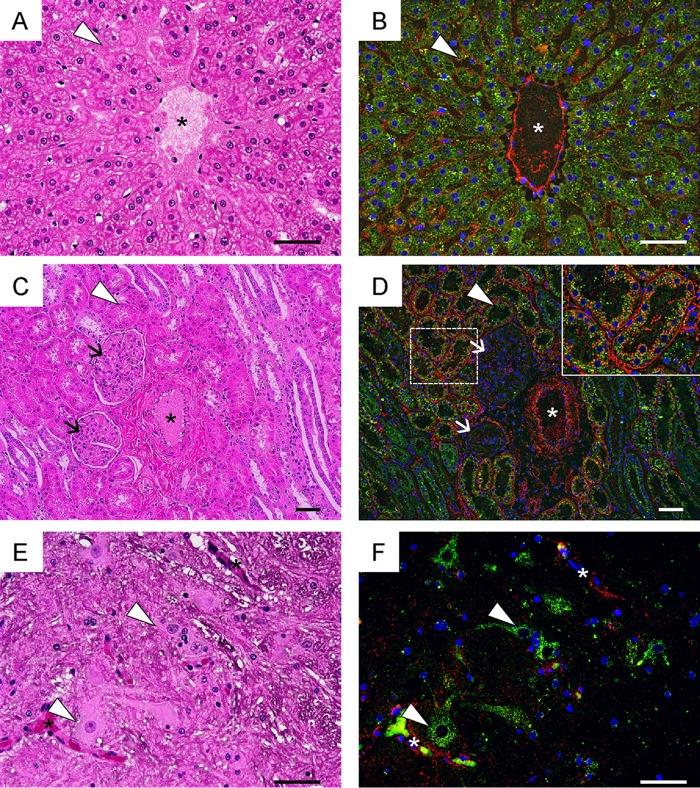
We hypothesized that HS acts as a decoy to specifically adsorb VP1-145G viruses (28). The results above suggested that this hypothesis is also true in cynomolgus monkeys. To test this, we examined the distributions of HS and SCARB2 in the livers, kidneys, and spinal cords of monkeys 5212 and 5254 (Fig. 5). In all tissues, HS was strongly expressed on vascular epithelial cells, where SCARB2 expression was undetectable (Fig. 5). In the liver, HS was detected mainly in sinusoidal endothelial cells and vascular endothelial cells, while SCARB2 was highly expressed in hepatocytes (Fig. 5A and B). In the kidney, HS was expressed in vascular endothelial cells, renal tubules, and basement membranes surrounding renal tubules, whereas SCARB2 was expressed mainly in renal tubules (Fig. 5C and D). In the spinal cord, SCARB2 was expressed in neuronal cells but not in the vascular endothelial cells (Fig. 5E and F). Therefore, HS is not expressed at high levels in SCARB2-positive neuronal cells. The distributions of HS and SCARB2 in monkeys are similar to those in hSCARB2 tg mice (28), suggesting that the HS attachment receptor does not contribute to productive infection mediated by SCARB2 in neuronal cells in vivo.
FIG 5.
HS expression in cynomolgus monkeys. Serial tissue sections of liver (A and B), kidney (C and D), and spinal cord (E and F) prepared from monkeys 5215 and 5254 were stained using H&E (A, C, and E) or subjected to immunofluorescence staining with anti-HS (red) and anti-hSCARB2 (green) (B, D, and F). Nuclei are stained with DAPI (blue). Blood vessels are indicated with asterisks (A to F). Sinusoidal endothelium (arrowheads in panels A and B), renal tubules (arrowheads in panels C and D), glomeruli (arrows in panels C and D), and neuronal cells (arrowheads in panels E and F) are labeled. The area surrounding the renal tubules marked by a dashed box in D is enlarged in the inset. Scale bars indicate 50 μm.
Production of neutralizing antibody against EV71.
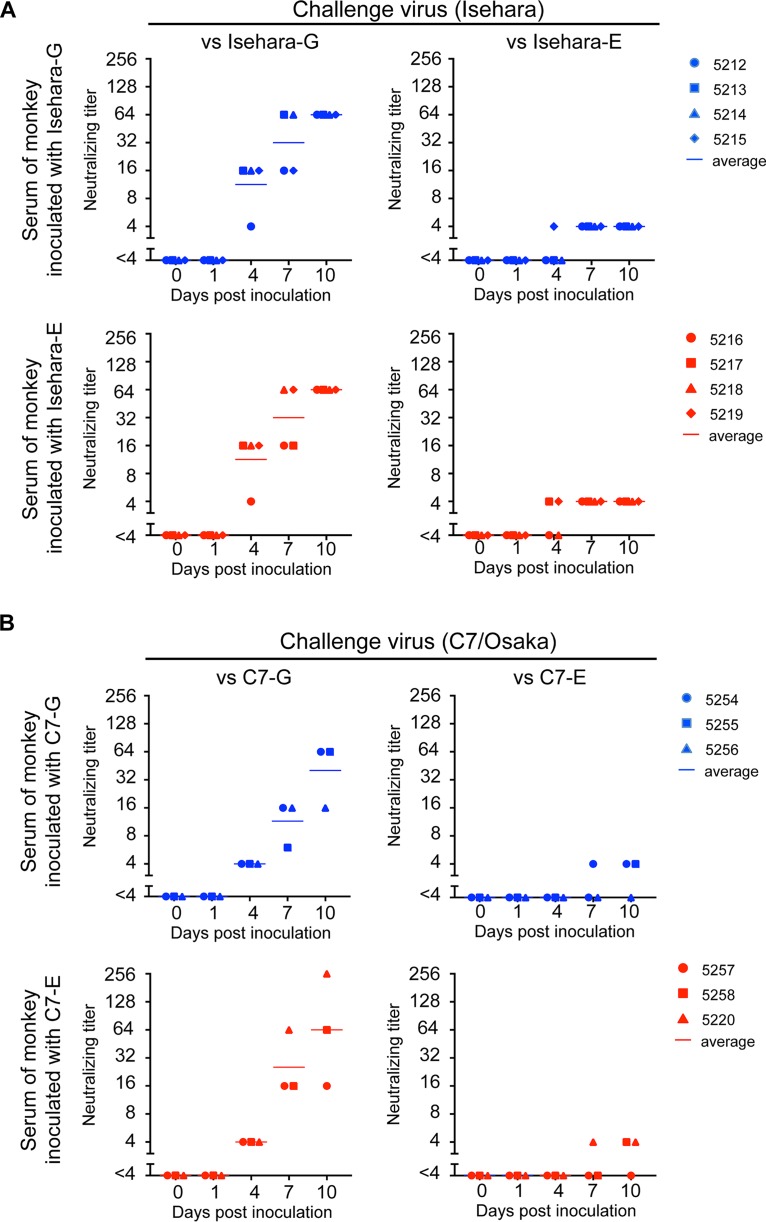
Serum neutralizing antibodies play an important role in preventing enterovirus infection and clinical symptoms (1, 35–37). We therefore examined whether the ability of VP1-145G and VP1-145E to induce neutralizing antibodies differs after intravenous infection. We initially used microneutralization assays to determine the neutralizing titer of the monkey sera (38). However, it was difficult to determine the endpoint of serum dilution because dose dependency was not clear. When we used Isehara-E or C7-E viruses as challenge viruses, we often found that cells that had been challenged with viruses mixed with a higher concentration of antiserum were not protected, whereas those that had been challenged with viruses with a lower concentration of the antiserum were protected. This phenomenon was observed when we performed microneutralization assays using several EV71 VP1-145E strains (data not shown). We suspected nonspecific virus aggregation. However, neither sonication nor nonionic detergent treatment (39) of the challenge viruses improved the situation (data not shown). These results suggested that aggregation is not the main factor contributing to this phenomenon and that VP1-145E viruses are relatively resistant to neutralizing antibodies and cannot be completely neutralized with a large amount of antibody. We therefore decided to determine the neutralizing titer by a plaque reduction neutralization test (PRNT) using VP1-145G and VP1-145E viruses as challenge viruses. We defined the neutralizing titer as the highest dilution at which plaque number reduction is more than 80%. In monkeys inoculated with Isehara-G and Isehara-E, neutralizing antibodies against Isehara-G appeared from 4 dpi (1:4 to 1:16), and the titer increased until 10 dpi (1:16 to 1:256) (Fig. 6A, versus Isehara-G). Neutralizing antibodies were also detected at 4 dpi and beyond when Isehara-E was used as the challenge virus, but the titers were much lower than those measured when Isehara-G was used as the challenge virus (Fig. 6A, versus Isehara-E). Similar results were obtained in the C7 virus-inoculated group (Fig. 6B). Both VP1-145G and VP1-145E viruses elicited similar levels of neutralizing antibodies with similar kinetics. These results suggested that infection by both VP1-145G and VP1-145E viruses was successful enough to elicit neutralizing antibodies and that the abilities of the two variants to induce neutralizing antibodies are not significantly different. However, the neutralizing antibody titer against VP1-145E viruses was lower than that against VP1-145G viruses; thus, Isehara-E and C7-E seem to be more resistant to neutralizing antibodies.
FIG 6.
Neutralization titer in sera of monkeys inoculated with the Isehara (A) or C7/Osaka (B) strain of EV71. Serum samples were 4-fold serially diluted (1:4 to 1:256), and PRNT was performed using corresponding VP1-145G and VP1-145E viruses as challenge viruses. The highest serum dilution that achieved more than 80% plaque reduction was defined as the neutralizing titer. The neutralizing titers against VP1-145G and VP1-145E are shown in the left (versus VP1-145G) and right (versus VP1-145E) panels, respectively.
Different resistant properties of EV71 strains to neutralizing antibodies.
To compare the resistance of VP1-145 variants in Isehara (subgenogroup C2), C7/Osaka (subgenogroup B4), and SK-EV006 (subgenogroup B2) backgrounds to neutralizing antibodies, we performed PRNT using three antibodies, α-1095, α-Y90-3205, and MAB979. These antibodies were chosen because they can be used to test the effects of a variety of epitopes that are homotypic or heterotypic with respect to VP1-145 variation and subgenogroup. We determined the 80% neutralization dose (ND80) of α-1095, α-Y90-3205, and MAB979 against Isehara-G and Isehara-E (Fig. 7A). The ND80 of antibodies against Isehara-E was significantly higher than that of antibodies against Isehara-G for all three different antibodies. Similar results were observed in C7/Osaka and SK-EV006 backgrounds (Fig. 7B and C). α-1095 is a rabbit polyclonal antibody raised against EV71-1095/Shiga (subgenogroup C2, VP1-145G), α-Y90-3205 is a rabbit polyclonal antibody against EV71-Y90-3205 (subgenogroup B2,VP1-145E), and MAB979 is a mouse anti-VP2 monoclonal antibody. The α-1095 and α-Y90-3205 antisera neutralized all three viruses belonging to homo- and heterotypic subgenogroups, suggesting that the neutralization observed in this experiment was not subgenogroup specific. The α-1095 antiserum should contain VP1-145G-specific antibodies as well as antibodies to epitopes in common to VP1-145E and VP1-145G variants, while the α-Y90-3205 antiserum should contain VP1-145E-specific antibodies and antibodies to common epitopes. The α-1095 antiserum neutralized homotypic VP1-145G viruses more efficiently than VP1-145E viruses. However, the α-Y90-3205 antiserum also neutralized heterotypic VP1-145G viruses more efficiently than VP1-145E viruses. This asymmetric cross-neutralization of the VP1-145G virus by the serum raised against VP1-145E virus suggested that the neutralization of the VP1-145G viruses by α-Y90-3205 antiserum was not due to VP1-145G strain-specific antibody but was due to antibodies common to both variants. Furthermore, MAB979 also neutralized VP1-145G viruses more efficiently than VP1-145E viruses. The results suggested that VP1-145E viruses are more resistant to neutralization than VP1-145G viruses irrespective of the antisera. Of note, the VP1-145E viruses were not completely neutralized even at the lowest dilution. These results indicated that VP1-145E viruses are more resistant to three different antibodies than VP1-145G viruses, suggesting that VP1-145E viruses have better survival chances in the presence of neutralizing antibodies.
FIG 7.
Neutralization sensitivity of VP1-145G and VP1-145E viruses. Dose-response neutralization profiles were generated by incubating VP1-145 variants of Isehara (A), C7/Osaka (B), or SK-EV006 (C) with serial dilutions of α-1095 rabbit antiserum, α-Y90-3205 rabbit antiserum, or MAB979 mouse monoclonal antibody for 2 h at 37°C, followed by plaque assay. The number of plaques was counted, and the resulting data were fit by nonlinear regression to determine the ND80. Error bars represent the standard error from two experiments. Dashed lines indicate the ND80. The means of ND80 ± standard error against VP1-145G and VP1-145E are shown in blue and red, respectively, below each panel. The significance of differences between ND80 against the VP1-145G and VP1-145E viruses was determined using Student's t test, and P values are indicated by asterisks (*, P < 0.5; **, P < 0.05).
DISCUSSION
To reveal how VP1-145 residues contribute to EV71 pathogenesis, we compared the virulence of VP1-145G and VP1-145E viruses using cynomolgus monkeys. VP1-145E, but not VP1-145G, viruses induced neuropathological changes in the CNS and neurological symptoms (Fig. 1 and 2). Viruses were frequently detected in tissues of monkeys infected with VP1-145E viruses (Fig. 3). Viruses were less frequently detected in VP1-145G virus-infected monkeys, and the detected viruses had a G-to-E mutation at VP1-145 (Fig. 3 and Table 1). VP1-145E viruses induced more extensive innate immune responses than VP1-145G viruses (Fig. 4). In addition to these results, Kataoka et al. showed that a non-PB strain (corresponding to VP1-145E) exhibits a more virulent phenotype than the PB strain (corresponding to VP1-145G) in cynomolgus monkeys (23). We conclude that VP1-145E viruses have significant advantages in replication and are more virulent than VP1-145G viruses in a nonhuman primate. Therefore, VP1-145 is a virulence determinant in a nonhuman primate.
Using an hSCARB2 tg mouse model, Kobayashi et al. (28) found that VP1-145G viruses bind to HS and that HS is expressed highly in cells that do not express SCARB2. HSs act as decoys that trap the VP1-145G viruses and prevent their spread in vivo. We hypothesize that this is a major mechanism of VP1-145G virus attenuation. Isehara-G viruses were not detected in throat swabs, rectal swabs, or blood (Fig. 3). C7-G viruses were detected in these clinical samples only at 1 dpi and then disappeared, being replaced by VP1-145E mutants (Fig. 3 and Table 1). This was probably because VP1-145G virus was adsorbed by HS as predicted from the results with hSCARB2 tg mice. HS, but not SCABR2, is highly expressed in monkey vascular endothelial cells. Infection of these cells is considered to be abortive. Renal tubules in the kidney are positive for both HS and SCARB2. However, these cells are not permissive for EV71 infection for a number of reasons, including the type I interferon response, as suggested by our previous work on poliovirus (40). It is clear that HS expression does not contribute to infection in neurons. Since the distributions of HS and SCARB2 are similar in hSCARB2 tg mice and cynomolgus monkeys (Fig. 5), the hypothesis may be applicable to other primate models and humans. Similar attenuation mechanisms have been reported for many other viruses (41–53). However, this is, to our knowledge, the first report using nonhuman primates.
VP1-145E viruses are more resistant to two different polyclonal antibodies than VP1-145G viruses We believe that this difference is not due to strain- or subgenogroup-specific antibodies dominantly present in the sera. Supporting this idea, VP1-145E viruses are also more resistant to the monoclonal antibody MAB979, which recognizes an epitope in VP2 conserved among the EV71 strains (54) (Fig. 7). Substitution of VP1-145 affects sensitivity to MAB979 (Fig. 7). While we are not certain why variations at VP1-145 affect antibody resistance, one possibility is that VP1-145 amino acid substitution induces allosteric structural alteration of a number of epitopes on the virion and changes sensitivity to neutralizing antibodies. A previous study using dengue virus showed that a single point mutation in the envelope protein affects sensitivity to a monoclonal antibody by changing the conformation of an epitope in another domain of the protein (55). In the case of norovirus, substitution of an amino acid located outside the epitope affects sensitivity to neutralizing antibodies (56, 57). Thus, VP1-145 of EV71 might be a key residue not only for surface charge around the 5-fold axis of the virion but also for the structural properties of the whole virion. We found that relatively high concentrations of antisera failed to neutralize the VP1-145E viruses completely (Fig. 7). When we performed microneutralization assays using VP1-145E viruses, neutralization was sometimes successful in some wells but not in other wells in the microplates due to the residual viruses. This may be the reason why endpoint of dilution was not clearly determined.
When monkeys are immunized with an attenuated strain of EV71, they produce sufficient amounts of neutralizing antibodies and are protected from challenge by VP1-145E strains (58). This result indicates the importance of neutralizing antibodies in clearing the infecting virus. In this study, the monkeys inoculated with EV71 began to produce neutralizing antibodies at 4 dpi before the onset of the clinical symptoms (Fig. 6). At the same time, VP1-145G viruses disappeared (Fig. 3). However, VP1-145E viruses were excreted in the feces and entered into the CNS even in the presence of neutralizing antibodies at 4 to 10 dpi (Fig. 3), suggesting that the VP1-145E viruses continued to replicate in the presence of low levels of neutralizing antibodies. VP1-145E viruses were more resistant to neutralizing antibodies than VP1-145G viruses in PRNTs (Fig. 7). This phenomenon was not specific to Isehara, C7/Osaka, or SK-EV006 strains. The resistance of other virulent EV71 strains, which are virulent in hSCARB2 tg mice, to neutralizing antibodies was also observed (data not shown). It is possible that a small amount of neutralizing antibody raised in the early phase of infection is not sufficient to block the dissemination of VP1-145E viruses. In addition to differences in HS adsorption, the different resistance of the VP1-145 variants to neutralizing antibodies might be another reason for the differences in in vivo viral fitness and EV71 neurovirulence.
In summary, VP1-145 influences the usage of attachment receptors. VP1-145G/Q viruses can use HS as an attachment receptor. VP1-145 may also influence the three-dimensional structure of the whole virion. Finally, VP1-145E viruses are resistant to neutralizing antibodies. Thus, VP1-145 is a key amino acid residue that determines EV71 virulence through these mechanisms.
MATERIALS AND METHODS
Ethics statement.
Animal experiments were conducted in compliance with Japanese legislation (Act on Welfare and Management of Animals, 1973, revised in 2012) and guidelines under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology, Japan (Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions, 2006). Animal care, housing, feeding, sampling, observation, and environmental enrichment were performed in accordance with the guidelines. Every possible effort was made to minimize suffering. The protocols of animal experiments were approved by the committee of biosafety and animal handling and the committee of ethical regulation of the National Institute of Infectious Diseases, Japan (authorization number 515001). Each animal was housed in a separate cage, received standard primate feed and fresh fruit daily, and had free access to water at the National Institute of Infectious Diseases, Japan. Animal welfare was observed on a daily basis. Sampling procedures were conducted under anesthesia (10 mg/kg ketamine intramuscularly). Animals were sacrificed under excess anesthesia.
Cells.
Vero, RD-A, and COS7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Nissui) supplemented with 5% fetal bovine serum (FBS). Generation of RD-SCARB2 cells was as described in the accompanying paper (28). RD-SCARB2 cells were cultured in DMEM supplemented with 5% FBS and 1 μg/ml puromycin.
Viruses.
Preparation of VP1-145 variants based on the Isehara, C7/Osaka, and SK-EV006 viruses (Isehara-E, Isehara-G, C7-G, C7-E, SK-G, and SK-E) was as described in the accompanying paper (28). Viral titers were determined by a microtitration assay using 96-well plates and Vero cells, as previously described (59). Viral titers were expressed as 50% tissue culture infectious dose (TCID50).
Animals.
Fourteen male cynomolgus macaques (Macaca fascicularis) imported from the Philippines were used in this study. All monkeys were 3 years old. The average weight was 4.2 kg (range, 2.6 to 5.16 kg). All procedures were performed in a biosafety level 2 containment facility. Serological testing revealed that all animals used in this study were free from infection with EV71.
Inoculations.
Eight monkeys were divided into two groups; one group received Isehara-G inoculations (monkeys 5212, 5213, 5214, and 5215), and the other group received Isehara-E inoculations (monkeys 5216, 5217, 5218, and 5219). The other six monkeys were divided into two groups; one group received C7-G inoculations (monkeys 5254, 5255, and 5256), and the other group received C7-E inoculations (monkeys 5257, 5258, and 5220). Virus inoculation and sampling procedures were conducted under anesthesia (10 mg/kg ketamine intramuscularly). Before inoculation of virus (0 dpi), manifestations were observed under anesthesia, including HFMD-like symptoms such as blisters within the oral cavity, on the palms, and on the soles of the feet, and whole blood, throat swab, and rectal swab samples were collected. One milliliter of EV71 virus solution containing 106 TCID50 of virus was intravenously inoculated into the right tibial vein. Following inoculation, monkeys were observed daily for 10 days to assess neurological manifestations. Clinical manifestations were observed in more detail under anesthesia for HFMD-like symptoms at 1, 4, 7, and 10 dpi. Whole blood, throat swab, and rectal swab samples were collected at these times for virological and immunological analyses (Fig. 1).
Monkeys were sacrificed under deep anesthesia at 10 dpi, and various parts of the CNS, main organs, and lymph nodes were collected for histopathological and virological analyses. Ten percent (wt/vol) homogenates of collected tissues were prepared in DMEM containing 5% FBS with MagNA Lyzer (Roche Life Science) before centrifugation at 10,000 × g for 5 min at 4°C to remove tissue debris. The supernatants were used for virus isolation in RD-SCARB2 cells and RNA extraction for viral RNA detection.
Histopathology and immunohistochemistry.
Brain and spinal cord samples were fixed in 10% formalin in PBS and embedded in paraffin. Paraffin-embedded sections were stained with hematoxylin and eosin (H&E). Immunohistochemical detection of the EV71 antigen was performed on paraffin-embedded sections using a labeled streptavidin-biotin method (Dako Denmark A/S). For antigen retrieval, sections were heated to 121°C for 10 min in an autoclave with 10 mM citrate buffer solution (pH 6.0). A polyclonal antibody raised against denatured virus particles of EV71-1095/Shiga (subgenogroup C2) was used as the primary antibody (33). Peroxidase activity was detected with 3,3-diaminobenzidine (Sigma-Aldrich), and sections were counterstained with hematoxylin.
Virus isolation.
For virus isolation, RD-SCARB2 cells inoculated with clinical sample preparations or tissue homogenates were observed for CPEs for 1 week, and then blind passage was performed for CPE-negative samples after freeze-thawing of the first-round passage. If no CPEs were observed during first- and second-round cultures, the result of virus isolation was recorded as negative.
Viral RNA detection and sequencing.
Before RNA extraction, swab samples were centrifuged at 2,000 rpm for 3 min, and the supernatants were used for RNA extraction. Viral genomic RNA was extracted from clinical samples or 10% tissue homogenates of autopsy samples using the HighPure viral RNA purification kit (Roche Life Science). Reverse transcription-PCR (RT-PCR) was performed using the PrimeScript II one-step RT-PCR kit (TaKaRa Bio). Nested PCR was performed using TaKaRa Taq Hot Start version (TaKaRa Bio). PCR products were purified using a PCR purification kit (Qiagen). The full length of the VP1 sequence was analyzed using the primers listed in Table 2.
TABLE 2.
Primers for viral RNA detection and sequencing
| Reaction | Name | Strand | Sequence |
|---|---|---|---|
| RT-PCR | con-2211F | Forward | 5′-ACCCTTGTAATACCATGGATCAGC-3′ |
| DG-3393R | Reverse | 5′-CACCCGTTGAADTCYCACCARTTGGCG-3′ | |
| Second PCR | Ise-2317F | Forward | 5′-TACGTGGTTCCAATTGGGGCACC-3′ |
| Ise-3391R | Reverse | 5′-GGCGGTTAACCACTCTAAAGTTGCCCAC-3′ | |
| C7-2328F | Forward | 5′-TCCAATTGGGGCACCCAACA-3′ | |
| C7-3345R | Reverse | 5′-GCCGAACTTTCCAAGGGTAGTAAT-3′ | |
| Sequencing | Ise-2317F | Forward | 5′-TACGTGGTTCCAATTGGGGCACC-3′ |
| C7-2328F | Forward | 5′-TCCAATTGGGGCACCCAACA-3′ | |
| DG-2757F | Forward | 5′-GCHAAYTGGGAYATAGACATAAC-3′ | |
| DG-3046F | Forward | 5′-GGTTTTAYGACGGRTAYCC-3′ | |
| DG-2780R | Reverse | 5′-CCDGTTATGTCTATRTCCC-3′ | |
| DG-3046R | Reverse | 5′-GGRTAYCCGTCRTAAAACC-3′ | |
| C7-3345R | Reverse | 5′-GCCGAACTTTCCAAGGGTAGTAAT-3′ | |
| Ise-3391R | Reverse | 5′-GGCGGTTAACCACTCTAAAGTTGCCCAC-3′ |
Cytokine and chemokine analyses.
To examine production of IFN-α2 in COS7 cells, 105 COS7 cells were infected with VP1-145 variants at 105 TCID50 (measured using Vero cells). After 48 h postinfection, culture supernatants were collected and stored at −80°C. IFN-α2 concentrations in culture supernatant or monkey serum were analyzed using the Cynomolgus/Rhesus IFN Alpha ELISA kit (PBL). Serum cytokine concentrations were analyzed using Cytokine Monkey Magnetic 29-Plex panels (Life Technologies) on a Luminex 200 (Luminex).
Immunofluorescence staining of monkey tissues.
Paraffin-embedded sections of cynomolgus monkeys were stained as described by Kobayashi et al. (28). Briefly, following the retrieval reaction at 121°C for 10 min in a 10 mM sodium citrate solution at pH 6.0, slides were blocked with 5% bovine serum albumin (BSA) in PBS at room temperature for 1 h. A goat anti-SCARB2 antibody (R&D Systems; 1:200 dilution) and mouse IgM anti-HS antibody F58-10E4 (Amsbio; 1:200 dilution) or isotype control mouse IgM MM-30 (BioLegend; 1:200 dilution) were incubated with the sections overnight at 4°C, followed by incubation with Alexa Fluor 488-conjugated donkey anti-goat IgG (Molecular Probes; 1:500 dilution) and Cy3-conjugated goat anti-mouse IgM (Jackson ImmunoResearch; 1:500 dilution) for 1 h at 37°C, respectively. The sections were mounted in Vectashield with DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories), and images were captured using a fluorescence microscope (BZ-X700; Keyence).
PRNT.
Serum samples were serially diluted in DMEM containing 5% FBS. The EV71 stock was diluted to a concentration of 500 PFU/100 μl, and then 100 μl of diluted serum was mixed with 100 μl of EV71 solution in 96-well plates and incubated for 2 h at 37°C. Following incubation, 200 μl of serum-virus mixtures was transferred to 6-well plates containing 106 RD-A cells and incubated for 1 h at 37°C. After incubation, serum-virus mixtures were removed, and RD-A cells were overlaid with minimum essential medium (MEM) containing 5% FBS and 1.2% methylcellulose. Neutralization titers were determined as the highest serum dilution that reduced the plaque number to less than 20% of the original number. Anti-EV71-1095/Shiga (subgenogroup C2) rabbit antiserum (α-1095 antiserum) was generated previously (33). Anti-EV71-Y90-3205 (subgenogroup B2) rabbit antiserum (α-Y90-3205 antiserum) was generated as follows. Y90-3205 was prepared as described previously (60). Virus was inactivated with 0.37% formalin at 37°C for 2 h. A rabbit (New Zealand White, specific pathogen free, 11-week-old female; Kumagai-shigeyasu Co., Ltd.) was subcutaneously injected with 1.6 ml virus fluid (approximately 107 to 108 TCID50) with the same volume of Freund incomplete adjuvant (Difco Laboratories) at an interval of 1 week. Three weeks after the fourth immunization, the rabbits were boosted by one subcutaneous injection of the virus with the adjuvant. Serum samples were collected 1 week later and stored at −70°C after heat inactivation (56°C for 30 min). Anti-VP2 monoclonal antibody MAB979 was purchased from Millipore. These antibodies were used for comparison of sensitivity to neutralizing antibodies. The ND80 for each neutralizing antibody was calculated by nonlinear regression analysis using Prism software (GraphPad Software).
ACKNOWLEDGMENTS
We thank A. Harashima, M. Fujino, and M. Ozaki (National Institute of Infectious Diseases) and T. Ito and N. Fukuda (Yamagata University) for excellent technical assistance. We thank Junjiro Horiuchi (Learning and Memory Project, Tokyo Metropolitan Institute of Medical Science) for critical reading of the manuscript.
This work was supported by JSPS KAKENHI (grant numbers 23390116 and 15K08514), MEXT KAKENHI (grant number JP24115006), and AMED (grant number JP15fk0108009).
REFERENCES
- 1.Pallansch M, Oberste MS, Whitton JL. 2013. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p 490–530. _In_Knipe D, Howley PM (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. 2010. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis 10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. 2012. Crystal structure of human enterovirus 71. Science 336:1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamayoshi S, Iizuka S, Yamashita T, Minagawa H, Mizuta K, Okamoto M, Nishimura H, Sanjoh K, Katsushima N, Itagaki T, Nagai Y, Fujii K, Koike S. 2012. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J Virol 86:5686–5696. doi: 10.1128/JVI.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamayoshi S, Ohka S, Fujii K, Koike S. 2013. Functional comparison of SCARB2 and PSGL1 as receptors for enterovirus 71. J Virol 87:3335–3347. doi: 10.1128/JVI.02070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P, Song Z, Qi Y, Feng X, Xu N, Sun Y, Wu X, Yao X, Mao Q, Li X, Dong W, Wan X, Huang N, Shen X, Liang Z, Li W. 2012. Molecular determinants of enterovirus 71 viral entry: cleft around GLN-172 on VP1 protein interacts with variable region on scavenge receptor B 2. J Biol Chem 287:6406–6420. doi: 10.1074/jbc.M111.301622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura Y, Shimojima M, Tano Y, Miyamura T, Wakita T, Shimizu H. 2009. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat Med 15:794–797. doi: 10.1038/nm.1961. [DOI] [PubMed] [Google Scholar]
- 9.Tan CW, Poh CL, Sam IC, Chan YF. 2013. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J Virol 87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang SL, Chou YT, Wu CN, Ho MS. 2011. Annexin II binds to capsid protein VP1 of enterovirus 71 and enhances viral infectivity. J Virol 85:11809–11820. doi: 10.1128/JVI.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su PY, Liu YT, Chang HY, Huang SW, Wang YF, Yu CK, Wang JR, Chang CF. 2012. Cell surface sialylation affects binding of enterovirus 71 to rhabdomyosarcoma and neuroblastoma cells. BMC Microbiol 12:162. doi: 10.1186/1471-2180-12-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su PY, Wang YF, Huang SW, Lo YC, Wang YH, Wu SR, Shieh DB, Chen SH, Wang JR, Lai MD, Chang CF. 2015. Cell surface nucleolin facilitates enterovirus 71 binding and infection. J Virol 89:4527–4538. doi: 10.1128/JVI.03498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du N, Cong H, Tian H, Zhang H, Zhang W, Song L, Tien P. 2014. Cell surface vimentin is an attachment receptor for enterovirus 71. J Virol 88:5816–5833. doi: 10.1128/JVI.03826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamayoshi S, Fujii K, Koike S. 2014. Receptors for enterovirus 71. Emerg Microbes Infect 3:e53. doi: 10.1038/emi.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarrazin S, Lamanna WC, Esko JD. 2011. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura Y, Lee H, Hafenstein S, Kataoka C, Wakita T, Bergelson JM, Shimizu H. 2013. Enterovirus 71 binding to PSGL-1 on leukocytes: VP1-145 acts as a molecular switch to control receptor interaction. PLoS Pathog 9:e1003511. doi: 10.1371/journal.ppat.1003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan CW, Sam IC, Lee VS, Wong HV, Chan YF. 2017. VP1 residues around the five-fold axis of enterovirus A71 mediate heparan sulfate interaction. Virology 501:79–87. doi: 10.1016/j.virol.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Chua BH, Phuektes P, Sanders SA, Nicholls PK, McMinn PC. 2008. The molecular basis of mouse adaptation by human enterovirus 71. J Gen Virol 89:1622–1632. doi: 10.1099/vir.0.83676-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Duo J, Liu J, Ma C, Zhang L, Wei Q, Qin C. 2011. A mouse muscle-adapted enterovirus 71 strain with increased virulence in mice. Microbes Infect 13:862–870. doi: 10.1016/j.micinf.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Arita M, Ami Y, Wakita T, Shimizu H. 2008. Cooperative effect of the attenuation determinants derived from poliovirus sabin 1 strain is essential for attenuation of enterovirus 71 in the NOD/SCID mouse infection model. J Virol 82:1787–1797. doi: 10.1128/JVI.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang SW, Wang YF, Yu CK, Su IJ, Wang JR. 2012. Mutations in VP2 and VP1 capsid proteins increase infectivity and mouse lethality of enterovirus 71 by virus binding and RNA accumulation enhancement. Virology 422:132–143. doi: 10.1016/j.virol.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Zaini Z, McMinn P. 2012. A single mutation in capsid protein VP1 (Q145E) of a genogroup C4 strain of human enterovirus 71 generates a mouse-virulent phenotype. J Gen Virol 93:1935–1940. doi: 10.1099/vir.0.043893-0. [DOI] [PubMed] [Google Scholar]
- 23.Kataoka C, Suzuki T, Kotani O, Iwata-Yoshikawa N, Nagata N, Ami Y, Wakita T, Nishimura Y, Shimizu H. 2015. The role of VP1 amino acid residue 145 of enterovirus 71 in viral fitness and pathogenesis in a cynomolgus monkey model. PLoS Pathog 11:e1005033. doi: 10.1371/journal.ppat.1005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Zou Q, Chen L, Zhang H, Wang Y. 2011. Molecular analysis of virulent determinants of enterovirus 71. PLoS One 6:e26237. doi: 10.1371/journal.pone.0026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang SC, Li WC, Chen GW, Tsao KC, Huang CG, Huang YC, Chiu CH, Kuo CY, Tsai KN, Shih SR, Lin TY. 2012. Genetic characterization of enterovirus 71 isolated from patients with severe disease by comparative analysis of complete genomes. J Med Virol 84:931–939. doi: 10.1002/jmv.23287. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Fu C, Wu S, Chen X, Shi Y, Zhou B, Zhang L, Zhang F, Wang Z, Zhang Y, Fan C, Han S, Yin J, Peng B, Liu W, He X. 2014. A novel finding for enterovirus virulence from the capsid protein VP1 of EV71 circulating in mainland China. Virus Genes 48:260–272. doi: 10.1007/s11262-014-1035-2. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Wu X, Huang K, Li L, Zheng L, Wan C, He ML, Zhao W. 2014. The variations of VP1 protein might be associated with nervous system symptoms caused by enterovirus 71 infection. BMC Infect Dis 14:243. doi: 10.1186/1471-2334-14-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi K, Sudaka Y, Takashino A, Imura A, Fujii K, Koike S. 2018. Amino acid variation at VP1-145 of enterovirus 71 determines attachment receptor usage and neurovirulence in human scavenger receptor B2 transgenic mice. J Virol 92:e00681-. doi: 10.1128/JVI.00681-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagiwara A, Yoneyama T, Hashimoto I. 1983. Isolation of a temperature-sensitive strain of enterovirus 71 with reduced neurovirulence for monkeys. J Gen Virol 64:499–502. doi: 10.1099/0022-1317-64-2-499. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto I, Hagiwara A. 1982. Pathogenicity of a poliomyelitis-like disease in monkeys infected orally with enterovirus 71: a model for human infection. Neuropathol Appl Neurobiol 8:149–156. doi: 10.1111/j.1365-2990.1982.tb00269.x. [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto I, Hagiwara A. 1982. Studies on the pathogenesis of and propagation of enterovirus 71 in poliomyelitis-like disease in monkeys. Acta Neuropathol 58:125–132. doi: 10.1007/BF00691653. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto I, Hagiwara A, Kodama H. 1978. Neurovirulence in cynomolgus monkeys of enterovirus 71 isolated from a patient with hand, foot and mouth disease. Arch Virol 56:257–261. doi: 10.1007/BF01317855. [DOI] [PubMed] [Google Scholar]
- 33.Nagata N, Shimizu H, Ami Y, Tano Y, Harashima A, Suzaki Y, Sato Y, Miyamura T, Sata T, Iwasaki T. 2002. Pyramidal and extrapyramidal involvement in experimental infection of cynomolgus monkeys with enterovirus 71. J Med Virol 67:207–216. doi: 10.1002/jmv.2209. [DOI] [PubMed] [Google Scholar]
- 34.Nagata N, Iwasaki T, Ami Y, Tano Y, Harashima A, Suzaki Y, Sato Y, Hasegawa H, Sata T, Miyamura T, Shimizu H. 2004. Differential localization of neurons susceptible to enterovirus 71 and poliovirus type 1 in the central nervous system of cynomolgus monkeys after intravenous inoculation. J Gen Virol 85:2981–2989. doi: 10.1099/vir.0.79883-0. [DOI] [PubMed] [Google Scholar]
- 35.Cellier C, Foray S, Hermine O. 2000. Regional enteritis associated with enterovirus in a patient with X-linked agammaglobulinemia. N Engl J Med 342:1611–1612. doi: 10.1056/NEJM200005253422113. [DOI] [PubMed] [Google Scholar]
- 36.Mease PJ, Ochs HD, Wedgwood RJ. 1981. Successful treatment of echovirus meningoencephalitis and myositis-fasciitis with intravenous immune globulin therapy in a patient with X-linked agammaglobulinemia. N Engl J Med 304:1278–1281. doi: 10.1056/NEJM198105213042107. [DOI] [PubMed] [Google Scholar]
- 37.Nathanson N, Bodian D. 1962. Experimental poliomyelitis following intramuscular virus injection. III. The effect of passive antibody on paralysis and viremia. Bull Johns Hopkins Hosp 111:198–220. [PubMed] [Google Scholar]
- 38.WHO. 2011. A guide to clinical management and public health response for hand, foot and mouth disease (HFMD). WHO, Geneva, Switzerland. [Google Scholar]
- 39.Gwaltney JM Jr, Calhoun AM. 1970. Viral aggregation resulting in the failure to correctly identify an unknown rhinovirus. Appl Microbiol 20:390–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ida-Hosonuma M, Iwasaki T, Yoshikawa T, Nagata N, Sato Y, Sata T, Yoneyama M, Fujita T, Taya C, Yonekawa H, Koike S. 2005. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J Virol 79:4460–4469. doi: 10.1128/JVI.79.7.4460-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernard KA, Klimstra WB, Johnston RE. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93–103. doi: 10.1006/viro.2000.0546. [DOI] [PubMed] [Google Scholar]
- 42.Lee E, Wright PJ, Davidson A, Lobigs M. 2006. Virulence attenuation of dengue virus due to augmented glycosaminoglycan-binding affinity and restriction in extraneural dissemination. J Gen Virol 87:2791–2801. doi: 10.1099/vir.0.82164-0. [DOI] [PubMed] [Google Scholar]
- 43.Lee E, Hall RA, Lobigs M. 2004. Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. J Virol 78:8271–8280. doi: 10.1128/JVI.78.15.8271-8280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee E, Lobigs M. 2002. Mechanism of virulence attenuation of glycosaminoglycan-binding variants of Japanese encephalitis virus and Murray Valley encephalitis virus. J Virol 76:4901–4911. doi: 10.1128/JVI.76.10.4901-4911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Pfeiffer JK. 2016. Emergence of a large-plaque variant in mice infected with coxsackievirus B3. mBio 7:e00119. doi: 10.1128/mBio.00119-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anez G, Men R, Eckels KH, Lai CJ. 2009. Passage of dengue virus type 4 vaccine candidates in fetal rhesus lung cells selects heparin-sensitive variants that result in loss of infectivity and immunogenicity in rhesus macaques. J Virol 83:10384–10394. doi: 10.1128/JVI.01083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nickells J, Cannella M, Droll DA, Liang Y, Wold WS, Chambers TJ. 2008. Neuroadapted yellow fever virus strain 17D: a charged locus in domain III of the E protein governs heparin binding activity and neuroinvasiveness in the SCID mouse model. J Virol 82:12510–12519. doi: 10.1128/JVI.00458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, Heinz FX. 2001. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J Virol 75:5627–5637. doi: 10.1128/JVI.75.12.5627-5637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva LA, Khomandiak S, Ashbrook AW, Weller R, Heise MT, Morrison TE, Dermody TS. 2014. A single-amino-acid polymorphism in chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J Virol 88:2385–2397. doi: 10.1128/JVI.03116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashbrook AW, Burrack KS, Silva LA, Montgomery SA, Heise MT, Morrison TE, Dermody TS. 2014. Residue 82 of the chikungunya virus E2 attachment protein modulates viral dissemination and arthritis in mice. J Virol 88:12180–12192. doi: 10.1128/JVI.01672-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner CL, Hritz J, Sun C, Vanlandingham DL, Song TY, Ghedin E, Higgs S, Klimstra WB, Ryman KD. 2014. Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: a model for rational arboviral vaccine design. PLoS Negl Trop Dis 8:e2719. doi: 10.1371/journal.pntd.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryman KD, Klimstra WB, Johnston RE. 2004. Attenuation of Sindbis virus variants incorporating uncleaved PE2 glycoprotein is correlated with attachment to cell-surface heparan sulfate. Virology 322:1–12. doi: 10.1016/j.virol.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Klimstra WB, Ryman KD, Johnston RE. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol 72:7357–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu CC, Chou AH, Lien SP, Lin HY, Liu SJ, Chang JY, Guo MS, Chow YH, Yang WS, Chang KH, Sia C, Chong P. 2011. Identification and characterization of a cross-neutralization epitope of enterovirus 71. Vaccine 29:4362–4372. doi: 10.1016/j.vaccine.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 55.Dowd KA, DeMaso CR, Pierson TC. 2015. Genotypic differences in dengue virus neutralization are explained by a single amino acid mutation that modulates virus breathing. mBio 6:e01559-. doi: 10.1128/mBio.01559-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolawole AO, Smith HQ, Svoboda SA, Lewis MS, Sherman MB, Lynch GC, Pettitt BM, Smith TJ, Wobus CE. 2017. Norovirus escape from broadly neutralizing antibodies is limited to allostery-like mechanisms. mSphere 2:e00334-. doi: 10.1128/mSphere.00334-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindesmith LC, Mallory ML, Debbink K, Donaldson EF, Brewer-Jensen PD, Swann EW, Sheahan TP, Graham RL, Beltramello M, Corti D, Lanzavecchia A, Baric RS. 2018. Conformational occlusion of blockade antibody epitopes, a novel mechanism of GII.4 human norovirus immune evasion. mSphere 3:e00518-17. doi: 10.1128/mSphere.00518-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arita M, Nagata N, Iwata N, Ami Y, Suzaki Y, Mizuta K, Iwasaki T, Sata T, Wakita T, Shimizu H. 2007. An attenuated strain of enterovirus 71 belonging to genotype a showed a broad spectrum of antigenicity with attenuated neurovirulence in cynomolgus monkeys. J Virol 81:9386–9395. doi: 10.1128/JVI.02856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, Koike S. 2009. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med 15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- 60.Mizuta K, Aoki Y, Suto A, Ootani K, Katsushima N, Itagaki T, Ohmi A, Okamoto M, Nishimura H, Matsuzaki Y, Hongo S, Sugawara K, Shimizu H, Ahiko T. 2009. Cross-antigenicity among EV71 strains from different genogroups isolated in Yamagata, Japan, between 1990 and 2007. Vaccine 27:3153–3158. doi: 10.1016/j.vaccine.2009.03.060. [DOI] [PubMed] [Google Scholar]