Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer's disease markers in subjects with mild cognitive impairment (original) (raw)
Abstract
Background
Alzheimer's disease (AD) prevalence is increasing, but its etiology remains elusive. Gut microbes can contribute to AD pathology and may help identifying novel markers and therapies against AD. Herein, we examine how the gut microbiome differs in older adults with mild cognitive impairment compared to cognitively normal counterparts, and whether and how a modified Mediterranean-ketogenic diet (MMKD) alters the gut microbiome signature in association with cerebrospinal fluid (CSF) AD biomarkers.
Methods
A randomized, double-blind, cross-over, single-center pilot study of MMKD versus American Heart Association Diet (AHAD) intervention is performed on 17 subjects (age: 64.6 ± 6.4 yr), of which 11 have mild cognitive impairment, while 6 are cognitively normal. Subjects undergo MMKD and AHAD intervention for 6-weeks separated by 6-weeks washout periods. Gut microbiome, fecal short-chain fatty acids (SCFAs), and markers of AD in CSF including amyloid β (Aβ)-40 and Aß-42, total tau, and phosphorylated tau-181 (tau-p181) are measured at before and after diet interventions.
Findings
At baseline, subjects with normal vs. impaired cognition show no notable difference in microbiome diversity but several unique microbial signatures are detected in subjects with mild cognitive impairment. Proteobacteria correlate positively with Aβ-42: Aβ-40 while fecal propionate and butyrate correlates negatively with Aβ-42 in subjects with mild cognitive impairment. Several bacteria are differently affected by the two diets with distinct patterns between cognitively normal and impaired subjects. Notably, the abundance of Enterobacteriaceae, Akkermansia, Slackia, Christensenellaceae and Erysipelotriaceae increases while that of Bifidobacterium and Lachnobacterium reduces on MMKD, while AHAD increases Mollicutes. MMKD slightly reduces fecal lactate and acetate while increasing propionate and butyrate. Conversely, AHAD increases acetate and propionate while reducing butyrate.
Interpretation
The data suggest that specific gut microbial signatures may depict the mild cognitive impairment and that the MMKD can modulate the gut microbiome and metabolites in association with improved AD biomarkers in CSF.
Keywords: Alzheimer, Dementia, Microbiota, Diet, Nutrition, Ketogenic, High fat, Short-chain fatty acids
Abbreviations: AD, Alzheimer's disease; AHAD, American Heart Association Diet; Aß40/42, amyloid ß40/42; ApoE ε-4, apolipoprotein-E ε-4 allele; CN, cognitively normal; KD, ketogenic diet; MCI, mildly cognitive impaired; MMKD, modified Mediterranean ketogenic diet; SCFAs, short-chain fatty acids; FFAR 2/3, free fatty acid receptor 2/3; LPS, lipopolysaccharide; PD, phylogenetic diversity; OTUs, operational taxonomic units; LDA, linear discrimination analysis; Lefse, LDA effect size
Research in context.
Evidence before this study
Alzheimer's disease (AD) and related dementia is rising worldwide but its etiology and therapies remain unknown. One in every three Alzheimer's cases involves certain modifiable risk elements and hence might be preventable. Emerging evidence suggest that the gut microbiome is associated with AD pathology. Diet is superlative modulators of brain health, and it is also one of the major modulators of the gut microbiome. Ketogenic diet (KD), a very low-carb and high-fat diet that effectively treats refractory epilepsy, is also known to modulate the gut microbiome as well as the brain function. Several mechanisms of KD might also involve to modulate AD pathology such as amyloid aggregation and tau hyperphosphorylation. However, novel gut microbiome and brain biomarkers are needed to pin down the mechanisms by which KD may be protective against AD. Hence, elucidation of gut microbiome during the preclinical stages of AD with no or mild cognitive impairment (MCI) can help identifying novel markers or therapies against AD.
Added value of this study
To our knowledge, this is the first study to demonstrate specific differences in the gut microbiome signature between older-adult MCI patients compared to cognitively normal (CN) counterparts, while also evaluating the impact of a modified Mediterranean-ketogenic diet (MMKD) on gut microbiome composition, short-chain fatty acids (SCFAs) levels, and AD biomarkers. The study identifies specific gut microbiome signatures that are associated with MCI and how these signatures correlate with AD biomarkers including the deposition of β-amyloid (Aβ)-40 and Aβ-42 and tau (total and phosphorylated) in the cerebrospinal fluid (CSF) of these subjects. In addition, the study demonstrates how MMKD distinctly influences the gut microbiome and SCFAs as well as their associations with the Alzheimer's CSF biomarkers in subjects with or without MCI.
Implications of all the available evidence
These findings provide important information to base future intervention and clinical studies to define the novel microbiome based markers to depict the MCI and understand diet-microbiome interactions that may be helpful to ameliorate AD pathology in high risk individuals.
Alt-text: Unlabelled Box
1. Introduction
Alzheimer's disease (AD) is the most common form of dementia in older adults, with clinical manifestations of progressive cognitive and functional impairment. Nearly 50 million people are living with dementia worldwide and this number is expected to increase to over 65 million by 2030 and 115 million by 2050. AD is a progressive, fatal disease that causes heavy burdens on patients, families, and societies. Despite years of research, the underlying etiology of AD remains unknown, with no available treatments or proven preventative interventions [1]. AD pathology is commonly characterized by extracellular accumulations of amyloid-β-peptides (Aβ) in senile plaques and intracellular depositions of hyperphosphorylated tau that forms neurofibrillary tangles [2]. Genetically, Apolipoprotein E (APOE) alleles (ε2, ε3 and ε4) carry different risks for developing AD. Individuals with the ε-4 allele have an increased risk compared with individuals with the more common ε3 allele, while the ε2 allele is related to decreased risk [3]. Recently, it has become clear that the pathogenesis of AD is far more complex and that amyloid deposition in the brain may precede clinical symptoms by 10–20 years [4]. There is clear evidence that AD is related to chronic inflammation both in the central nervous system and in the periphery, and that the amyloid plaques are associated with years of inflammatory antimicrobial responses [[5], [6], [7]]. Researchers have recently proposed a potential role for gut microbiome in the initiation and exacerbation of AD pathology and revealed that Alzheimer's patients have compositionally distinct and less diverse microbiota from control sex- and age-matched individuals [8]. In AD, these alterations in the gut microbiome included a greater abundance of pro-inflammatory bacterial taxa along with less anti-inflammatory taxa [9].
Gut microbiota produce numerous metabolites that directly or indirectly affect brain functions. Among these, short-chain fatty acids (SCFAs) including acetate, propionate and butyrate are known to beneficially modulate the peripheral and central nervous systems. Acetate crosses the blood-brain barrier (BBB) and modulates brain signals to regulate food intake [10]. Acetate also impacts microglial cells and reduces blood-brain barrier permeability [10,11]. Butyrate is a multifaceted compound that is not only the preferred energy source of intestinal cells but also inhibits histone deacetylases to change the expression of several genes and proteins in intestinal and neuronal cells [12]. Butyrate feeding significantly improves learning and memory by amplifying the expression of learning associated genes in AD mouse models and restoring histone acetylation [13]. This evidence indicates that restoring the production of SCFAs in the gut may help in preventing or ameliorating AD pathology. Additionally, the gut microbiome is associated with changes in neurotransmitter signaling, synaptic protein expression, long-term potentiation, and myelination [14]. Therefore, examination of gut microbiome could present potential avenues and opportunities to discover and develop specific microbial signatures that are associate with the prognosis of AD progression. In addition, interventions that modulate the gut microbiome and enrich beneficial bacteria and bacterial metabolites may be helpful in ameliorating AD.
Though the precise etiology of AD remains elusive, it is recognized that dietary patterns may play a role in the pathology of AD. Increased risk of AD and dementia is associated with dietary patterns high in saturated fat and simple carbohydrates, also termed ‘Western’ diets, while diets high in mono- and poly-unsaturated fats, vegetables, fruits, and lean proteins are associated with reduced risk [[15], [16], [17]]. Prompted by these evidences, a move toward a Mediterranean-style diet has been exemplified as not only a prudent lifestyle choice but also as a scientifically accepted mechanism able to confer benefits for prevention and management of several disease pathologies and an overall health and well-being. In 2013, the Mediterranean diet was enrolled on the “Representative List of the Intangible Cultural Heritage of Humanity” by the United Nations Educational, Scientific and Cultural Organization (UNESCO) [18]. A Mediterranean-style diet typifies a nutritionally balanced diet, characterized by a high intake and frequency of fiber, olive oil, fruit, nuts, vegetables, and cereals; a moderate consumption of fish and poultry; a low intake of dairy products, red meat, processed meats, and sweets; and wine in moderation, consumed with meals [19]. Diet is not only a strong modulator of the gut microbiota but is also a powerful modulator of the brain health. Higher adherence to Mediterranean-style diet has been found to be associated with reduced incidences of several chronic diseases, such as obesity, type-2 diabetes, gastrointestinal cancer, cardiovascular diseases, and neurodegenerative diseases including Alzheimer's disease [17,20,21]. Although the underlying mechanisms remain largely undetermined, recent evidence suggests modulation of gut microbiome and microbial metabolites as one of the possible factors mediating the health effects of Mediterranean diet [20,22]. However, data on Mediterranean diet-induced microbiome modulation in context to AD pathology are lacking. Emerging evidence indicates that specific dietary profiles and nutrients modulate the gut microbiota composition and are associated with the production or aggregation of amyloid proteins [[15], [16], [17]]. Therefore, diets that modulate gut microbiome may be an effective strategy to impact the development of AD and other forms of dementia.
The very low carbohydrate and high fat ketogenic diet (KD) has been used for the treatment of refractory epilepsy. The KD is remarkably effective for this purpose: 50–70% of patients have >50% seizure reduction and 30% have >90% seizure reduction [23]. In recent years, several adaptations of the KD have been developed that improve compliance and reduce health risks associated with prolonged high intake of saturated fats. One such adaptation, the modified Mediterranean KD (MMKD), has comparable efficacy to the original KD, but allows slightly higher carbohydrate consumption to permit increased intake of vegetables and fruits, while emphasizing fats and proteins derived from healthy sources such as olive oil and fish [17]. Several mechanisms of action of KD, including reduction of neuronal hyperexcitability, enhancement of mitochondrial metabolism, reduced oxidative stress, and inhibited mammalian target of rapamycin, could also be a modulator of AD pathological processes such as amyloid aggregation and tau hyperphosphorylation. In addition, the KD has also been shown to modulate the gut microbiome in mice, while ketone production is also related to the gut microbiome alterations [24]. Propelled by these ever-mounting data connecting the gut microbiome to metabolism, neural activity, and behavior, and also considering that the KD can modulate the gut microbiome composition, we hypothesized that an intervention of MMKD would improve the gut microbiome by maintaining/ restoring the homeostasis of gut microbiota (beneficial and fiber-degrading gut commensal bacteria) and microbial metabolites (e.g., butyrate) along with dementia/AD biomarkers. Herein, we show how the gut microbiome signature differs in participants with mild cognitive impairment (MCI) compared to cognitively normal (CN) counterparts. In addition, the MMKD alters the gut microbiome signature and SCFAs, and these changes are associated with improved cerebrospinal fluid (CSF) AD biomarkers in older adults.
2. Methods
2.1. Study participants
Stool samples used for analyses were collected as part of a pilot study of a 6-week MMKD or American Heart Association Diet (AHAD) intervention in older adults at risk for AD due to baseline cognitive impairment (MCI) or cogni/subjective memory complaints. The study was a randomized, double-blind, crossover, single-center pilot trial and included 17 participants (mean age: 64.6 ± 6.4 yr) of which 11 participants were diagnosed with MCI (diagnosed using ADNI-2 criteria for early MCI (http://www.adni-info.org) and 6 participants were cognitively normal (CN). The detailed flowchart of the study design and recruitment process is provided in Suppl. Fig. 1, and the detailed metadata of the study participants are provided in supplementary material (Suppl. Table 1). Simple randomization was performed by the study dietician at study onset. The study was originally planned for the recruitment of 30 participants for the diet study, with at least 20 completing both diets at study completion, thereby yielding a greater sample size than that used in recent studies reporting the beneficial effect of a 6-week ketogenic intervention on verbal memory in MCI subjects [25] and thus also increasing the power to detect the intervention's effect in the study groups. Additionally, the crossover design of the study enhanced the power and reduced the variability in that each participant served as their own control for the individual effects of the diets. The present study included only those subjects (n = 17) that provided stool specimens at each time-point of the two dietary arms. The trial was registered before the recruitment started (Clinical Trials # NCT02984540). The study was approved by the Institutional Review Boards of the Wake Forest School of Medicine and was conducted in the Wake Forest Clinical Research Units. Written informed consent was obtained from all participants and/or their legally designated representatives. Diagnoses and eligibility were determined by expert physicians and neuropsychologists based upon the data of cognitive testing, evaluation of medical history, physical examination, and clinical laboratory. Participants were free from alcoholism, head trauma, hypoxia, neurologic disorders except MCI, renal or hepatic disorders, chronic obstructive pulmonary disease, unstable cardiac disease, and psychiatric diseases except mild controlled depression. All study personnel collecting data remained blinded to status or intervention assignment throughout the study. Participants with recent history of use of statins or anti-diabetic medications or agents with prominent CNS effects (except anti-depressants and AD medications) were excluded.
The study was approved by the Institutional Review Boards of the Wake Forest School of Medicine and was conducted in the Wake Forest Clinical Research Units. All protocols related to the cohorts involved in the study have been reviewed and approved by the Institutional Review Board of the Wake Forest School of Medicine. All experiments and samplings were carried out in accordance with ethical and biosafety protocols approved by the Institutional guidelines.
2.2. Study design
Participants were randomly assigned to either a MMKD or an AHAD for 6-weeks, followed by a 6-week washout, and then a 6-week intervention with the second diet (Suppl. Fig. 1). All participants underwent a lumbar puncture (LP) at baseline of diet 1, at the end of diet 1, and again at the end of diet 2. Fasting blood collection and collection of stool samples occurred before and at the end of each diet.
2.3. Intervention
A registered dietitian developed daily meal plans for each study participant based upon their food preferences and caloric needs as determined by a food diary and resting metabolic rate assessment. Participants had weekly in-person or phone visits for meal planning and adherence assessment. Participants maintained a food record that was reviewed at these visits. Blood ketones were measured weekly to ensure compliance on the MMKD. Participants supplied their own food based upon a daily meal plan, food list, and other provided material. There were various recipes (created for the study) and used as a reference for participants to make. Diets were constructed with the following framework: MMKD: The low-carbohydrate diet consisted of a meal plan of <20 g of carbohydrates per day to be consumed over 6 weeks. Foods that provide high levels of healthy fats (preferably low in saturated fats) were generously included in the diet plan. Various lean meats, fish, and nutrient rich foods that meet the requirement of <20 g total carbohydrates per day were included in the meal plans. Carbohydrates were expected to make up <10% of total caloric intake. Participants in the low-carbohydrate diet group received a supply of extra virgin olive oil at study visits to incorporate into their individualized meal plans. AHAD: The low-fat diet consisted of a low-fat, higher-carbohydrate meal plan to be consumed over 6 weeks. Participants were encouraged to limit their amount of fat intake to <40 g per day, while eating plentiful fruits, vegetables, and carbohydrates containing adequate fiber. Various lean meats and other sources of protein were included in the diet plan. Carbohydrates were expected to make up 50–60% of total caloric intake. Overall, the target macronutrient composition (% of total calories) for the MKMD was <10% carbohydrate, 60–65% fat, and 30–35% protein, and for the AHAD was 55–65% carbohydrate, 15–20% fat, and 20–30% protein. While on the MKMD, participants were supplied with 2 L of extra virgin olive oil, and were encouraged to eat fish, lean meats, and nutrient rich foods. While on the AHAD, participants were instructed to limit their fat intake to <40 g/day, while eating fruits, vegetables, and fiber-containing carbohydrates. Throughout the study, participants received a daily multivitamin and restricted use of supplements with antioxidant or ketone-inducing effects (i.e., resveratrol, coenzyme Q10, fish oil, coconut oil, MCT supplements).
2.4. Lumbar puncture, CSF biomarkers assays, and ApoE ε-4 genotyping
Concentrations of CSF Aβ-42, Aβ-40, tau, and phospho-tau (tau-p181) were measured according to the methods described previously [26]. Briefly, CSF (25 mL) was collected in the morning after overnight fast as per the standardized best practice guidelines of the National Alzheimer's Coordinating Center using the 20 gauge Sprotte spinal needles and polypropylene tubes (https://www.alz.washington.edu/BiospecimenTaskForce.html). CSF was transferred in 0.2 mL aliquots into pre-chilled polypropylene tubes, frozen immediately on dry ice, and stored at −80 °C until assay. AD biomarkers (Aβ-42, Aβ-40, tau, tau-p181) were measured using the AlzBio3 multiplex assay (FujiRebio Europe). Subjects were stratified based on the carriage of the ApoE ε-4 allele, a well-known risk factor for AD that predicts insulin response, as per our previously described method [27].
2.5. Fecal microbiome analysis
16S rRNA gene sequencing was performed as previously described [[28], [29], [30], [31], [32]]. Briefly, genomic DNA was extracted from 200 mg fecal samples using QiaAmp PowerFecal DNA kit (Qiagen, CA, USA) per manufacturer's instructions. The V4 hypervariable regions of the 16S rDNA gene were amplified using the universal primers 515F (individually barcoded) and 806R; the resulting amplicons were purified with AMPure® magnetic purification beads (Agencourt); the purified products were quantified using the Qubit-3 fluorimeter (InVitrogen); and the amplicon library was generated according to methods of Caporaso et al. [33] The purified PCR product was pooled in equal molar concentrations and sequenced on an Illumina MiSeq platform and 2x300bp reagent kit (Miseq reagent kit v3; Illumina Inc.) for paired-end sequencing. Sequences were demultiplexed and quality filtered using QIIME (Quantitative Insights into Microbial Ecology; version 1.9.1) software package per default parameters [34]. Operational taxonomic units (OTUs) were chosen by open reference OTU picking based on 97% sequence similarity to the Greengenes database [34]. Taxonomy assignment and rarefaction were performed within QIIME. To avoid bias due to different sequencing depth, the OTU tables were rarefied to the lowest number of sequences per sample (12,151 sequences per sample) for computing alpha-diversity metrics within QIIME using the determination of the number of observed OTUs. Beta diversity of the microbiome was analyzed using principal coordinate analysis (PCoA) of the unweighted and weighted Unifrac distance (using EMPeror version 0.9.3-dev). LEfSE (Linear discriminatory analysis [LDA] Effect Size) was used to identify bacterial taxa that drive differences between MCI or diet groups [35]. To explore the metabolic and other functional activities of the gut bacterial communities in CN vs MCI subjects, an open source bioinformatics tool PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) was used [36]. The OTUs generated from the 16S amplicon sequencing data were entered into PICRUSt software and were analyzed for the prediction of functional genes of the classified members of the gut microbiota resulting from reference-based OTU picking against Greengenes database. Subsequently, the inferred gene families were annotated against KEGG (Kyoto encyclopedia of genes and genomes) orthologs (Kos) and then collapsed into KEGG pathways to generate the functional pathway. The functions were finally categorized and compared at levels 2 and 3 [36].
2.6. Lactate and SCFAs analyses
Fecal organic acids were measured as per the previously described method [28,29,31]. Briefly, an approximately 100 mg aliquot of feces was aseptically mixed with 900 μL sterile PBS buffer (pH 7.4) in a sterile tube and vortexed for one minute or until uniformly suspended. The suspension was centrifuged at 12,000 ×g for 10 min and the supernatant was filtered through a 0.45 μm membrane-filter. Final concentrations (μmol per gram of fecal sample) of lactate, acetate, propionate, and butyrate in fecal specimen were determined using a high-performance liquid chromatography (HPLC; Waters-2695 Alliance HPLC system, Waters Corporation, Milford, MA, United States) with DAD detector at 210 nm, equipped with a Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA, United States). Ten μL sample was injected on the HPLC system; and sulphuric acid (0.005 N) was used to elute the column with a flow rate of 0.6 mL/min at 35 °C.
2.7. Statistical analyses
Bacterial diversity, taxon abundance, and SCFAs levels between groups were compared using non-parametric tests in R statistical software package (version 3.4.3; https://www.r-project.org/). Alpha-diversity indices, bacterial abundance, and fecal organic acid levels at the baseline between MCI and CN participants was compared using two-tailed unpaired Student's _t_-test. Statistically significant differences between different groups during the dietary interventions were calculated by Kruskal-Wallis test followed by Dunn's post-hoc analysis. Hierarchical clustering and heat-maps depicting the patterns of abundance and log values were constructed within R using the ‘heatmap.2’ package. Correlation between bacterial abundance and CSF biomarkers was estimated by using Spearman's rank correlation coefficient test (GraphPad Prism software system, version 6.0). Unless otherwise stated, all the values presented herein are means ± SEM. P < .05 was considered statistically significant unless specified.
3. Results
The recruitment process of study participants continued from January 2015 to April 2017. The study was registered before the recruitment started (Clinical Trials # NCT02984540). Of 30 subjects originally planned for recruitment, 27 subjects met the eligibility criteria and hence were enrolled in the study. Of these, four subjects dropped or were excluded and remaining 23 subjects were finally included for the randomization in the two dietary groups. Finally, only those subjects (n = 17) that provided the fecal specimens at each arm of the two interventions were included for the microbiome and related analysis. The details of the study design and recruitment process are provided in Suppl. Fig. 1, and the general characteristics of the study participants are provided in Suppl. Table 1.
3.1. MCI status is associated with gut microbiome signature
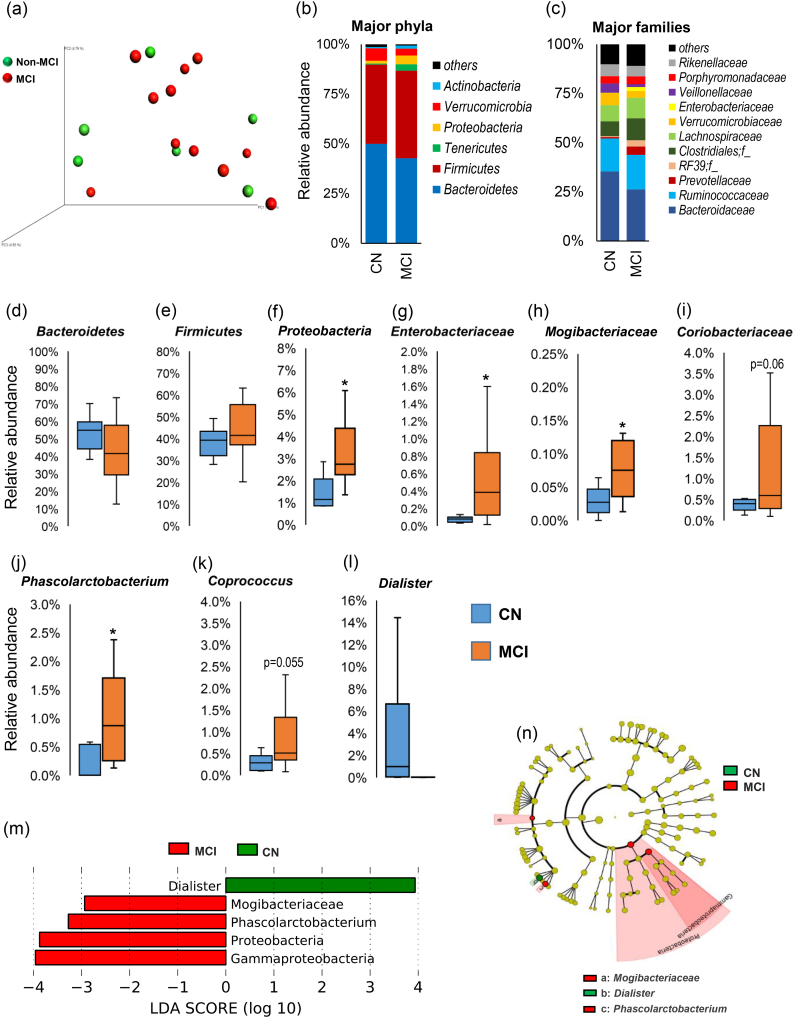
The analysis of gut microbiome diversity at baseline demonstrate no notable difference in terms of beta-diversity (weighted unifrac analysis; Fig. 1a) or alpha-diversity (i.e., phylogenetic diversity, observed OTUs, Chao1 index, and Shannon index; Suppl. Fig. 2a) between CN and MCI participants. However, the taxonomical grouping and differential abundance of OTUs at the phylum, family, and genus levels of bacterial taxonomic classification reveal differences in the relative abundance of several taxa between CN and MCI participants (Fig. 1b-n). At phylum level, MCI participants show slightly higher abundance of phylum Firmicutes and slightly lower abundance of phylum Bacteroidetes (Fig. 1b, d, e) as compared to CN participants. In addition, MCI participants have significantly higher abundance of Proteobacteria (Fig. 1f, m), while having slightly higher abundance of Tenericutes and slightly lower abundance of Verrucomicrobia (Fig. 1b). At family level, MCI participants show significantly higher abundance of families Enterobacteriaceae and Mogibacteriaceae and insignificantly higher abundance of family Coriobacteriaceae (Fig. 1g-i). The genus level comparison and the Linear Discriminatory Analysis (LDA) effect size (Lefse) analysis reveal that the abundance of genera Phascolarctobacterium and Coprococcus was higher, while that of Dialister was lower in MCI participants (Fig. 1j-l). The analysis of fecal concentration of lactate and SCFAs viz. acetate, propionate, and butyrate reveal no significant difference between CN and MCI participants, although MCI participants appear to have slightly lower levels of acetate and propionate (Fig. 2a-d). In addition, the analysis of PICRUSt-inferred functional categorization of the gut microbiome show higher abundance of several disease-related pathways and lower abundance of several metabolism-related pathways in MCI subjects compared with CN subjects (Suppl. Fig. 3).
Fig. 1.
Differences in the gut microbiome between subjects clinically diagnosed with mild cognitive impairment (MCI) versus cognitively normal (CN) counterparts. (a) Microbiome beta-diversity in terms of weighted unifrac distance, and (b-c) gut microbiome composition at the level of major phyla (b) and families (c) in CN (n = 6) and MCI subjects (n = 11). (d-l) Differences in the relative abundance of bacterial taxa in the gut microbiome of CN versus MCI subjects (*p < .05); (m-n): Linear discriminant analysis (LDA) effect size (Lefse) plot and cladogram representing the unique bacterial signatures identified in CN and MCI subjects.
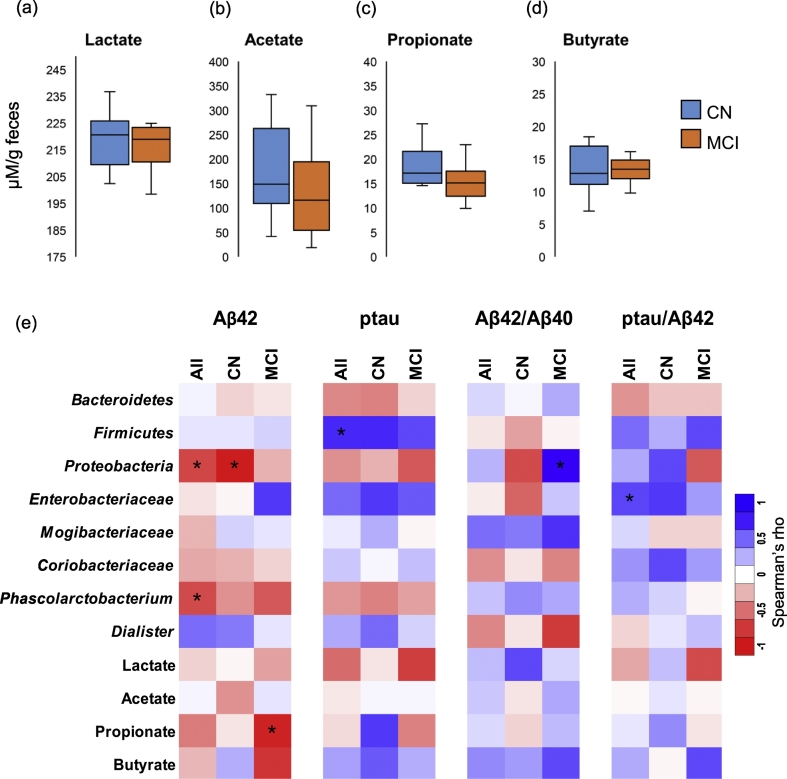
Fig. 2.
Gut bacterial taxa and fecal organic acids correlate with cerebral spinal fluid (CSF) biomarkers of Alzheimer's disease (AD) differently in subjects clinically diagnosed with mild cognitive impairment (MCI) versus cognitively normal (CN) counterparts. (a-d) Fecal concentration of organic acids including lactate and short-chain fatty acids in CN (n = 6) versus MCI subjects (n = 11). (e) Correlation (Spearman R; *p < .05) of gut bacterial taxa and fecal organic acids with CSF biomarkers of AD in CN versus MCI subjects.
3.2. Specific gut bacterial taxa correlate with CSF biomarkers of AD at baseline
We further performed correlation analyses to find relationships between any taxa and metabolites with CSF biomarkers including Aß42 and tau-p181 (Fig. 2e). The results show that the phylum Firmicutes is positively correlated with tau-p181 (Fig. 2e). Further, Proteobacteria are negatively correlated with Aß42 mainly in CN participants; whereas in MCI participants, these correlate positively with Aß42/Aß40 ratio (Fig. 2e). Family Enterobacteriaceae is positively associated with tau-p181 and tau-p181/Aß42 ratio, while Mogibacteriaceae is positively correlated with Aß42/Aß40 ratio (Fig. 2e). Among fecal organic acids, propionate correlates negatively with Aß42 particularly in MCI participants, while sharing positive relationship with tau-p181 in CN participants (Fig. 2e). In addition, several other bacterial taxa are also found to be associated positively or negatively with these biomarkers (Suppl. Fig. 2b).
3.3. MMKD influences gut microbiome and organic acids distinctly in participants with or without MCI
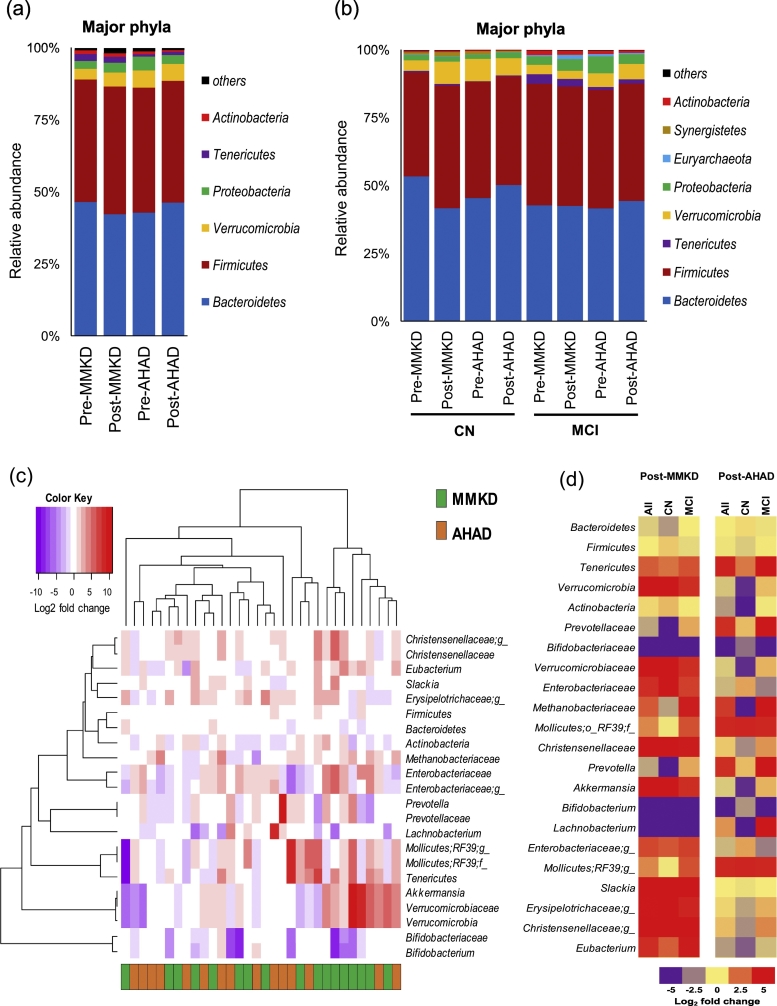
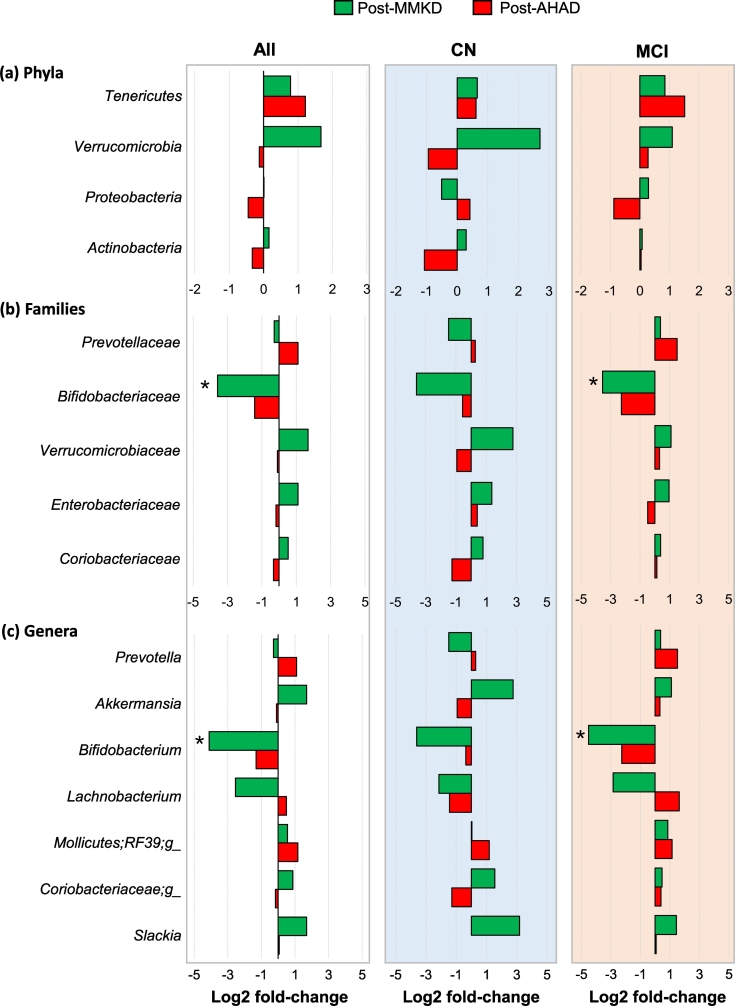
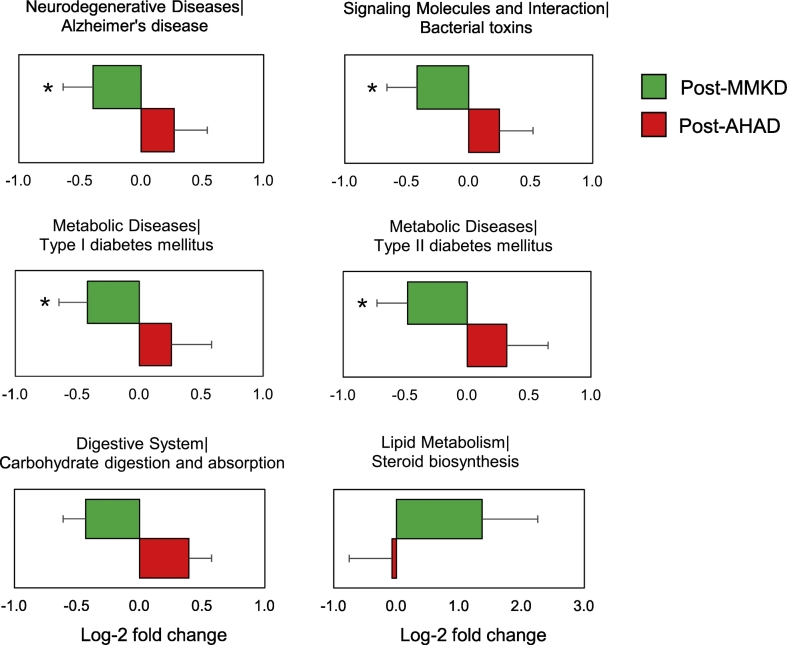
Six weeks of MMKD and AHAD do not show a strong effect on the overall microbiome in terms of the α- and β-diversity indices, which are not significantly different between the two groups. Similarly, major phyla including Firmicutes, Bacteroidetes and Proteobacteria do not demonstrate any significant change during the two dietary interventions in CN and MCI participants (Fig. 3a-b). However, at family and genus level, several taxa were found to be affected by the two diets (Fig. 3c) with patterns differing between CN and MCI participants (Fig. 3d). In particular, family Bifidobacteriaceae and genus Bifidobacterium were significantly reduced on MMKD wherein the reduction was more prominent in MCI participants (Fig. 4b-c; Suppl. Fig. 4). A slight decline in bifidobacteria was seen during AHAD intervention but only in MCI participants (Fig. 3d; 4b-c). On the other hand, the genus Akkermansia from phylum Verrucomicrobia and family Verrumicrobiaceae was insignificantly increased on MMKD but not on AHAD (Fig. 3d; 4a-c). Both diets lead to a slight increase in the abundance of phylum Tenericutes in both of the groups (Fig. 3d; 4a). In addition, MMKD but not AHAD lead to a decline in the abundance of genus Lachnobacterium while increasing that of genus Slackia (Fig. 4c). Interestingly, the PICRUSt-inferred predictions of metagenome function reveal that MMKD but not AHAD decreases the abundance of gene families annotated to the Alzheimer's disease (Fig. 5). In addition, the abundance of KEGG pathways associated with type-1 diabetes, type-2 diabetes, and bacterial toxins is also decreased (p < .05) following MMKD but not AHAD. In terms of metabolism-related pathways, the abundance of gene families associated with carbohydrate digestion and absorption is slightly reduced on MMKD (plausibly due to the ketogenic nature of MMKD) and slightly increased on AHAD, whereas the pathway related to the lipid metabolism i.e., the steroid biosynthesis is slightly increased on MMKD but not on AHAD (Fig. 5).
Fig. 3.
Mediterranean-style ketogenic diet (MMKD) and American Heart Association Diet (AHAD) differently influence gut microbiome in subjects clinically diagnosed with mild cognitive impairment (MCI) versus cognitively normal (CN) counterparts. (a-b) Phyla-level gut microbiome composition at baseline and endpoint of 6-weeks MMKD and AHAD intervention in all participants (n = 17) (a) and separately in CN (n = 6) versus MCI subjects (n = 11) (b). (c) Hierarchical clustering heat-map of gut bacterial taxa that showed >1.0 or < −1.0 Log2-fold increase or decrease in relative abundance during MMKD or AHAD intervention. (d) Heat-map summarizing the differential patterns of diet-induced alterations (mean Log2-fold change in relative abundance) in gut bacterial taxa that showed >1.0 or < −1.0 Log2-fold increase or decrease in relative abundance during MMKD or AHAD intervention in CN and MCI subjects.
Fig. 4.
Mediterranean-style ketogenic diet (MMKD) and American Heart Association Diet (AHAD) induce specific changes in the gut microbiome of subjects clinically diagnosed with mild cognitive impairment (MCI) versus cognitively normal (CN) counterparts. Mean Log2-fold change in the relative abundance of major phyla (a), families (b) and genera (c) in CN (n = 6) versus MCI subjects (n = 11) (*p < .05).
Fig. 5.
Differential effects of Mediterranean-style ketogenic diet (MMKD) and American Heart Association Diet (AHAD) on the functional analysis of the gut microbiome. Functional content of the gut microbiota was inferred by using PICRUSt based on the baseline and endpoint 16S amplicon sequencing data. Changes in the functional KEGG pathways during each dietary intervention were calculated by comparing the relative abundance of the KEGG orthologs and pathways (Level 2 and 3) at baseline vs. endpoint data and were interpreted in the form of mean Log2-fold change.
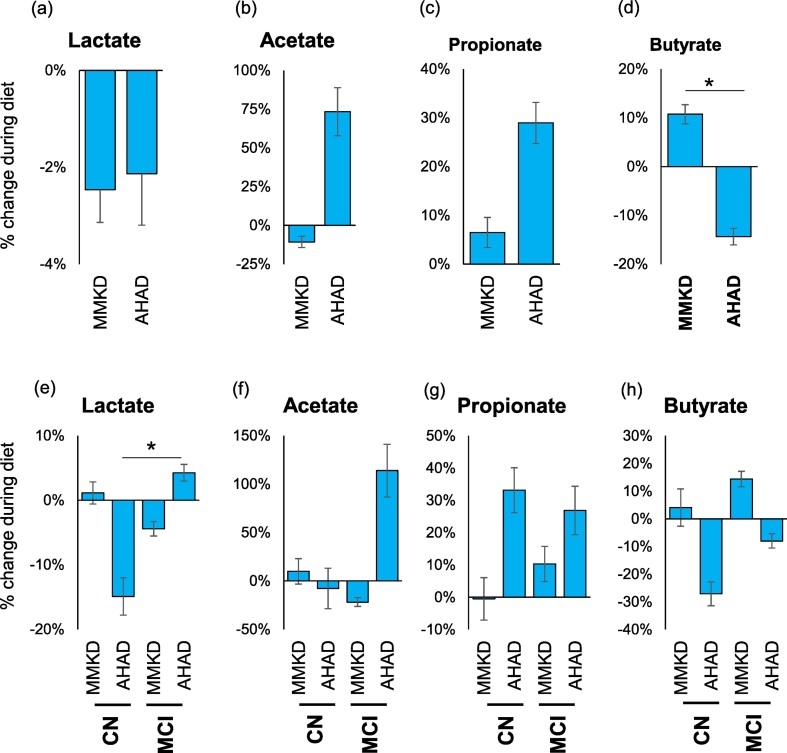
The two dietary interventions also influence the fecal levels of organic acids including lactate and short-chain fatty acids (acetate, propionate and butyrate) with patterns being different between the diets or between CN and MCI participants (Fig. 6; Suppl. Fig. 5). Overall, both diets reduced lactate (Fig. 6a) and increased propionate (Fig. 6c). Acetate was increased considerably on AHAD but was slightly reduced on MMKD (Fig. 6b). On the other hand, butyrate was increased after MMKD but was reduced after AHAD (Fig. 6d), thereby leading to a significant difference between the effects of the two diets. Further analyses show several differential effects of the two diets on fecal organic acids between CN vs. MCI participants (Fig. 6e-h; Suppl. Fig. 5). For instance, after MMKD, lactate was slightly increased in CN participants, whereas in MCI subjects, it was slightly reduced (Fig. 6e; Suppl. Fig. 5). On the other hand, after AHAD, lactate was significantly reduced in CN participants in contrast to a slight increase in MCI participants (Fig. 6e; Suppl. Fig. 5). Acetate remained largely unaffected by the two diets in CN participants but in MCI participants its levels were increased after AHAD but not after MMKD (Fig. 6f; Suppl. Fig. 5). Propionate shows consistent effect wherein its levels were increased in both CN and MCI participants but only after AHAD (Fig. 6g; Suppl. Fig. 5). Butyrate was slightly increased in both CN and MCI participants after MMKD and slightly reduced after AHAD in both groups (Fig. 6h; Suppl. Fig. 5).
Fig. 6.
Mediterranean-style ketogenic diet (MMKD) and American Heart Association Diet (AHAD) differently influence gut microbial metabolites in subjects clinically diagnosed with mild cognitive impairment (MCI) versus cognitively normal (CN) counterparts. Mean percent change in the fecal concentration of intestinal organic acids in all participants (n = 17) (a-d) and separately in CN (n = 6) versus MCI subjects (n = 11) (e-h) (*p < .05).
3.4. Diet-induced changes in gut microbiome and organic acids correlate with changes in the CSF biomarkers of AD
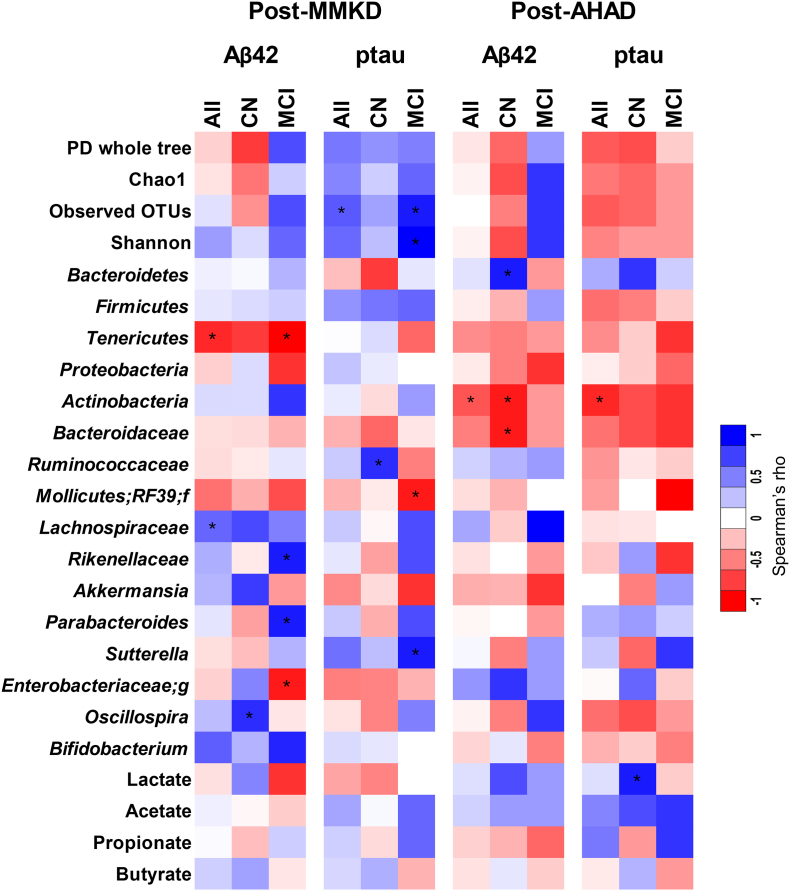
Analysis of correlation of changes occurring in the abundance of gut bacterial taxa and fecal levels of SCFAs with changes in the levels of CSF AD biomarkers (Aß42, Aß40, total tau and tau-p181) reveal several distinct patterns of positive and negative association of biomarkers with several gut bacterial OTUs and SCFAs. These patterns are presented in Fig. 7 in the form of a color-based heat-map. For example, phylum Tenericutes, which was increased by both diets particularly in MCI participants, was negatively correlated with changes in the CSF levels of Aß42, particularly in MCI participants after MMKD (Fig. 7). In addition, family Enterobacteriaceae, which was increased after MMKD in both participant groups, was also negatively correlated with Aß42 but only in MCI participants on MMKD. Family Lachnospiraceae was positively associated with Aß42 in both diet groups. Family Rikenellaeae and genus Parabacteroides were positively associated with Aß42 but only in MCI participants after MMKD, while genus Oscillospira was positively correlated with CN participants after MMKD. Alpha-diversity indices and genus Sutterella were positively associated, while Mollicutes was negatively correlated with tau-p181 in MCI after MMKD. Family Ruminococcaceae was positively associated with tau-p181 but only in CN participants after MMKD. Several similar patterns of correlation of gut microbial taxa with CSF biomarkers Aß40 and total tau and the ratio of Aß42/Aß40 and tau-p181/Aß42 were also seen (see Suppl. Fig. 6).
Fig. 7.
Diet-induced changes in gut microbiome and fecal organic acids are associated with changes in cerebral spinal fluid (CSF) biomarkers of Alzheimer's disease (AD) in subjects clinically diagnosed with mild cognitive impairment (MCI) versus cognitively normal (CN) counterparts. Heat-map depicting the correlation patterns (Spearman R; *p < .05) of changes (percent-fold) in gut bacterial taxa and fecal organic acids with changes (percent-fold) in CSF biomarkers of AD in CN (n = 6) versus MCI subjects (n = 11) (e-h).
4. Discussion
AD is a progressive age-related neurodegenerative disease with no disease-modifying therapy or validated preventative strategy currently available. Early diagnosis and interventions are prerequisites to prevent or at least slow down the progression of AD. Early diagnosis of AD by molecular markers is not reliably consistent; hence, investigation of novel biomarkers may help strengthen the diagnosis and ultimately the prognosis of AD. Considering their emerging role in brain function and pathology [[37], [38], [39]], the gut microbiome and SCFAs may serve as potential candidates for AD biomarkers. Herein, we identify the bacterial taxa that differ between older-adults with MCI and cognitively normal counterparts. Our results suggest that certain bacterial groups show MCI-specific profile and hence may be used as additional and corroborating biomarkers for the detection of MCI.
The overall microbiome diversity in terms of α- and β-diversity indices appears to be comparable between the two groups; however, we note that the MCI subjects have higher abundance of Firmicutes and lower abundance of Bacteroidetes compared to CN counterparts. Notably, increased Firmicutes and decreased Bacteroidetes populations are commonly associated with dysbiotic microbiome signatures and negative health outcome, suggesting that the microbiome in MCI participants is in a dysbiotic state. Although this difference in Firmicutes and Bacteroides was not statistically different apparently which might be because of the small sample size; however, our results concur well with those of a recent study that also reported a similar spectrum of gut dysbiosis in AD patients characterized by reduced abundance of bacterial families and genera within the phylum Firmicutes and lower microbiome diversity [8]. AD patients also had higher abundance of Bacteroidetes [8]. This disparity from early reports of microbiome composition in AD patients [8] may be due to differences in the geographical background, dietary habits, and age of subjects. In addition, the participants in our study are likely to be in the earlier stages of AD spectrum (MCI) compared to Vogt et al. [8] that assessed adults with AD. Hence, considering that MCI is an earlier stage of AD spectrum, the MCI-specific microbiome differences seen in our study cohort suggest that the changes in the gut microbiome composition are linked in neuropathology of AD. Whereas, adults with AD dementia may also have several other health- and lifestyle-related issues that may induce stage-specific microbiome alterations. However, we still observe different patterns of several bacterial groups between MCI and CN subjects, indicating an intricate relationship of host-microbiome interactions and clearly highlights the importance of more comprehensive investigations of the microbiome in AD to establish and validate microbial signatures throughout the spectrum of AD pathology.
The higher abundance of phylum Proteobacteria, class Gammaproteobacteria, and family Enterobacteriaceae (all of which comprise gram-negative bacterial groups) in MCI versus CN participants is intriguing. A wide variety of pathogens belong to the Proteobacteria phylum, while Gammaproteobacteria is also comprised of several pathobionts including Enterobacteriaceae, Vibrionaceae and Pseudomonacaceae families. Notably, the cell wall of these pathobionts is enriched with highly inflammatory components such as lipopolysaccharides (LPS) that are known to induce low-grade systemic inflammation and aggravate AD pathology [40]. Interestingly, higher abundance of gram-negative bacteria in AD patients compared to normal participants has also been reported previously [41]. In addition, we found that MCI participants have higher Dialister and lower Phascolarctobacterium abundance compared to CN participants, a finding that also concurs with previous report [41] and suggests that these genera may be used as a unique biomarker in understanding the progression of MCI to AD dementia. Hence, given the biological applicability of these associations, it would be a worthwhile topic for further studies with larger cohorts to validate and establish these signatures.
Interestingly, the abundance of Mogibacteriaceae family, which is also a periodontal environment-linked bacterial group, is significantly higher in MCI vs. CN subjects, hinting that the gastrointestinal tract of MCI subjects may be invaded by microbes living in other bodily niches [42,43]. Additionally, we find higher abundance of the order Gamellales and family Gamellaceae in ApoE ε-4+ve subjects, suggesting that Gamella might somehow be associated with ε-4 allele and hence could be used to determine the MCI and/or AD risk in ε-4+ve individuals. This finding also concurs with previous study by Vogt et al. [41] that also reported higher abundance of Gamellaceae and Gamella in AD patients. In our study, five out of eleven MCI participants were of APOE ε-4+ve. However, no significant difference in gut bacterial diversity, the abundance of specific bacterial groups, or the fecal SCFAs levels was observed between MCI participants with versus without ε-4+ve genotype (Suppl. Fig. 7). Further, no consistent or significant pattern was seen in the effect of dietary interventions on bacterial taxa, SCFAs, or their correlation with CSF biomarkers (Suppl. Fig. 8).
MCI is an early stage of AD pathology, and accumulating evidence suggests that a substantial proportion of MCI participants remain stable for years [44], indicating that MCI can be targeted preferably by non-pharmacological interventions including dietary and nutritional elements.
The role of diet and gut microbiome in the pathophysiology of neurodegenerative diseases, e.g., AD, has recently received considerable attention [[45], [46], [47]]. Gut microbes may contribute to the modulation of several neurochemical and neuro-metabolic pathways, through complex gut-brain interaction [48,49]. Recent evidence suggests that gut microbiome dysbiosis may affect the synthesis and secretion of several brain-derived neurotrophic factors widely associated with cognitive decline and dementia [47,48], and that modulation of the gut microbiome may induce positive effects on neuronal pathways that can slow down the progression of AD [50]. Given that there are undefined amounts of amyloids in the human gut, it is plausible that the human gut microbiome might play a role in the etiopathogenesis of neurological disorders characterized by amyloidogenic features, such as AD [48,51,52]. Further, the contribution of gut microbiota to amyloid formation and dissemination becomes particularly important during aging, when both the gut-blood barrier and the blood-brain barrier become more permeable to various small molecules [53,54]. Moreover, during aging, the gut microbiome composition undergoes considerable changes which are influenced by different dietetic regimens in addition to several lifestyle- and senescence-related factors [55]. Propelled by these notion, we herein tested the impact of MMKD and AHAD on CSF biomarkers of AD as well as on the gut microbiome signature and SCFAs levels in MCI and CN participants. While there were no significant differences in the α- and β-diversity of the gut microbiome between participants after MMKD and AHAD interventions, we did notice that several gut bacterial phyla, families, and genera are differentially altered after MMKD and AHAD intervention in MCI versus CN subjects. For example, the phylum Actinobacteria, family Bifidobacteriaceae, and genus Bifidobacterium are significantly decreased in MCI group after MMKD as compared to CN counterparts and AHAD group participants. This pattern coincides with a specific decrease in fecal lactate and acetate levels in MCI-MMKD group, suggesting that the effects of specific diets may differ between MCI versus CN subjects. In addition, this also indicates that the MMKD possibly causes a slight increase in the beneficial SCFAs including butyrate. Because these SCFAs content was measured in the feces, the precise source/origin of these SCFAs remains unknown. However, we speculate that either these might have originated in the liver and were subsequently secreted into the gut (because the participants were on a ketogenic state), or the ketogenic diet might have promoted the intestinal SCFAs production by supplying plant-based fermentable fibers to be fermented by the gut bacteria.
Apart from the observed changes in microbial abundances, diet can also influence the features of host metabolism and immune function by affecting various genes, for e.g., those involved in carbohydrate and lipid metabolism, endocrine functions, inflammatory responses, energy balance, etc. Using PICRUSt-inferred predicted metagenomes of gut microbiome followed by the Clusters of Orthologous Groups (COG) classification, we observe that there are several functional pathways affected differently by the two diets. Most notably, the finding of decreased abundance of KEGG pathways annotated to the Alzheimer's disease as well as the type-1 and -2 diabetes following MMKD but not AHAD is interesting and hints toward the dietary influences beyond the lower gut. While MMKD is already known to be beneficial for metabolic health and related diseases including obesity and diabetes, these data further indicate that MMKD can confer benefits to the neurodegenerative diseases as well. In addition, the reduction in the abundance of pathways associated with bacterial toxins following MMKD further underpins the beneficial influence of MMKD on checking the population of toxigenic pathobionts and/or opportunistic pathogens in the gut thereby helping in maintaining/ restoring the gut microbial homeostasis. No doubt, these functional analyses are helpful in understanding the relationship between the host diet, microbiome and functional capacities, it must, however, be noted that these predictions are inferred only from the known functions of the gut microbial communities and hence might be over- or underestimated in specific milieus. Future studies employing the whole genome shotgun sequencing and not just the 16S data might provide more insightful data on these functionalities of the gut microbiome.
Recently studies have shown that abnormally higher level of lactate in the gut is associated with brain fogginess [56], a transient condition characterized by the combination of symptoms of mental confusion, impaired judgment, poor short-term memory, and difficulty with concentration. Hence, the decreased lactate levels in MCI-MMKD group may be associated with improved memory and cognition in MCI participants. On the other hand, increased butyrate after MMKD may be beneficial in terms of reduced/ homeostatic gut leakiness and restrained LPS diffusion. Butyrate is well known to possess neuroprotective actions and improve brain health [57,58]. Butyrate is used as a preferential source of energy by the enterocytes, which results in improved/ homeostatic functions of the gut epithelial and enteroendocrine cells (for the release of gut hormones), goblet cells (for the release of mucus), enterochromaffin cells (for sensory functions), enteric neuronal functions, and mucosal immune cells (to remain in controlled/ homeostatic inflammatory state), as well as enhanced expression of tight junction proteins (required for homeostatic gut permeability). In AD mouse models, butyrate treatment profoundly improves learning and memory ability in unison with improved expression of learning associated genes, that are associated with restoring histone acetylation [13,[59], [60], [61], [62]]. Altogether, these studies as well as our data of specific MCI-associated patterns of microbial modulation accompanied by increased butyrate levels and their correlations with AD biomarkers hint that MMKD may be beneficial for participants with MCI.
To our knowledge, this is the first report to demonstrate specific differences in the gut microbiome signature between older adults with MCI versus cognitively normal counterparts, while also evaluating the impact of a modified Mediterranean-ketogenic diet on intestinal microbiome composition, SCFAs levels, and AD biomarkers. Despite these exciting results, our study is not without limitations. For instance, our cohort may be limited in sample size to find statistically robust differences in gut microbiome and metabolite profiling; however, it still represents an ideal model study to investigate the microbiome and its metabolites as biomarkers for MCI versus CN subjects and should propel and facilitate further larger and more inclusive studies to validate and further elucidate these findings. Indeed, the data are interesting and might pave way for a new paradigm shift in the prediction, progression and management of MCI and AD in prospective broader and comprehensive studies. Although we find interesting differences in the gut microbiome upon dietary interventions that are differentially modulated in MCI versus CN participants, the duration of intervention is relatively short. Further long-term studies of MMKD or similar interventions would help validate these or similar differences in gut microbiome, SCFAs and AD biomarkers in MCI, as well as the progression of MCI to AD dementia. Factors such as gender and ethnicity may also influence the microbiome composition but our small cohort was predominated by females (female/male: 12/5) and white Americans (White/African-American: 12/5) and hence was quite homogenous in terms of gender, ethnicity and age while at the same time not providing enough sample size to discriminate the influence of these factors on the gut microbiome diversity and composition. Nevertheless, our results encouragingly suggest that specific gut microbiome signatures and organic acids might be used as biomarkers for the spectrum of AD pathology and hence would facilitate prospective intervention and clinical studies exploring novel nutrition- and microbiome-related biomarkers and therapeutic targets that could be helpful for precision medicine approaches against MCI, dementia and AD risk.
Funding
The work was supported by National Institutes of Health grant P30AG049638; R01AG055122; Hartman Family Foundation (SC), the Department of Defense funding W81XWH-18-1-0118; W81XWH-19-1-0236 (HY), as well as the funds and services provided from Center for Diabetes, Obesity and Metabolism, Wake Forest Baptist Medical Center and National Center for Advancing Translational Sciences (NCATS), National Institutes of Health funded Wake Forest Clinical and Translational Science Institute (WF CTSI) through Grant Award Number UL1TR001420. The funders had no role in the conceptualization, study design, data collection and analyses, decision to publish, or preparation of the manuscript.
Author contributions
RN: performed microbiome measurement, analyzed data, drafted the manuscript; BJN, SC: coordinated in collecting human feces from participants enrolled in respective studies, and revising drafts of manuscript; SW: measured fecal organic acids; SC and HY: conceived the idea, supervised the study, helped in data interpretations, revised drafts of manuscript. All authors reviewed and approved the final version of the manuscript.
Declaration of Competing interest
The authors declare no competing interests.
Acknowledgements
The authors gratefully acknowledge all the study participants and study staff for their help and cooperation during the study, and Mr. Mohammad Kawas (Neuroscience Graduate Program, Department of Radiology, Wake Forest School of Medicine) for feedback and inputs during writing this manuscript.
Footnotes
Contributor Information
Suzanne Craft, Email: suzcraft@wakehealth.edu.
Hariom Yadav, Email: hyadav@wakehealth.edu.
Appendix A. Supplementary data
Supplementary material
References
- 1.Association, A. s Alzheimer's disease facts and figures, an annual report. https://www.alz.org/alzheimers-dementia/facts-figures
- 2.Hardy J., Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol Sci. 1991;12(10):383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 3.Liu C.C., Liu C.C., Kanekiyo T., Xu H., Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penke B., Bogar F., Fulop L. Beta-amyloid and the Pathomechanisms of Alzheimer's disease: a comprehensive view. Molecules (Basel, Switzerland) 2017;22:10. doi: 10.3390/molecules22101692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Page A., Dupuis G., Frost E.H., Larbi A., Pawelec G., Witkowski J.M. Role of the peripheral innate immune system in the development of Alzheimer's disease. Exp Gerontol. 2018;107:59–66. doi: 10.1016/j.exger.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 6.Bronzuoli M.R., Iacomino A., Steardo L., Scuderi C. Targeting neuroinflammation in Alzheimer's disease. Journal of inflammation research. 2016;9:199–208. doi: 10.2147/JIR.S86958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagan B.L., Jang H., Capone R., Teran Arce F., Ramachandran S., Lal R. Antimicrobial properties of amyloid peptides. Mol Pharm. 2011;9(4):708–717. doi: 10.1021/mp200419b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogt N.M., Kerby R.L., Dill-McFarland K.A., Harding S.J., Merluzzi A.P., Johnson S.C. Gut microbiome alterations in Alzheimer's disease. Sci Rep. 2017;7(1):13537. doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattaneo A., Cattane N., Galluzzi S., Provasi S., Lopizzo N., Festari C. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging. 2017;49:60–68. doi: 10.1016/j.neurobiolaging.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Frost G., Sleeth M.L., Sahuri-Arisoylu M., Lizarbe B., Cerdan S., Brody L. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun. 2014;5:3611. doi: 10.1038/ncomms4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deelchand D.K., Shestov A.A., Koski D.M., Ugurbil K., Henry P.G. Acetate transport and utilization in the rat brain. J Neurochem. 2009;109(Suppl. 1):46–54. doi: 10.1111/j.1471-4159.2009.05895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh M.E., Bhattacharya A., Sataranatarajan K., Qaisar R., Sloane L., Rahman M.M. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell. 2015;14(6):957–970. doi: 10.1111/acel.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govindarajan N., Agis-Balboa R.C., Walter J., Sananbenesi F., Fischer A. Sodium butyrate improves memory function in an Alzheimer's disease mouse model when administered at an advanced stage of disease progression. J Alzheimers Dis. 2011;26(1):187–197. doi: 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- 14.Vuong H.E., Yano J.M., Fung T.C., Hsiao E.Y. The microbiome and host behavior. Annu Rev Neurosci. 2017;40(1):21–49. doi: 10.1146/annurev-neuro-072116-031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris M.C., Tangney C.C. Dietary fat composition and dementia risk. Neurobiol Aging. 2014;35(Suppl. 2):S59–S64. doi: 10.1016/j.neurobiolaging.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Y., Scarmeas N. Dietary patterns in Alzheimer's disease and cognitive aging. Curr Alzheimer Res. 2011;8(5):510–519. doi: 10.2174/156720511796391836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chianese R., Coccurello R., Viggiano A., Scafuro M., Fiore M., Coppola G. Impact of dietary fats on brain functions. Curr Neuropharmacol. 2018;16(7):1059–1085. doi: 10.2174/1570159X15666171017102547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bach-Faig A., Berry E.M., Lairon D., Reguant J., Trichopoulou A., Dernini S. Mediterranean diet pyramid today. Science and cultural updates. Public Health Nutr. 2011;14(12A):2274–2284. doi: 10.1017/S1368980011002515. [DOI] [PubMed] [Google Scholar]
- 19.Willett W.C., Sacks F., Trichopoulou A., Drescher G., Ferro-Luzzi A., Helsing E. Mediterranean diet pyramid: a cultural model for healthy eating. Am J Clin Nutr. 1995;61(6):1402S–1406S. doi: 10.1093/ajcn/61.6.1402S. Suppl. [DOI] [PubMed] [Google Scholar]
- 20.Tosti V., Bertozzi B., Fontana L. Health benefits of the Mediterranean diet: metabolic and molecular mechanisms. J Gerontol A Biol Sci Med Sci. 2018;73(3):318–326. doi: 10.1093/gerona/glx227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sofi F., Abbate R., Gensini G.F., Casini A. Accruing evidence on benefits of adherence to the Mediterranean diet on health: an updated systematic review and meta-analysis. Am J Clin Nutr. 2010;92(5):1189–1196. doi: 10.3945/ajcn.2010.29673. [DOI] [PubMed] [Google Scholar]
- 22.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payne N.E., Cross J.H., Sander J.W., Sisodiya S.M. The ketogenic and related diets in adolescents and adults--a review. Epilepsia. 2011;52(11):1941–1948. doi: 10.1111/j.1528-1167.2011.03287.x. [DOI] [PubMed] [Google Scholar]
- 24.Ma D., Wang A.C., Parikh I., Green S.J., Hoffman J.D., Chlipala G. Ketogenic diet enhances neurovascular function with altered gut microbiome in young healthy mice. Sci Rep. 2018;8(1):6670. doi: 10.1038/s41598-018-25190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krikorian R., Shidler M.D., Dangelo K., Couch S.C., Benoit S.C., Clegg D.J. Dietary ketosis enhances memory in mild cognitive impairment. Neurobiol Aging. 2012;33(2) doi: 10.1016/j.neurobiolaging.2010.10.006. 425 e19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craft S., Claxton A., Baker L.D., Hanson A.J., Cholerton B., Trittschuh E.H. Effects of regular and long-acting insulin on cognition and Alzheimer's disease biomarkers: a pilot clinical trial. J Alzheimers Dis. 2017;57(4):1325–1334. doi: 10.3233/JAD-161256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claxton A., Baker L.D., Wilkinson C.W., Trittschuh E.H., Chapman D., Watson G.S. Sex and ApoE genotype differences in treatment response to two doses of intranasal insulin in adults with mild cognitive impairment or Alzheimer's disease. J Alzheimers Dis. 2013;35(4):789–797. doi: 10.3233/JAD-122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmadi S., Nagpal R., Wang S., Gagliano J., Kitzman D.W., Soleimanian-Zad S. Prebiotics from acorn and sago prevent high-fat-diet-induced insulin resistance via microbiome-gut-brain axis modulation. J Nutr Biochem. 2019;67:1–13. doi: 10.1016/j.jnutbio.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagpal R., Wang S., Ahmadi S., Hayes J., Gagliano J., Subashchandrabose S. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-30114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagpal R., Shively C.A., Appt S.A., Register T.C., Michalson K.T., Vitolins M.Z. Gut microbiome composition in non-human primates consuming a Western or Mediterranean diet. Front Nutr. 2018;5:28. doi: 10.3389/fnut.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagpal R., Wang S., Solberg Woods L.C., Seshie O., Chung S.T., Shively C.A. Comparative microbiome signatures and short-chain fatty acids in mouse, rat, non-human primate, and human feces. Front Microbiol. 2018;9:2897. doi: 10.3389/fmicb.2018.02897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagpal R., Newman T.M., Wang S., Jain S., Lovato J.F., Yadav H. Obesity-linked gut microbiome dysbiosis associated with derangements in gut permeability and intestinal cellular homeostasis independent of diet. J Diabetes Res. 2018;2018 doi: 10.1155/2018/3462092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fung T.C., Olson C.A., Hsiao E.Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20(2):145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghaisas S., Maher J., Kanthasamy A. Gut microbiome in health and disease: linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol Ther. 2016;158:52–62. doi: 10.1016/j.pharmthera.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 40.Kitazawa M., Oddo S., Yamasaki T.R., Green K.N., LaFerla F.M. Lipopolysaccharide-induced inflammation exacerbates tau pathology by a cyclin-dependent kinase 5-mediated pathway in a transgenic model of Alzheimer's disease. J Neurosci. 2005;25(39):8843–8853. doi: 10.1523/JNEUROSCI.2868-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogt N.M., Kerby R.L., Dill-McFarland K.A., Harding S.J., Merluzzi A.P., Johnson S.C. Gut microbiome alterations in Alzheimer's disease. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lourenvarsigmao T.G.B., Spencer S.J., Alm E.J., Colombo A.P.V. Defining the gut microbiota in individuals with periodontal diseases: an exploratory study. J Oral Microbiol. 2018;10(1) doi: 10.1080/20002297.2018.1487741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biagi E., Franceschi C., Rampelli S., Severgnini M., Ostan R., Turroni S. Gut microbiota and extreme longevity. Curr Biol. 2016;26(11):1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Koepsell T.D., Monsell S.E. Reversion from mild cognitive impairment to normal or near-normal cognition: risk factors and prognosis. Neurology. 2012;79(15):1591–1598. doi: 10.1212/WNL.0b013e31826e26b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larroya-Garcia A., Navas-Carrillo D., Orenes-Pinero E. Impact of gut microbiota on neurological diseases: diet composition and novel treatments. Crit Rev Food Sci Nutr. 2018:1–15. doi: 10.1080/10408398.2018.1484340. [DOI] [PubMed] [Google Scholar]
- 46.Pistollato F., Iglesias R.C., Ruiz R., Aparicio S., Crespo J., Lopez L.D. Nutritional patterns associated with the maintenance of neurocognitive functions and the risk of dementia and Alzheimer's disease: a focus on human studies. Pharmacol Res. 2018;131:32–43. doi: 10.1016/j.phrs.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Szablewski L. Human gut microbiota in health and Alzheimer's disease. J Alzheimers Dis. 2018;62(2):549–560. doi: 10.3233/JAD-170908. [DOI] [PubMed] [Google Scholar]
- 48.Junges V.M., Closs V.E., Nogueira G.M., Gottlieb M.G.V. Crosstalk between gut microbiota and central nervous system: a focus on Alzheimer's disease. Curr Alzheimer Res. 2018;15(13):1179–1190. doi: 10.2174/1567205015666180904155908. [DOI] [PubMed] [Google Scholar]
- 49.Burokas A., Moloney R.D., Dinan T.G., Cryan J.F. Microbiota regulation of the mammalian gut-brain axis. Adv Appl Microbiol. 2015;91:1–62. doi: 10.1016/bs.aambs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Bonfili L., Cecarini V., Berardi S., Scarpona S., Suchodolski J.S., Nasuti C. Microbiota modulation counteracts Alzheimer's disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci Rep. 2017;7(1):2426. doi: 10.1038/s41598-017-02587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pistollato F., Sumalla Cano S., Elio I., Masias Vergara M., Giampieri F., Battino M. Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr Rev. 2016;74(10):624–634. doi: 10.1093/nutrit/nuw023. [DOI] [PubMed] [Google Scholar]
- 52.Zhao Y., Lukiw W.J. Microbiome-generated amyloid and potential impact on amyloidogenesis in Alzheimer's disease (AD) J Nat Sci. 2015;1:7. [PMC free article] [PubMed] [Google Scholar]
- 53.Marques F., Sousa J.C., Sousa N., Palha J.A. Blood-brain-barriers in aging and in Alzheimer's disease. Mol Neurodegener. 2013;8:38. doi: 10.1186/1750-1326-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran L., Greenwood-Van Meerveld B. Age-associated remodeling of the intestinal epithelial barrier. J Gerontol A Biol Sci Med Sci. 2013;68(9):1045–1056. doi: 10.1093/gerona/glt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagpal R., Mainali R., Ahmadi S., Wang S., Singh R., Kavanagh K. Gut microbiome and aging: physiological and mechanistic insights. Nutr Healthy Aging. 2018;4(4):267–285. doi: 10.3233/NHA-170030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao S.S.C., Rehman A., Yu S., Andino N.M. Brain fogginess, gas and bloating: a link between SIBO, probiotics and metabolic acidosis. Clin Transl Gastroenterol. 2018;9(6):162. doi: 10.1038/s41424-018-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bourassa M.W., Alim I., Bultman S.J., Ratan R.R. Butyrate, neuroepigenetics and the gut microbiome: can a high fiber diet improve brain health? Neurosci Lett. 2016;625:56–63. doi: 10.1016/j.neulet.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathewson N.D., Jenq R., Mathew A.V., Koenigsknecht M., Hanash A., Toubai T. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016;17(5):505–513. doi: 10.1038/ni.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matt S.M., Allen J.M., Lawson M.A., Mailing L.J., Woods J.A., Johnson R.W. Butyrate and dietary soluble Fiber improve Neuroinflammation associated with aging in mice. Front Immunol. 2018;9:1832. doi: 10.3389/fimmu.2018.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimazu T., Hirschey M.D., Newman J., He W., Shirakawa K., Le Moan N. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science (New York, NY) 2013;339(6116):211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van der Auwera I., Wera S., Van Leuven F., Henderson S.T. A ketogenic diet reduces amyloid beta 40 and 42 in a mouse model of Alzheimer's disease. Nutrition & metabolism. 2005;2:28. doi: 10.1186/1743-7075-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin J.X., Maalouf M., Han P., Zhao M., Gao M., Dharshaun T. Ketones block amyloid entry and improve cognition in an Alzheimer's model. Neurobiol Aging. 2016;39:25–37. doi: 10.1016/j.neurobiolaging.2015.11.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material