Modulation of RNA Condensation by the DEAD-Box Protein eIF4A (original) (raw)
. Author manuscript; available in PMC: 2021 Feb 6.
SUMMARY
Stress granules are condensates of non-translating mRNAs and proteins involved in the stress response and neurodegenerative diseases. Stress granules form in part through intermolecular RNA-RNA interactions, and to better understand how RNA-based condensation occurs, we demonstrate that RNA is effectively recruited to the surfaces of RNA or RNP condensates in vitro. We demonstrate that, through ATP-dependent RNA binding, the DEAD-box protein eIF4A reduces RNA condensation in vitro and limits stress granule formation in cells. This defines a function for eIF4A to limit intermolecular RNA-RNA interactions in cells. These results establish an important role for eIF4A, and potentially other DEAD-box proteins, as ATP-dependent RNA chaperones that limit the condensation of RNA, analogous to the function of proteins like HSP70 in combatting protein aggregates.
Graphical Abstract
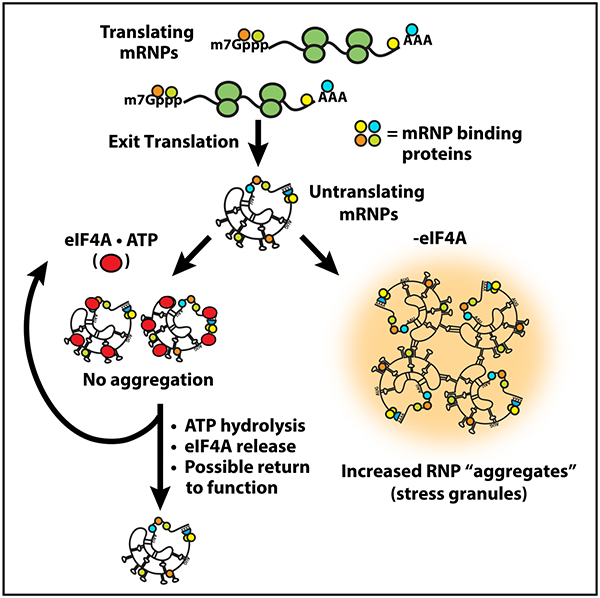
In Brief
RNA-RNA interactions promote formation of RNP condensates, which can be counteracted by DEAD-box proteins to curb excessive granule formation.
INTRODUCTION
Eukaryotic cells contain ribonucleoprotein (RNP) granules in the nucleus and cytosol, including P-bodies (PBs) and stress granules (SGs) (Anderson and Kedersha, 2006; Banani et al., 2017). SGs are cytosolic condensates composed of non-translating RNPs that are involved in the stress response, neurodegeneration, and viral infection (Protter and Parker, 2016; Ivanov et al., 2019). SGs typically form in response to translation shutoff induced by noxious stimuli such as arsenite, heat shock, and endogenous inflammatory molecules like prostaglandins, which all lead to phosphorylation of eIF2α and the activation of the integrated stress response (Aulas et al., 2017; Tauber and Parker, 2019). SGs can also form independently of eIF2α phosphorylation in response to inhibition of the eIF4F complex or osmotic stress (Aulas et al., 2017). SGs and other RNP condensates are thought to form in part through multimeric RNA binding proteins crosslinking RNPs into larger networks (Banani et al., 2017; Shin and Brangwynne, 2017).
Recent evidence suggests that intermolecular RNA-RNA interactions are involved in SG formation. SGs require a substantial pool of non-translating RNA to form (Protter and Parker, 2016). Thus, elevating the non-translating RNA concentration by injecting exogenous RNA induces SGs (Mahadevan et al., 2013). Furthermore, certain RNAs can seed foci in human cell lysates that recruit many SG proteins (Fay et al., 2017). Strikingly, modest concentrations of yeast total RNA readily condense in physiological salt and polyamine conditions, recapitulating the SG transcriptome in a protein-free context (Khong et al., 2017; Van Treeck et al., 2018), arguing that trans RNA-RNA interactions contribute to SG formation. Messenger RNPs (mRNPs) are recruited to SGs in a biphasic manner, first engaging in transient docking interactions with the SG surface, which then transition into stable locking interactions that leave the RNA immobile within the granule (Moon et al., 2019), implying that surface recruitment of RNAs to SGs is a precursor to a more stable RNP assembly.
Roles for RNA in forming, maintaining, and organizing RNP condensates are not limited to SGs. For example, nuclear paraspeckles require the long non-coding RNA (lncRNA) NEAT1 to form (Fox and Lamond, 2010; West et al., 2016). Moreover, specific RNA-RNA interactions between NEAT1 domains may be important for paraspeckle formation and integrity (Lu et al., 2016; Lin et al., 2018). Similarly, the formation of RNA foci of repeat expansion RNAs has been suggested to occur through an RNA-driven condensation (Jain and Vale, 2017). In the Drosophila embryo, homotypic intermolecular base pairing between oskar or bicoid mRNAs determines whether the RNAs will localize to RNP granules in the anterior or the posterior of the embryo, promoting polarity establishment (Ferrandon et al., 1997; Jambor et al., 2011). Specific trans interactions between mRNAs may also be important for their recruitment to polarized RNP granules in Ashbya gossypii (Langdon et al., 2018). Despite their relevance to RNP condensate formation in cells, the properties of condensed RNA are not well understood.
The diversity of intermolecular RNA-RNA interactions relevant to RNP granules and the low barriers for RNA condensation in vitro (Van Treeck and Parker, 2018) are consistent with RNA-based assemblies playing multiple cellular roles but also create a need for the cell to regulate the processes of RNA condensation and recruitment. One possible modulatory mechanism would be the activity of RNA helicases—ATPases that can unwind RNA-RNA interactions and could thereby limit RNA condensation in the cell (Jarmoskaite and Russell, 2014). A major class of RNA helicases are members of the “DExD/H-box” family that typically show highly cooperative binding to ATP and RNA but display low affinities for RNA when complexed with ADP, effectively making them ATP-dependent RNA binding proteins (Andreou and Klostermeier, 2013; Putnam and Jankowsky, 2013). Modulation of their ATPase activity can therefore control the rearrangement of RNPs (Hodge et al., 2011; Noble et al., 2011; Jankowsky et al., 2001; Putnam and Jankowsky, 2013). DEAD-box proteins typically disrupt structures in RNAs in part through ATP-dependent RNA binding, with ATP hydrolysis acting as a switch to release the protein from the RNA (Putnam and Jankowsky, 2013). Virtually all RNP granules contain DEAD-box proteins (Charroux et al., 1999; Saitoh et al., 2004; Dias et al., 2010; Hubstenberger et al., 2013; Calo et al., 2015; Tu and Barrientos, 2015; Jain et al., 2016; Hubstenberger et al., 2017; Markmiller et al., 2018; Youn et al., 2018), and these are often conserved across eukaryotes (Linder and Fuller-Pace, 2013; Jarmoskaite and Russell, 2014). For example, SGs contain multiple conserved helicases, including eIF4A/Tif1 and DDX3/Ded1, in both mammals and yeast, respectively (Jain et al., 2016). Similarly, DEAD-box proteins localize to bacterial RNP granules as well (Al-Husini et al., 2018; Hondele et al., 2019).
Herein, we examine RNA condensation in vitro and SG formation in cells and how those processes are modulated by the essential translation initiation factor, SG component, and archetypal DEAD-box protein eIF4A. We show that RNA and RNP condensates stably interact with other RNAs or RNA-based condensates at their surfaces, promoting RNA condensate assembly. In contrast, we demonstrate that eIF4A limits the recruitment of RNAs to SGs, combating this process. Moreover, we show that the ATP-dependent binding of eIF4A to RNA, separate from its role in translation, can prevent the formation of SGs in cells, limit the interactions of PBs and SGs, and reduce the condensation of RNA in physiological conditions in vitro. Consistent with the mechanism of RNA unwinding by DEAD-box proteins, we show that RNA binding of eIF4A can be sufficient to limit RNA condensation or SG formation but that ATP hydrolysis may increase the effectiveness of eIF4A in limiting RNA condensation by allowing multiple cycles of RNA binding. Together, these data show that eIF4A functions as an ATP-dependent RNA binding protein to limit inappropriate intermolecular RNA-RNA interactions. Such an ATP-dependent RNA chaperone function is analogous to the role of protein chaperones like HSP70 in limiting inappropriate intermolecular protein-protein interactions.
RESULTS
To examine the nature of RNA condensation and how that process might affect RNP granule formation, we examined model RNA condensate systems in vitro with synthetic or purified RNAs. A wide variety of RNAs form RNA condensates in vitro, including all four homopolymers (Langdon et al., 2018; Van Treeck et al., 2018; Jain and Vale, 2017; Aumiller et al., 2016), demonstrating that RNA condensation can occur through non-Watson-Crick interactions. We examined the co-assembly of pairwise combinations of the homopolymer RNAs polyU, polyC, or polyA, which individually condense into RNA droplets of increasing viscosity (Van Treeck et al., 2018). The differential recruitment of short fluorescent RNA oligos into homopolymer assemblies (Figures S1A and S1B) allowed us to distinguish between homopolymer condensates. The robustness of oligo recruitment coincided with the relative strengths of the RNA-RNA interactions between the oligos and the homopolymer scaffolds, with the most efficient recruitment occurring when the oligo could form Watson-Crick base pairs with the condensate scaffold. Some non-complementary RNAs were robustly recruited to condensate surfaces instead, such as oligoG with polyA or PTR with polyC (Figures S1A and S1B). These RNAs have some capacity for homotypic trans interactions; for example, oligoG could in principle form intermolecular G-quadruplexes, suggesting that surface recruitment may occur when trans RNA-RNA interactions between the client RNAs on the surface are preferred over interactions with the homopolymer scaffold or solvent.
Examination of the condensation of mixed homopolymers revealed that increasing the net RNA-RNA interaction strength through extensive base pairing between polyA and polyU leads to gelatinous aggregates containing both homopolymers (Figure 1A). In contrast, mixtures of polyC and polyU spontaneously self-organize into patterned networks of alternating, immiscible polyC and polyU droplets (Figure 1A). A similar, but less organized, flocculation of polyA and polyC assemblies occurred (Figure 1A). These observations demonstrate that RNA-RNA interaction strengths determine the material phase of RNA condensates and that differential interaction strengths can lead to the homotypic clustering and specific compartmentalization of RNAs. Such sequence-specific organization of condensed RNAs may be relevant to the homotypic clustering and/or organization of RNAs in RNP condensates in cells, such as the Drosophila germ granule (Trcek et al., 2015).
Figure 1. RNAs Are Recruited to and Self-Organize on RNA Condensate Surfaces.
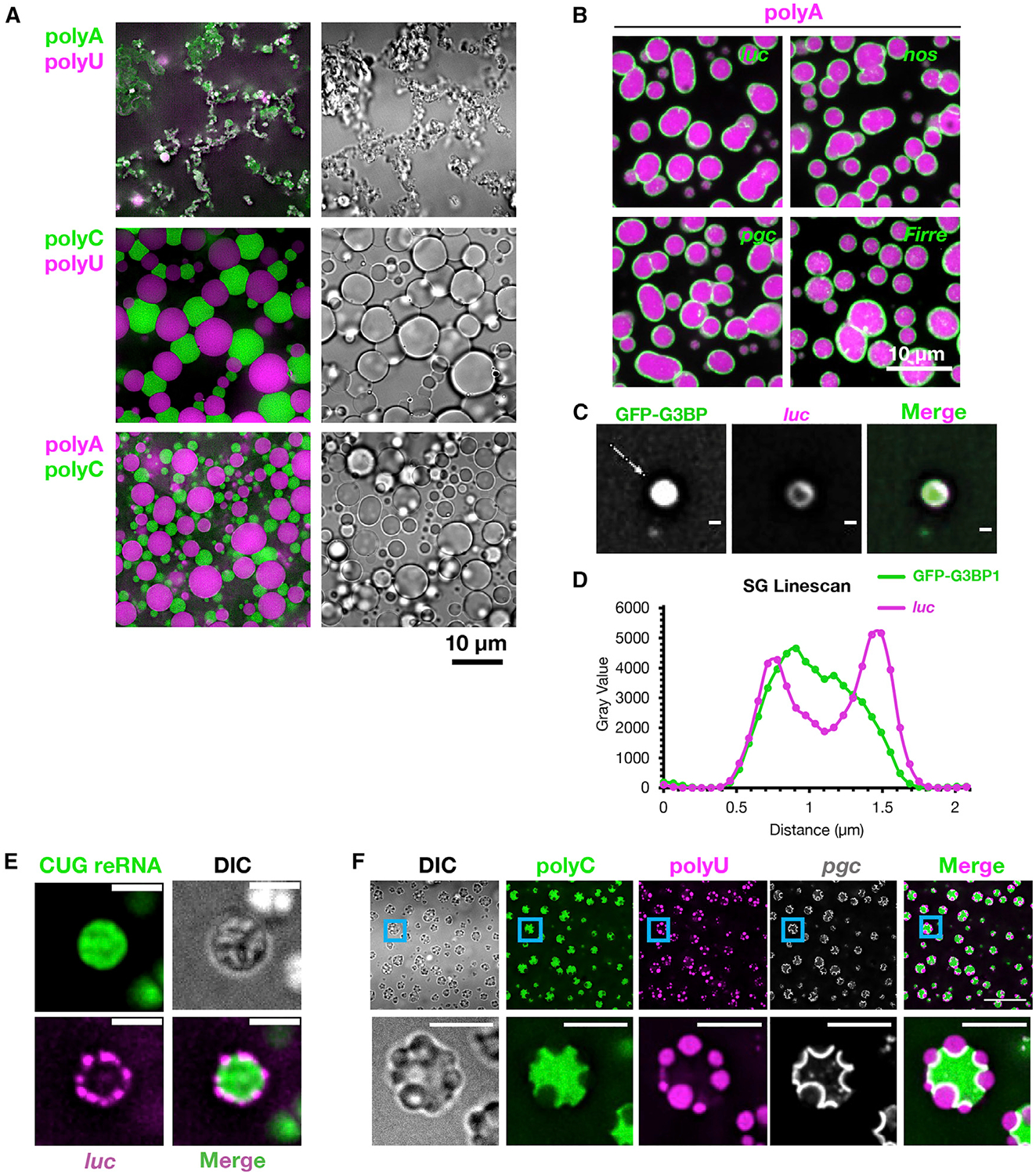
(A) Pairwise combinations of homopolymers were condensed together and visualized with fluorescent antisense oligos.
(B) PolyA assemblies (labeled by oligoU) were condensed with fluorescent in vitro transcribed RNAs.
(C) Purified SG cores containing GFP-G3BP1 were incubated with fluorescent luc RNA. Scale bars, 500 nm.
(D) Line profile of (C) along the line denoted by the white arrow.
(E) A fluorescent myotonic dystrophy repeat RNA (reRNA) containing exons 5–11 of DMPK and ~590 CUG repeats was condensed with fluorescent luc. Scale bars, 2 μm.
(F) Fluorescent pgc was condensed with polyC and polyU (and the corresponding fluorescent antisense oligos), localizing to the polyC/polyU interface. Scale bars, 5 μm.
See also Figure S1.
Multiple observations indicate that RNA condensates associate with one another at interfaces to minimize the total Gibbs free energy of their surfaces, which is the energetic cost of forming an interface between two phases (equivalent to the integral of the interfacial tensions with respect to area; Rowlinson and Widom, 1982). First, the polyC/polyU interface is elongated, deforming the droplets from the typically favorable spherical shape and forming regions with all three phases in contact (Figure 1A). This implies that the polyC/polyU interaction reduces the total Gibbs surface free energy of each individual droplet pair and the whole system in comparison to an equivalent system of dispersed droplets lacking the interfacial interactions (Rowlinson and Widom, 1982). The similar docking of polyA and polyC condensates (Figure 1A) is also consistent with interfacial interactions reducing the total surface free energy. In addition, time-lapse microscopy shows that polyC and polyU condensates are associated from an early time point and appear to nucleate off of each other (Video S1), indicating that their interfacial association is not merely due to droplet crowding. Finally, polyC and polyU droplets remain associated with each other following mechanical disruption (Figure S1C), indicating a physical interaction between the two condensate types.
The interactions of heterotypic RNA condensates suggested that the recruitment of RNAs to RNA/RNP condensate surfaces would also occur. To test this possibility, we condensed polyA, polyC, or polyU in the presence of fluorescently labeled mRNAs. We observed that most mRNAs localized to the surfaces of all three homopolymer condensates, though some RNAs were internalized in polyC condensates (Figures 1B and S1D). Similarly, we observed that SGs isolated from mammalian cells (Figures 1C and 1D), and RNA assemblies formed from a myotonic dystrophy associated CUG repeat RNA (known to form RNA condensates in cells; Jain and Vale, 2017), recruit RNA to their surfaces in vitro (Figure 1E). Thus, a diversity of RNA and RNP assemblies recruit RNAs to their surfaces.
We also observed that the condensation of pgc RNA with both polyC and polyU led to the formation of polyC/polyU multi-phase assemblies (Figure 1F), with pgc RNA localized robustly to the polyC/polyU interface. This observation argues that specific RNAs might act similarly to surfactants or interfacial shells between distinct RNA/RNP condensate phases and thereby stabilize multiphase RNP condensates like the nucleolus (Feric et al., 2016).
Three observations demonstrate that the recruitment of RNAs to the surface of an RNA condensate leads to enhanced interaction between those RNAs and the formation of an RNA shell of enhanced stability enveloping the underlying RNA condensate. First, the surface ring of pgc mRNAs recruited to polyA condensates is more stable following dilution than the underlying polyA assembly, persisting for several minutes longer (Figures 2A and 2B; Video S2). Second, while the internal polyA droplet (visualized by fluorescent oligoU) is somewhat dynamic by fluorescence recovery after photobleaching (FRAP) analysis (mobile fraction [m.f.] at 15 min recovery, = 0.29 ± 0.02), the pgc shell shows virtually no recovery (m.f. = 0.02 ± 0.02), implying that the shell is immobile, which we confirmed by photobleaching only half the condensate and observing no pgc diffusion (Figure S2). Third, using 4′-aminomethyltrioxsalen, which crosslinks RNA duplexes in UV light (Frederiksen and Hearst, 1979), we observed that intermolecular _pgc_-pgc crosslinking, analyzed on denaturing gels, increased when assembled on the surface of polyA condensates as compared to pgc in dilute solution or to homotypic, self-assembled pgc gels (Figures 2C–2E).
Figure 2. RNA Condensate Surface Localization Stabilizes Intermolecular RNA-RNA Interactions.
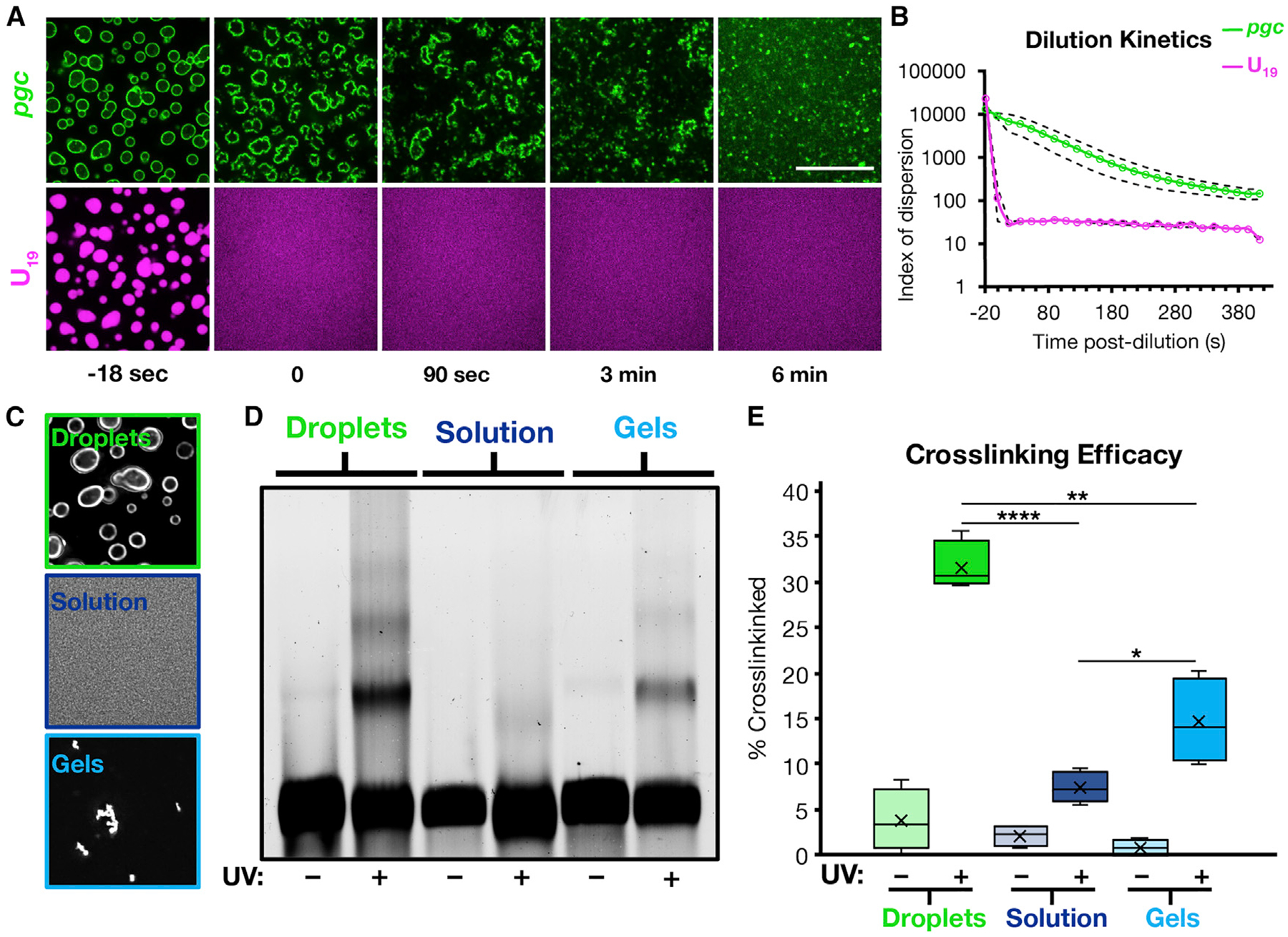
(A) Fluorescent pgc was condensed with polyA and fluorescent U19, and the condensates were subjected to 1:10 dilution in TE buffer. Scale bar, 20 μm.
(B) Quantification of (A) as an index of dispersion, showing the persistence of pgc shell assemblies over time. Dashed lines are 95% confidence intervals. n = 6 replicates.
(C) Images of the crosslinking conditions showing pgc on droplets, in solution, and as gels.
(D) Representative fluorescence denaturing gel.
(E) Quantification of (D), showing that RNA condensate surfaces enhance intermolecular RNA interactions compared to solvated RNA or RNA gels alone. X represents the mean. *p < 0.05, **p < 0.01, ****p < 10−4. n = 4 replicates.
See also Figure S2.
These observations argue that the surfaces of RNA assemblies recruit RNAs. Moreover, the binding of RNAs onto the surface of an RNP granule can concentrate RNAs/RNPs, thereby promoting additional intermolecular RNA interactions, further stabilizing the condensate. This raises the possibility that recruitment of RNAs to the surfaces of RNP granules in cells will be a robust process, and therefore, mechanisms must exist to limit the recruitment of RNAs onto the surfaces of RNP granules. We hypothesized that one or more DEAD-box proteins would act as ATP-dependent RNA chaperones in cells by limiting or dissociating the trans RNA-RNA interactions that promote RNP condensation.
eIF4A Limits RNP Partitioning into SGs
To examine the role of DEAD-box proteins functioning as RNA chaperones in limiting RNA condensation, we focused on SGs. Multiple DEAD-box proteins partition into SGs (Chalupníková et al., 2008; Hilliker et al., 2011; Jain et al., 2016; Markmiller et al., 2018) with eIF4A1, an essential, highly conserved component of the eIF4F translation initiation complex, being the most abundant in both U2OS and HeLa cell lines (Figure 3A; Beck et al., 2011; Itzhak et al., 2016). We hypothesized that eIF4A1 would have additional functions since eIF4A1 is present at ~10x the concentration of other eIF4F components (Itzhak et al., 2016; Beck et al., 2011; Pause et al., 1994), with between 5 and 50 molecules of eIF4A1 per mRNA in U2OS and HeLa cells (see STAR Methods).
Figure 3. eIF4A Limits RNA Recruitment to SGs.
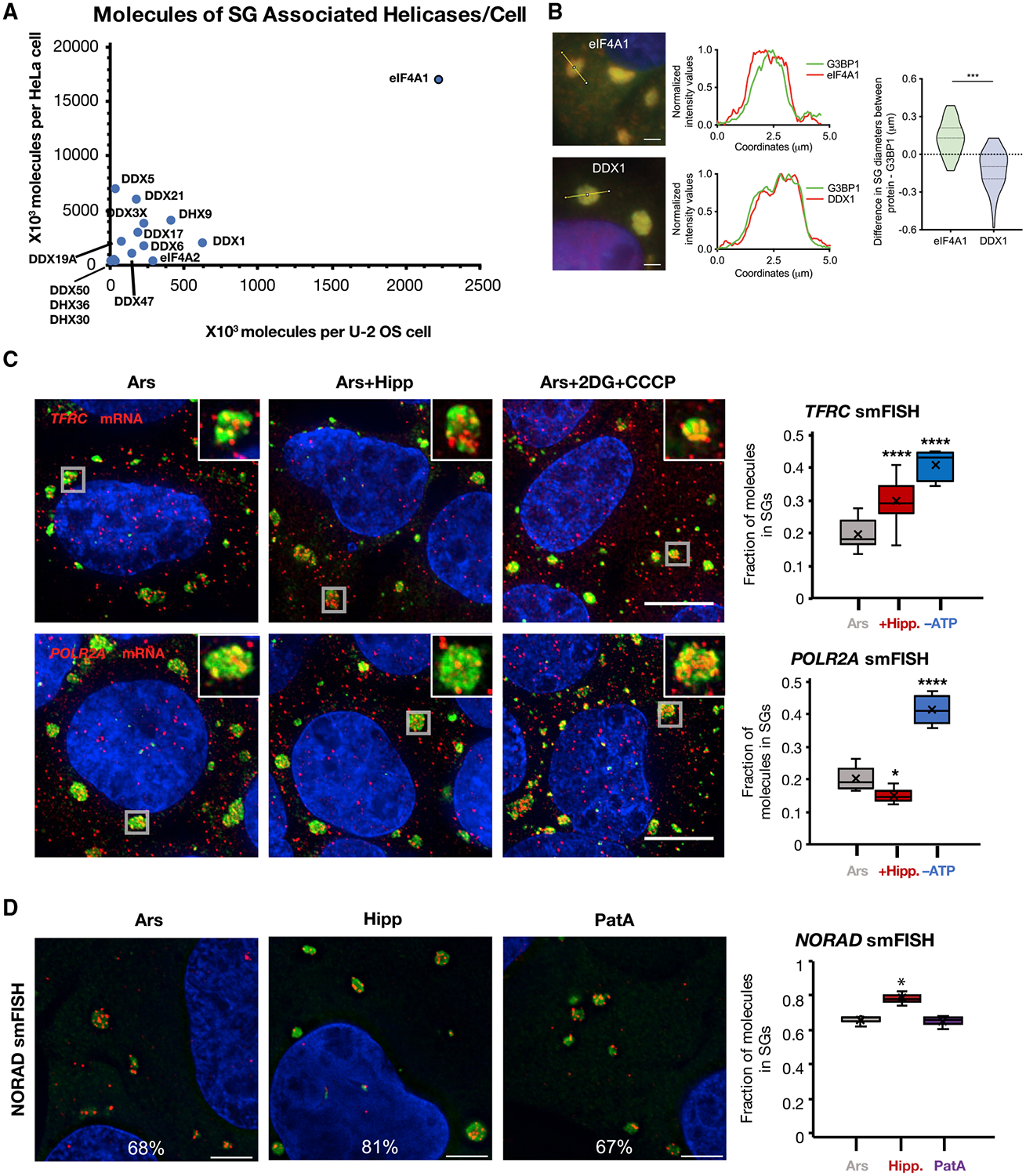
(A) Scatterplot of SG-associated helicase abundance in U2OS versus HeLa cells.
(B) IF of eIF4A1 in SGs. eIF4A1 extends past the SG periphery as compared to DDX1, which co-extends with G3BP1. Scale bars, 2 μm. n = 3 replicates.
(C) smFISH images and quantification of SG enrichment for TFRC and POLR2A mRNAs in U2OS cells treated with 60 min arsenite, then 30 min DMSO, Hipp, or 2DG and CCCP. Gray boxes denote the SGs in the insets. SGs are visualized by anti-PABPC1 IF. Scale bars, 20 μm. n ≥ 5 frames per condition. x represents the mean in our quantifications.
(D) NORAD lncRNA smFISH images and quantification in U2OS cells treated with arsenite, Hipp, or PatA. SGs are visualized by anti-G3BP1 IF. Scale bars, 5 μm. n = 3 replicates. *p < 0.05, **p < 0.01, ***p < 10−3, ****p < 10−4. x represents the mean in our quantifications.
See also Figure S3.
eIF4A1 partitions into SGs during arsenite stress and on average extends further into the cytosol than the canonical SG marker G3BP1 (Figure 3B), indicating eIF4A1 is enriched at the periphery of SGs. In contrast, DDX1, another SG RNA helicase, is uniformly distributed within the granule and overlaps with G3BP1 (Figure 3B). The peripheral concentration of eIF4A1 in SGs is consistent with eIF4A1 modulating the interactions of RNAs with the surfaces of SGs.
If eIF4A limits the condensation of RNAs into SGs, then eIF4A inhibition should increase RNA partitioning into SGs. To separate the effects of eIF4A inhibition on translation initiation from its helicase function, we first treated U2OS cells with arsenite to inhibit bulk translation initiation through eIF2α phosphorylation then added hippuristanol (Hipp) for 30 min, which specifically inhibits the helicase activity of eIF4A by preventing ATP-dependent RNA binding (Bordeleau et al., 2006a; Lindqvist et al., 2008). We then examined the partitioning of mRNAs into SGs by single molecule fluorescence in situ hybridization (smFISH).
We observed that the fraction of TFRC, but not POLR2A, mRNAs associated with SGs increased after Hipp treatment (Figure 3C). We also observed that bulk mRNAs (assessed by oligo(dT) FISH) and DYNH1C1, AHNAK, and PEG3 mRNAs, but not the MCM2 mRNA, partition more strongly into SG in the presence of arsenite and Hipp as compared to arsenite alone (Figure S3B). Since there are additional helicases in SGs, we also examined mRNA partitioning into arsenite-induced SGs following ATP depletion, which would inhibit all ATP-dependent RNA helicases. After ATP depletion, we observed both TFRC and POLR2A mRNAs increased their partitioning into SGs, consistent with other ATP-dependent mechanisms, including additional RNA helicases, limiting mRNA condensation into SGs (Figure 3C). We note the caveat that ATP depletion affects many cellular processes; hence, there may be indirect effects from ATP depletion. However, given the effects of other DEAD-box proteins on RNP granules (see below), it is highly probable that the increase in RNA partitioning we observe is due in part to inhibition of other ATP-dependent RNA chaperones.
To examine the role of eIF4A helicase activity on RNA partitioning into SGs independently of its role in translation in a second context, we also measured the partitioning of the SG-enriched NORAD lncRNA (Khong et al., 2017), which is not thought to be translated, in the presence of arsenite, Hipp, or pateamine A (PatA), which inhibits eIF4A’s function in translation while stimulating eIF4A RNA binding and helicase activity (Bordeleau et al., 2005; 2006b). We observed that NORAD RNA increased partitioning to SGs during Hipp treatment, as compared to arsenite or PatA (Figure 3D). Similarly, we observed that bulk mRNA and the abundant RNA binding protein G3BP1 partitioned more robustly to SGs in Hipp-treated cells as assessed by oligo(dT) FISH or immunofluorescence (IF), respectively, while average SG sizes where slightly decreased with Hipp or PatA (Figures S3B and S3C). This suggests that the density of SGs is increased.
These observations demonstrate that the RNA helicase activity of eIF4A1, independent of its role in translation, limits the accumulation of RNPs in SGs. We suggest that RNAs not affected by eIF4A, such as POLR2A or MCM2, might be limited from entering SGs by other RNA chaperones or might be primarily targeted to SGs by proteins. For example, DEAD-box proteins can preferentially bind, target, and/or unwind differently structured RNAs in vitro and in vivo (Murat et al., 2018; Ribeiro de Almeida et al., 2018; Chen et al., 2018), and these preferences could extend to the ability of DEAD-box proteins to limit the condensation of specific RNAs.
eIF4A Limits SG Formation
We hypothesized that limiting intermolecular RNA interactions through eIF4A would affect not only the recruitment of mRNPs to SGs but also trans RNA-RNA interactions between mRNPs that contribute to SG assembly. To test this possibility, we took advantage of the observation that during arsenite stress, SGs do not form in cells lacking the abundant RNA binding proteins G3BP1/G3BP2, which provide protein-protein interactions to assist SG assembly through G3BP dimerization (Tourrière et al., 2003; Kedersha et al., 2016). We hypothesized that inhibition of eIF4A would compensate for the absence of G3BP in SG formation by promoting increased levels of intermolecular RNA-RNA interactions. Thus, we examined SG formation in wild-type (WT) and ΔΔG3BP1/2 cell lines during arsenite stress, where we altered eIF4A function after 30 min of arsenite stress with either Hipp, blocking RNA and ATP binding by eIF4A, or PatA as a negative control. Puromycin labeling revealed that translation was repressed similarly in all three conditions, indicating that any effects are not due to additional translational repression (Figure S4A).
Importantly, Hipp treatment partially restored SG formation in the ΔΔG3BP1/2 cell lines as assessed by PABPC1 IF, but PatA treatment did not (Figures 4A and 4B). These PABPC1 foci are SGs since they are sensitive to cycloheximide (Figure S5A), which blocks SG formation by trapping mRNAs in polysomes (Kedersha et al., 2000); they do not colocalize with PBs; and they contain multiple SG proteins, polyadenylated RNA, and the SG-enriched RNAs AHNAK and NORAD (Figure S5B). Small interfering RNA (siRNA) depletion of eIF4A1 (Figure S4B), the predominant form of eIF4A in U2OS cells (Figure 3A), also led to the restoration of SGs in the ΔΔG3BP1/2 cell line during arsenite treatment (Figures 4B and 4C). This demonstrates that eIF4A limits SG formation separately from promoting translation.
Figure 4. eIF4A Limits SG Formation.
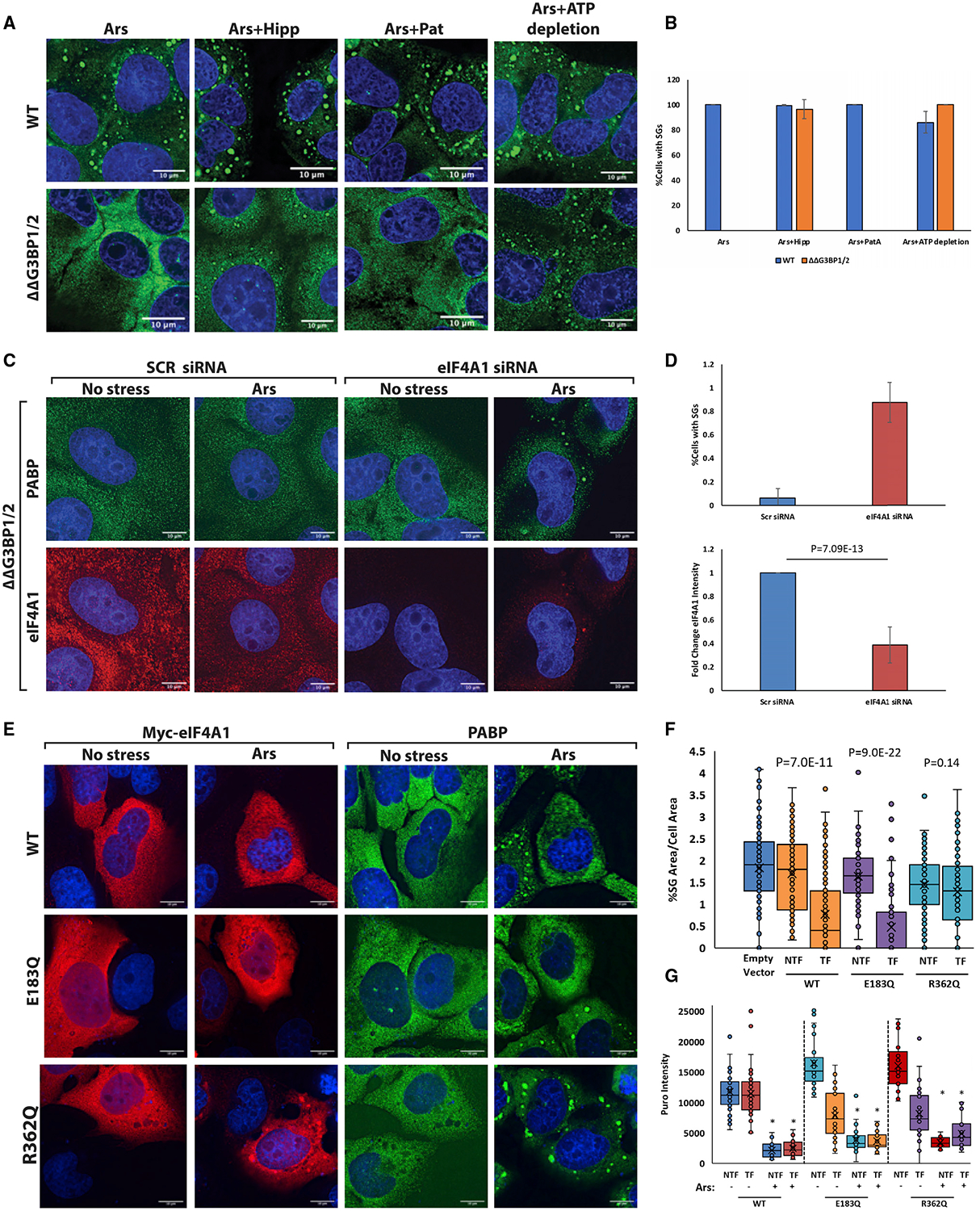
(A) Images displaying SG formation (assessed by PABPC1 IF) in both WT and ΔΔG3BP1/2 U2OS cells with the indicated treatments. Hipp treatment or ATP depletion following arsenite induces SG formation in ΔΔG3BP1/2 cells.
(B) Quantification of images in (A). n = 3. Error bars, SD.
(C) siRNA knockdown of eIF4A1 in ΔΔG3BP1/2 cells restores SGs upon addition of arsenite. n = 3. Error bars, SD.
(D) Quantification of (C) as fold change.
(E) Overexpression of eIF4A1 in WT U2OS cells prevents SG formation in cells expressing Myc-tagged eIF4A1, compared to non-transfected neighbor cells or control transfections. ATPase mutant E183Q was able to prevent SG formation when overexpressed, but RNA binding mutants R362Q and T158Q were not.
(F) Quantifications of (E) as %SG area/cell area of Myc-eIF4A1 expressing (TF) or non-transfected (NTF) cells. x represents the mean of all cells analyzed. n = 3 replicates with all replicates pooled.
(G) Quantifications of translation between NTF and TF WT and mutant versions of Myc-eIF4A1 as assessed by puromycin intensity. Translation is repressed equally in all conditions when arsenite is added, indicating that effects on SG formation are not due to increased translation. *p ≤ 0.05. Error bars, SD with n ≥ 3 replicates.
See also Figures S4 and S5.
In principle, eIF4A could limit SG formation by limiting intermolecular RNA-RNA interactions or by increasing the off-rate of RNA binding proteins contributing to SG assembly. However, inhibition of eIF4A helicase activity with Hipp does not alter the exchange rates of G3BP1 (Figure S4H), consistent with eIF4A altering SG formation by promoting the dissociation of trans RNA-RNA interactions.
Since SGs were only partially restored with Hipp, we also depleted cells of ATP after 30 min of arsenite stress, when ATP is no longer necessary to release mRNAs from polysomes (Jain et al., 2016; Khong and Parker, 2018), to inhibit all ATP-dependent DEAD-box proteins. Depletion of ATP restored SG formation more robustly than Hipp in ΔΔG3BP1/2 cells (Figure 4A). We interpret these observations to signify that in addition to eIF4A, other ATP-dependent mechanisms limit SG formation, although we cannot rule out indirect effects of ATP depletion on cell physiology that also lead to enhanced SG formation. The observation that ATP depletion following translational arrest promotes SG formation is consistent with RNP condensation into SGs being an energetically favored process.
Since inhibiting eIF4A increased SG formation independent of its role in translation, we predicted that overexpression of eIF4A would limit intermolecular RNA-RNA interactions and thereby limit SG formation. To test this, we overexpressed Myc-tagged eIF4A1 in WT U2OS cells by transient transfection (Figure S4C). We then examined whether cells with overexpressed eIF4A1 could still form SGs in response to arsenite treatment. We observed that cells with overexpression of eIF4A1 demonstrated defects in SG formation, although those cells still robustly repressed translation (Figures 4E–4G and S4D), consistent with eIF4A1 limiting RNA condensation into SGs.
In principle, eIF4A could limit RNA condensation in two related manners. First, when bound to ATP, it could bind RNA and thereby compete with RNA-RNA interactions. Alternatively, it could utilize the energy of ATP hydrolysis to limit RNA condensation. Since eIF4A is known to promote RNA duplex unwinding by binding RNA in an ATP-dependent manner (Rogers et al., 1999, 2001), these are related mechanisms. To determine the mechanism by which eIF4A limits RNP condensation, we examined how mutations that inhibit either RNA binding or ATP hydrolysis affected the ability of eIF4A to limit SG formation when overexpressed in U2OS cells.
We observed that overexpression of an RNA binding mutant of eIF4A (R362Q; Pause et al., 1994) did not prevent SG formation (Figures 4E and 4F). This indicates that RNA binding by eIF4A is required to inhibit SG formation. In contrast, we observed that overexpression of an ATPase inactive mutant (E183Q) that can still bind ATP and RNA (Pause et al., 1994; Svitkin et al., 2001; Oguro et al., 2003) repressed SG formation similarly to WT eIF4A (Figures 4E and 4F). This indicates that, at least when overexpressed, eIF4A does not require ATP hydrolysis to limit SG formation, suggesting a model whereby eIF4A acts as an ATP-dependent RNA binding protein to inhibit SG formation, with ATP hydrolysis serving to release the RNA from eIF4A. These effects are independent of translation repression since cells with any of the eIF4A variants overexpressed repressed translation equally in response to arsenite treatment (Figure 4G).
Since other RNA helicases likely function to limit RNA condensation, we tested whether a closely related DEAD-box protein, DDX19A/DBP5 (Cencic and Pelletier, 2016), could prevent SG formation in an analogous manner to eIF4A1. Hochberg-Laufer et al. (2019) reported that DDX19A overexpression inhibits the formation of SGs induced by the transcriptional inhibitor tubercidin by an unknown mechanism. We reasoned that DDX19A could also limit RNA condensation since its helicase core sequence is highly similar to eIF4A. Overexpression of mCherry-DDX19A inhibited arsenite-induced SG formation to a similar extent as eIF4A, while overexpression of mCherry alone did not decrease SG formation (Figures S4E and S4G). Similar to eIF4A, overexpression of RNA-binding-deficient DDX19A fails to inhibit SG formation, while overexpression of an ATP hydrolysis mutant still inhibits SG formation (Figures S4E and S4G). This demonstrates that multiple ATP-dependent RNA chaperones can limit RNP condensation, consistent with ATP depletion having a larger effect on SG formation than eIF4A inhibition alone (Figure 4A).
eIF4A Limits Docking of P-bodies with SGs
Heterotypic RNP granules, like SGs and PBs (Kedersha et al., 2005) or nuclear speckles and paraspeckles (Fox et al., 2002), often dock in cells. Since heterotypic RNA condensates minimize their surface free energy through RNA-RNA docking interactions (Figure 1A), we hypothesized that PB/SG interfaces might occur through intermolecular RNA interactions. If so, the docking of PBs and SGs would be predicted to be modulated by eIF4A. To test this possibility, we treated cells with arsenite, either alone or with Hipp or PatA, and then quantified PB/SG interfaces. We normalized the docking frequency of PBs and SGs to either the total amount of PBs or SGs or to the total PB or SG area to control for the possibility that drug additions affected the amount or sizes of either RNP granule. We observed that the docking frequency of PBs and SGs increased with Hipp addition compared to arsenite or arsenite and PatA, regardless of normalization (Figure 5). When normalized to PB area, the difference between Hipp and PatA was not statistically significant, probably because PatA led to a decrease in total PB area (Figure 5F). These observations suggest that eIF4A can limit docking interactions between PBs and SGs, although whether this is directly due to altering trans RNA-RNA interactions between PB and SG RNAs remains to be established.
Figure 5. eIF4A Limits the Docking of P-bodies with SGs.
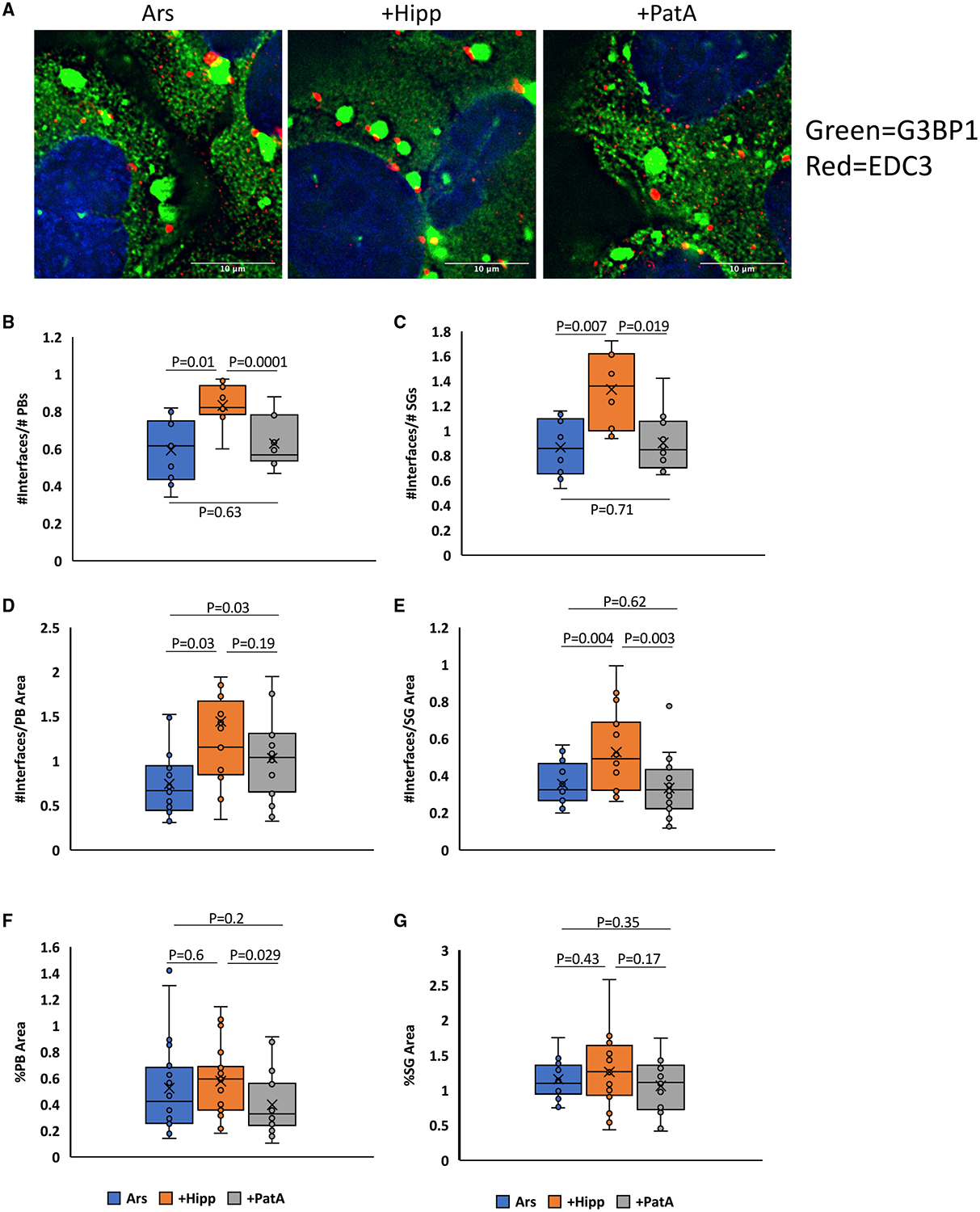
(A) The number of PB/SG interfaces increase in the presence of arsenite in combination with Hipp compared to combinations of arsenite with PatA or arsenite alone.
(B–E) This effect holds true when interface quantities are normalized to the total amount or area of PBs (B and D) or SGs (C and E). PBs and SGs are visualized by EDC3 and G3BP1 IF, respectively. n ≥ 5 replicates. x represents the mean.
(F and G) Hipp addition produces a slight, but not significant, increase in PB number and area (F) but not in SG area (G).
eIF4A1 and ATP Are Sufficient to Limit RNA Condensation In Vitro
To determine whether eIF4A was sufficient to limit RNA condensation, we examined how recombinant eIF4A1 affected the formation of RNA condensates in vitro. We assessed RNA condensate formation by observing RNA condensates stained with SYTO RNASelect, an RNA-specific fluorescent dye. RNA condensates were formed from total yeast RNA in physiological K+ and polyamine concentrations in the presence of polyethylene glycol to simulate the crowded cellular environment (Zimmerman, 1993; Ellis, 2001; Delarue et al., 2018). We performed these experiments with 10 μM eIF4A1, which is its approximate cytosolic concentration (Itzhak et al., 2016), and with RNA at 150 μg/mL, which we estimate is the concentration of exposed open reading frame RNA during acute translational inhibition (Van Treeck et al., 2018). We then examined the formation of RNA condensates over time in the presence or absence of recombinant eIF4A1 with ATP or the non-hydrolyzable ATP analog adenylyl-imidodiphosphate (ADPNP). We observed that ATP alone had a small effect on RNA condensation, perhaps because it is essentially a monovalent RNA and could compete for RNA-RNA interactions; ADPNP had a similar effect (Figure S6).
An important result was that RNA condensation, as assessed by fluorescent labeling was strongly reduced by the addition of eIF4A1 and ATP as compared to ATP alone (Figures 6A and 6B). eIF4A1 in the absence of ATP had no effect on RNA condensation (Figure S6). This demonstrates that eIF4A1 in the presence of ATP can limit RNA condensation. The substitution of the non-hydrolyzable ADPNP for ATP still allowed eIF4A1 to limit RNA condensation, but less effectively (Figures 6A and 6B). Since eIF4A retains its ability to bind RNA when bound to ADPNP (Lorsch and Herschlag, 1998a), this is consistent with ATP-dependent RNA binding being sufficient to limit RNA condensation and that process being increased in efficacy by ATP hydrolysis allowing eIF4A to perform multiple rounds of RNA binding.
Figure 6. Recombinant eIF4A Is Sufficient to Limit RNA Condensation In Vitro.
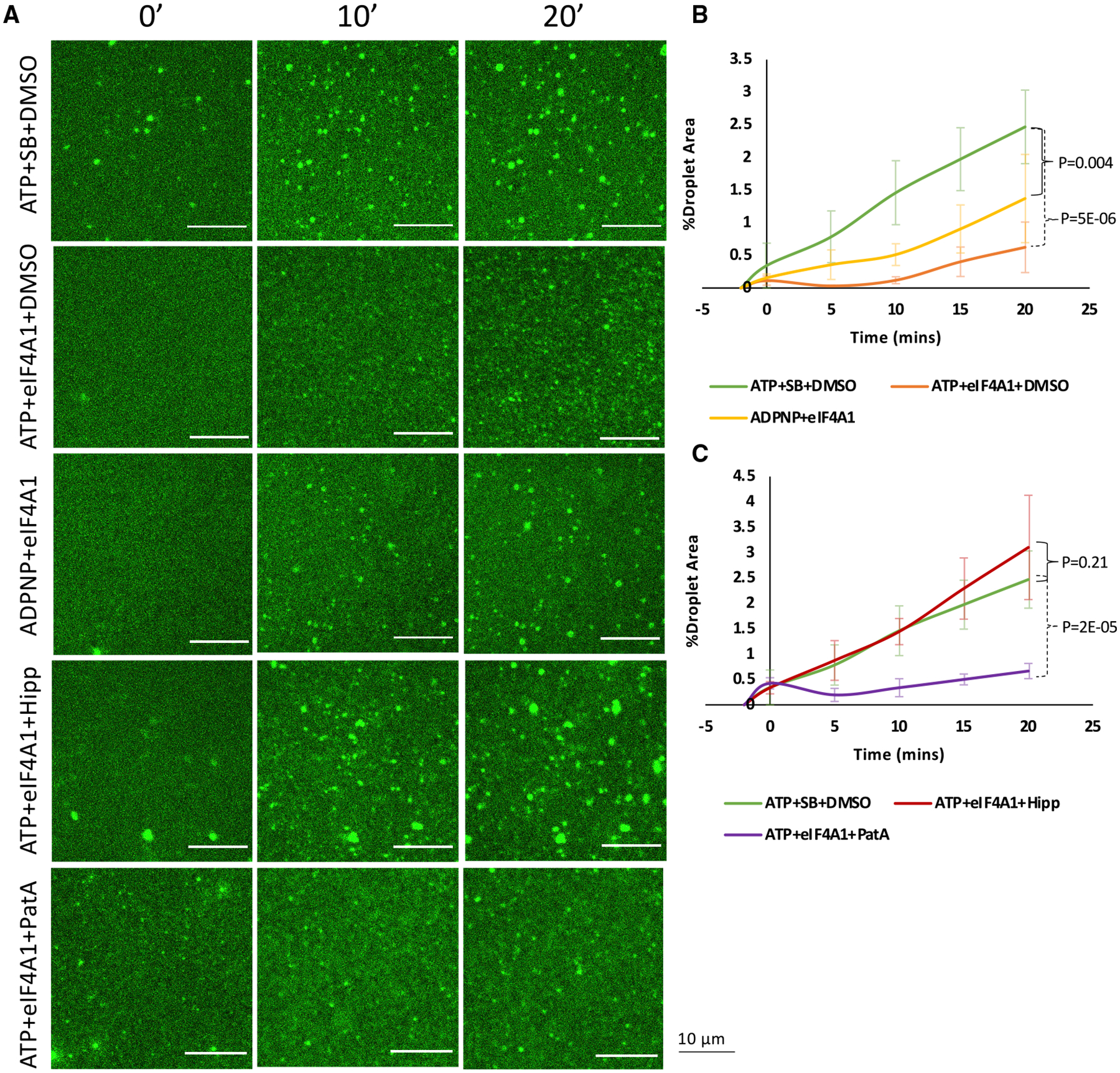
(A) Formation of fluorescent total RNA droplets was monitored over a period of 20 min and is inhibited by eIF4A + ADPNP and more drastically by eIF4A + ATP. Hipp inhibits eIF4A1 catalytic function and restores droplet formation, while PatA does not.
(B and C) Quantification of RNA condensation kinetics comparing ATP + eIF4A1 and ADPNP + eIF4A1 (B) and Hipp addition compared to PatA (C), as assessed by the percent of total frame area occupied by droplets (%Droplet area) over time. n = 3 replicates. SB, protein storage buffer. Error bars, SD.
See also Figure S6.
To further address the mechanism of RNA condensation inhibition by eIF4A1 in vitro, we tested how Hipp and patA affected RNA condensation. Without eIF4A1, Hipp or PatA in the presence of ATP had no effects on condensate formation (Figure S6). However, eIF4A1 incubated with Hipp before RNA addition did not prevent condensate formation (Figures 6A and 6C). Since Hipp prevents eIF4A from binding ATP and RNA, this argues that ATP and RNA binding are required for eIF4A to limit RNA condensation in vitro. In contrast, preincubation with PatA, which slightly increases the catalytic efficiency of eIF4A1 (Low et al., 2005), did not limit eIF4A1’s ability to prevent RNA condensate formation (Figures 6A and 6C). These data confirm our in vivo observations that Hipp, but not PatA, addition following translation inhibition in G3BP null cells can rescue SG formation by preventing eIF4A’s function in limiting RNA condensation.
DISCUSSION
Several observations argue that RNA condensation into RNP granules is a thermodynamically favored process that is countered by energy-consuming processes in the cell. First, multiple distinct RNAs are robust at self-assembly in vitro, including under physiological conditions of salts and polyamines (Figures 1 and S1; Aumiller et al., 2016; Jain and Vale, 2017; Langdon et al., 2018; Van Treeck et al., 2018). Moreover, RNA generally requires lower concentrations to condense in vitro than intrinsically disordered proteins (Van Treeck and Parker, 2018). Second, we observe that condensates of different RNA composition can interact to reduce the surface free energy of the RNA condensates (Figure 1). Third, multiple RNA condensates or purified SGs recruit RNAs to their surface in vitro (Figure 2). Furthermore, in cells, ATP depletion promotes the mRNP condensation into SGs (Figure 3C), even in the absence of protein factors normally required for SG formation (Figure 4A), demonstrating that cells utilize energy to limit SG formation.
In vitro, we observe that the recruitment of RNAs to the surface of an RNA condensate creates a high local concentration, which promotes the formation of additional interactions between the molecules to further stabilize the RNA assembly (Figure 2). This creates a positive feedback loop in RNA condensation. Thus, RNA condensation has the potential to create an “RNA entanglement catastrophe,” wherein extensive RNA condensation would limit the proper distribution and function of RNAs. An implication of these observations is that cells must contain mechanisms to limit the intermolecular interactions and condensation of RNA.
We present several lines of evidence arguing that eIF4A functions to limit cytosolic RNA condensation. First, inhibition of eIF4A1 function by knockdown or Hipp treatment, but not PatA, restores SG formation in cells lacking G3BP without changes in translational repression (Figures 4A and 4B). Second, overexpression of eIF4A1 limits arsenite-induced SG formation, although translation remains repressed (Figures 4E–4G and S4D). Third, inhibition of the RNA binding and ATPase activity of eIF4A by Hipp treatment during arsenite stress enriches bulk and specific RNAs in SGs (Figures 3 and S3). Fourth, inhibition of eIF4A RNA binding and ATPase activity with Hipp increases PB/SG docking events (Figure 5). Finally, recombinant eIF4A1 is able to limit RNA condensation in vitro in an ATP-dependent manner (Figure 6). Since inhibition of eIF4A helicase activity with Hipp does not alter the exchange rates of G3BP1 (Figure S4H), we interpret these observations to suggest that eIF4A limits RNA condensation by the disruption of intermolecular RNA-RNA interactions. Since RNA binding, but not ATP hydrolysis, is required for limiting SG formation when eIF4A1 is overexpressed (Figures 4E and 4F), RNA binding is the critical feature of eIF4A’s ability to limit RNA condensation.
These results synergize well with biochemical data on the eIF4A helicase mechanism. RNA duplex unwinding by eIF4A and other DEAD-box proteins is coupled not so much to ATP hydrolysis as to the ATP-dependent binding of eIF4A to the RNA substrate, which kinks RNA to locally destabilize the duplex, which may then dissociate thermally (Rogers et al., 1999, 2001; Liu et al., 2008; Putnam and Jankowsky, 2013). Hence, productive unwinding events are determined in part by the relative thermostability of the RNA$eIF4A$ATP complex compared to the structured RNA, limiting eIF4A’s ability to efficiently resolve duplexes more stable than ~10–15 bp (Δ_G_ approximately −20–25 kcal/mol; Rogers et al., 1999, 2001). This may provide insight into the nature of the trans RNA interactions that drive RNA condensation and recruitment to condensates, namely that they are individually rather weak, despite their apparent thermostability in summation. Thus, ATP serves as a molecular switch to increase the affinity of the protein for RNA, while ATP hydrolysis and Pi release cause conformational changes to lower the affinity of eIF4A for RNA, effectively making eIF4A function as an ATP-dependent RNA binding protein (Lorsch and Herschlag, 1998a, 1998b; Sun et al., 2014). Taken together, we suggest that while the binding of eIF4A to RNA is the key step in limiting RNA condensation, the ATPase-driven cycling of eIF4A on and off transcripts increases its ability to limit trans interactions by allowing multiple RNA binding events (Figure 7A). Accordingly, eIF4A limits RNA condensation as an ATP-dependent RNA binding protein, providing the cell with a controllable means of buffering intermolecular RNA interactions.
Figure 7. eIF4A Limits RNA Condensation as an ATP-Dependent RNA Chaperone, Analogous to Heat Shock Proteins.
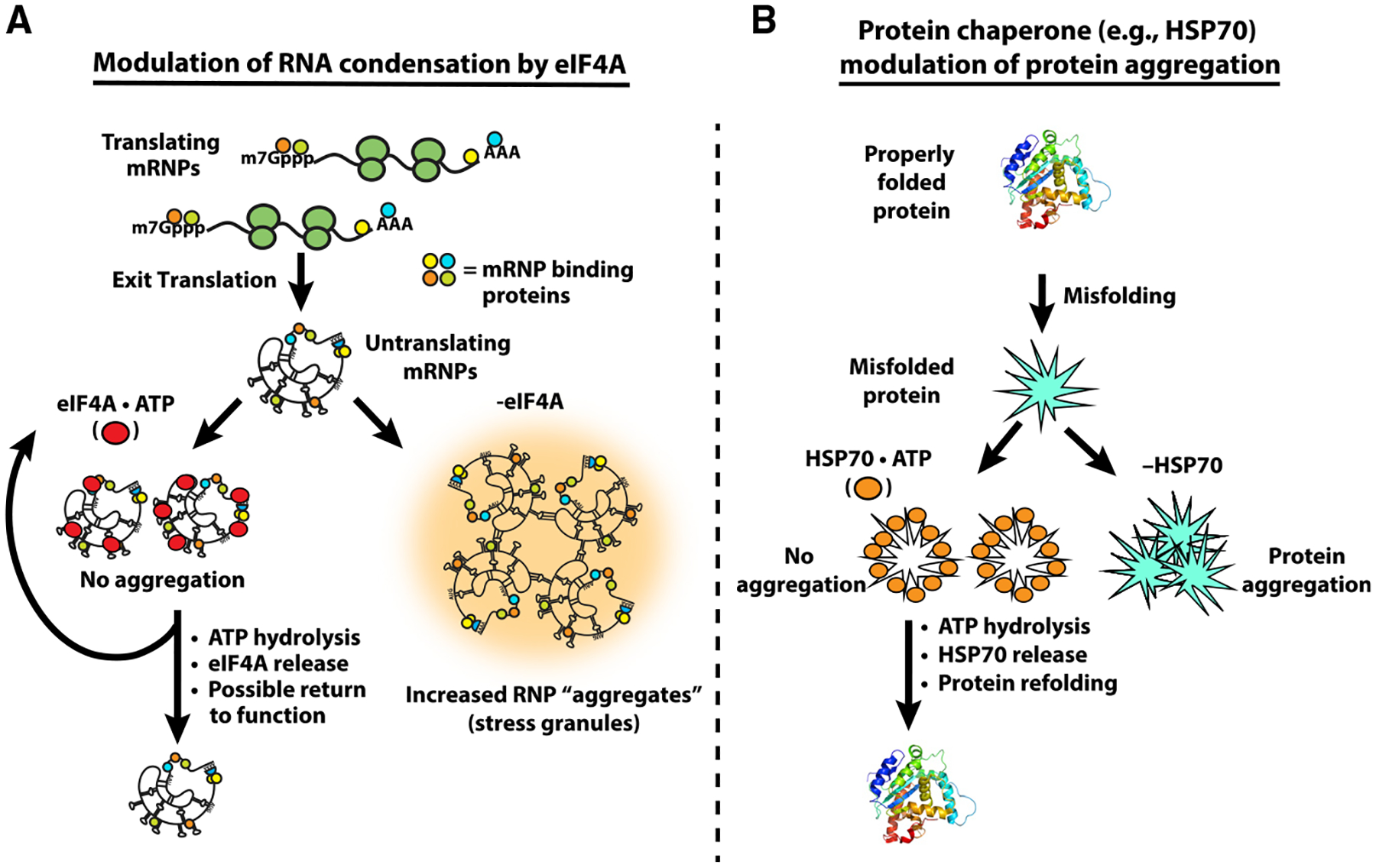
(A) The general mechanism utilized by chaperones like HSP70 to resolve aberrant protein-protein interactions. Misfolded proteins are bound by HSP70·ATP, which limits aggregation while ATP hydrolysis liberates the protein.
(B) Model of eIF4A’s function to resolve aberrant RNA-RNA interactions. ATP-dependent binding of eIF4A to RNA limits the multivalent RNA-RNA interactions driving RNA condensation, while ATP hydrolysis facilitates eIF4A release to re-enter the catalytic cycle.
We anticipate that the ATP-dependent RNA binding of eIF4A, and possibly other DEAD-box proteins, will also modulate the formation of intramolecular RNA duplexes. Such a mechanism of limiting or buffering RNA secondary structure in cells could explain why predicted local RNA secondary structure does not strongly affect the ability of the PUM2 protein to bind target sites in mRNAs (Jarmoskaite et al., 2019).
Since overexpressing ATPase-defective DEAD-box mutants was sufficient to prevent SGs (Figures 4 and S5), it is likely that high concentrations of monovalent RNA binding proteins can also reduce the condensation of RNA. This is consistent with the observation that overexpression of the abundant RNA binding protein YB-1 can limit the formation of SGs and the accumulation of certain transcripts in them, consistent with its RNP chaperone abilities in vitro (Bounedjah et al., 2014; Tanaka et al., 2014). Cells appear to have adapted to make use of this mechanism across domains of life; for instance, the bacterial cold shock protein CspA is present at high intracellular concentrations (~30 μM) and ameliorates global RNA structure during the cold shock response, possibly including trans RNA-RNA interactions (Zhang et al., 2018).
A general role for eIF4A in limiting RNA condensation in the cytosol provides an answer to the decades-old question of why eIF4A is present at ~10x higher concentrations than other translation initiation factors (Figure 3A; Pause et al., 1994). Since ATP depletion is more effective at rescuing SG formation in cell lines lacking G3BP (Figure 4A) and has a larger effect on mRNA recruitment to SGs during arsenite stress (Figure 3C), we suggest that additional ATP-dependent mechanisms, potentially including other SG-associated DEAD-box proteins (Figure 3A), also limit RNP condensation into SGs. Consistent with other DEAD-box proteins limiting SG formation, we demonstrate that overexpression of DDX19A, which is closely related to eIF4A (Cencic and Pelletier, 2016), can limit SG formation without affecting translation (Figures S4E–S4G). Interestingly, knockout of the helicase DHX36 leads to increased SG formation, but this appears to be due to constitutive activation of the eIF2α kinase PKR and increased translation repression (Sauer et al., 2019). An important area of future work will be identifying additional helicases that limit RNA condensation and determining whether they act in RNA-specific manners.
The generality of RNA condensation predicts it will be relevant to other RNP granules. Indeed, observations in the literature are consistent with other RNA helicases functioning to limit RNA retention in RNA condensates. For example, knockdown of the RNA helicase UAP56/DDX39B (which is related to eIF4A; Cencic and Pelletier, 2016), leads to increased accumulation of polyA+ RNA and spliced β-globin mRNA in nuclear speckles (Dias et al., 2010), which are RNP granules formed near sites of transcription. Similarly, catalytic UAP56 mutations lead to retention of influenza M mRNAs in speckles (Hondele et al., 2019). Additionally, knockdown of the UPF1 RNA helicase leads to retention of nascent transcripts in nuclear foci with DNA (Singh et al., 2019), where a high local concentration of nascent RNA might lead to RNA entanglements. These results suggest that RNA chaperones may be generally required to prevent the trapping of RNAs in energy wells at RNP granules. Additional mechanisms cells could utilize to limit RNA condensation include ribosome association, modulating RNA concentrations through RNA decay (Burke et al., 2019) or synthesis rates, and RNA modifications that alter the stability of RNA-RNA interactions.
DEAD-box proteins play two additional roles in modulating the formation and fate of mRNPs that will impact RNP granule assembly. First, in contrast to eIF4A1, many DEAD-box proteins have N- and C-terminal extensions that can play additional roles in promoting individual mRNP or mRNP granule assembly through additional protein-protein interactions (Hilliker et al., 2011; Mugler et al., 2016; Rao and Parker, 2017; Sachdev et al., 2019). In some cases, these N- and C-terminal extensions might contribute to RNP granule assembly by promoting RNP phase transitions (Hondele et al., 2019). In addition, ATP hydrolysis by DEAD-box proteins can also promote rearrangements in mRNPs, allowing the mRNA to undergo subsequent processing, translation, or degradation events that will alter the ability of the mRNP to assemble into an RNP granule (Hilliker et al., 2011; Sheth and Parker, 2006; Mugler et al., 2016). Thus, DEAD-box proteins can either promote or limit RNP granule assembly in a context-dependent manner.
A role for eIF4A, and other general RNA helicases, in limiting RNA condensation can be considered analogous to protein chaperones, such as HSP70, limiting the aggregation of misfolded proteins (Figure 7). Multiple protein chaperones, including HSP70 proteins, bind to protein aggregates to disassemble aberrant interactions, thereby allowing for aggregate solubilization and protein refolding, using ATP hydrolysis as a switch for binding (Mogk et al., 2018). We suggest that RNA condensation and inappropriate aggregation occurs when the amount of exposed RNA in the cell exceeds the capacity of the cellular machinery limiting RNA condensation. Thus, the intrinsic aggregation properties of both proteins and RNAs are countered by abundant cellular machinery to keep these macromolecules correctly folded and dispersed for proper function.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Roy Parker (roy.parker@colorado.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and growth conditions
Cell lines used in this study include WT parental human U2OS cells, ΔΔG3BP1/2 double knockout U2OS cells, and G3BP1-GFP U2OS cells. U2OS cells are female. All cell lines were grown and maintained at 37°C and 5% CO2. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin mix.
Yeast and bacterial strains and growth conditions
For total RNA extraction, Saccharomyces cerevisiae strain BY4741 (Dharmacon) was cultured in yeast extract peptone dextrose (YEPD) media at 30°C. For plasmid isolation, XLI-Blue supercompetent cells were grown in lysogeny broth (LB) at 30°C.
METHOD DETAILS
Plasmid Isolation and Site-Directed Mutagenesis
Plasmids were amplified by transformation into XL1-Blue supercompetent cells (Agilent Technologies) by following manufacturer’s instructions. Bacteria was then grown overnight in 5 mL cultures in LB media with respective selectable marker antibiotic. Plasmids were then isolated using ZymoPURE plasmid mini-prep kit by following manufacturer’s instructions. Plasmid concentrations were determined via absorbance at 260 nm using a Nanodrop 2000 device (Thermo Scientific). Site directed mutagenesis was performed on pcDNA3.1-Myc-eIF4A1 by using the Quick-change Site Directed mutagenesis kit (Agilent Technologies) according to manufacturer’s instructions. Mutations were verified via commercial sanger sequencing (Genewiz).
In vitro transcription of RNAs
Drosophila nos, pgc, ccr4, and cycB T7 transcription plasmids were procured from the Drosophila Genomics Research Center (DGRC; Bloomington, IN). The m_Firre_3.1 T7 transcription plasmid was generously provided by the John Rinn Lab at CU Boulder. The CUG repeat RNA construct was generously provided by the Matt Disney Lab (Scripps Research Institute). Plasmids were linearized via restriction digestion with BamHI-HF for the Drosophila RNAs, Afe1 for luciferase, and Kpn1-HF for Firre (enzymes from New England Biolabs (NEB)). The CUG repeat construct was linearized with HindIII-HF (NEB). Restriction digestion reactions were performed according to the manufacturers’ recommendations. Following linearization, plasmid DNA was recovered with ethanol precipitation followed by resuspension in TE buffer (10 mM Tris HCl pH 8.0, 1 mM EDTA).
In vitro transcription of fluorescent RNAs was performed using the T7 MEGAscript kit (Ambion) according to the manufacturer’s recommendations with the following modification: Addition of unlabeled UTP was reduced by 25% and an equal amount of fluorescently labeled UTP (fluorescein-12-UTP or cyanine-5-UTP, Enzo Life Sciences) was added into the reaction mixture. Following transcription, DNA was removed by treating with TURBO DNase I (Ambion) according to the manufacturer’s recommendations. RNA was recovered by sequential acid phenol/chloroform and chloroform extractions, followed by precipitation in isopropanol and NH4-OAc. RNA recovery was quantified via UV-Vis at 260 nm using a Nanodrop 2000 device (Thermo-Fischer). Proper RNA sizes were validated through either denaturing gel electrophoresis or Agilent TapeStation RNA ScreenTape analysis (performed by the BioFrontiers Next-Generation Sequencing Core Facility). Recovered RNA was resuspended in RNase-free water (Invitrogen) or TE and stored at −80°C. Working 100 μg/mL stocks were created as needed by diluting appropriately into RNase-free water and kept at −80°C.
Homopolymer condensation
Homopolymer RNAs were purchased as salts from Amersham Pharmacia Biotech, Inc. and Sigma-Aldrich and made into 25 mg/mL stock solutions in TE or RNase-free water, then kept at −80°C. Working 5 mg/mL stock solutions were prepared by diluting into RNase-free water. These working stocks were stored at −80°C or −20°C. The fluorescently-labeled oligonucleotides were purchased from IDT and resuspended, from which 2 μM stock solutions were made and stored at −80°C. An initial aliquot of the fluorescently-labeled PTR (_p_olypyrimidine _t_ract _R_NA) was purchased from IDT and was stored at −80°C as a 2 μM stock solution.
To form homopolymer condensates for the short RNA localization experiments, homopolymer RNA was condensed as described previously (Van Treeck et al., 2018), with the addition of 200 nM fluorescent RNA oligos in the condensation mix. Briefly, 500 μg/mL homopolymer RNA were denatured at 95°C for 2 min, cooled on ice for 30 s, then mixed with 750 mM NaCl, 10% w/v PEG 3350. To localize in vitro transcribed RNAs, homopolymer RNA was condensed with the transcribed RNA in the same manner as above, but instead mixed with 150 mM NaOAc, 600 mM NaCl, 1 mM MgCl2, and 10% w/v PEG 3350 (all in RNase-free water) to a final homopolymer concentration of 0.4 mg/mL and a final mRNA/lncRNA concentration of 10 μg/mL, yielding an approximate pH of 7 as assessed by pH paper.
Mechanical disruption of polyC/polyU networks
To mechanically disrupt polyC/polyU condensate networks, condensates were first prepared in glass bottom 96-well plates and incubated for an h. Thereafter, condensates were pipetted into adjacent empty wells, and pipetted up and down quickly five times. Afterward, they were immediately imaged.
Repeat RNA (reRNA) condensation
CUG repeat foci were prepared by condensing 200 μg/mL of fluorescein-labeled _DMPK_Exons 11–15-CUG590 RNA as in Jain and Vale (2017), in the presence or absence of 10 μg/mL Cy5-labeled in vitro transcribed RNAs or 200 nM PTR-Cy3. Briefly, RNAs were mixed together in a buffer of 10 mM MgCl2, 10 mM Tris HCl pH 6.8, and 25 mM NaCl, and subsequently denatured for 3 min at 95°C then cooled at ~4°C/min to 37°C in a thermocycler. Afterward, condensates were immediately imaged.
Stress granule isolation and RNA recruitment
To test the ability of SGs to recruit in vitro transcribed RNAs, SGs were purified as follows: First, GFP-G3BP1 U2OS cells were seeded at ~40% confluency and then grown to ~80% confluency, after which the media was replaced and the cells were stressed with 500 μM arsenite for 60 min. Afterward, the cells were washed, pelleted, and flash-frozen in liquid N2, then stored at −80°C for up to a week. SGs were isolated as in Khong et al. (2018) to produce SG-enriched fractions. Briefly, the cell pellet was thawed on ice and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.4], 100 mM KOAc, 2 mM MgOAc2, 0.5 mM DTT, 50 μg/mL heparin, complete mini EDTA protease inhibitor [Sigma-Aldrich 11836170001], 1 U/μl RNasin Plus RNase Inhibitor [Promega N2615]) after which cells were lysed by passing them through a 25G 5/8 needle several times. Afterward, cellular debris was removed by a 1000 x g spin (5 min), and SGs were enriched by two 18,000 x g spins (20 min each) followed by resuspension in lysis buffer. SG-enriched fractions were isolated through a final 850 x g spin (2 min). No affinity purification was performed. Other cellular debris was removed from the SG-enriched supernatant through both vigorous pipetting and by passing the supernatant through a 25G 5/8 needle, accompanied by quick spins. RNA recruitment to SGs was assessed by incubating 10 μg/mL Cy5-labeled luciferase RNA with SGs for 10 min at room temperature, followed by imaging.
RNA condensate dilution assays
To test the stability of RNA assemblies under dilution, _pgc_-decorated polyA droplets were prepared as above and incubated 2 h. Thereafter, they were subjected to spinning disk confocal time-lapse microscopy. Every 18 s, an image was taken in each channel in a 4×4 grid of adjacent frames using the Nikon Elements software “Large Image” tool. These images were automatically stitched together. Dilutions were performed while imaging was ongoing. In between imaging time points, TE buffer was quickly added drop-wise (so as to only minimally disturb assemblies) to a tenfold final dilution. The very next imaging time point was taken to be t = 0 for quantitative analysis.
RNA crosslinking assays
To observe RNA-condensate-associated RNA-RNA interactions, 4’- aminomethyltrioxsalen (AMT) UV crosslinking was performed (Frederiksen and Hearst, 1979). AMT was procured from Santa Cruz Biotechnology and solubilized to a final concentration of 1 mg/mL in DMSO. AMT stocks were stored at −20°C. Since AMT crosslinks pyrimidine residues, condensation-crosslinking experiments were performed using polyA condensates to avoid crosslinking the client RNA to the homopolymer scaffold.
In condensation-crosslinking experiments, RNA was demixed in the appropriate condition, but with the addition of 100 μg/mL AMT. Thus, droplets and gels were crosslinked in 600 mM NaCl, 150 mM NaOAc, 1 mM MgCl2, 10% PEG MW 3350, 0.1 mg/mL AMT. Crosslinking in solution conditions was performed in TE with 0.1 mg/mL AMT added. Following incubation, RNA was crosslinked by irradiating 15 min with 366 nm UV light. Afterward, samples were diluted 1:1.7 in TE and RNA was recovered by precipitation in 2 M LiCl. If RNA sample concentrations were low (e.g., gel conditions), 0.4 mg/mL polyA homopolymer RNA was added post-crosslinking and pre-precipitation as a carrier in order to promote efficient recovery.
Denaturing gel electrophoresis
Denaturing gel electrophoresis was performed to analyze fluorescent and crosslinked RNAs. RNA samples were mixed with either 6x formaldehyde loading dye (Ambion) or 2x formamide loading dye (95% deionized formamide, 0.025% w/v bromophenol blue, 5 mM EDTA) and denatured by incubation at 70°C for 15 min, followed by incubation on ice for 3 min. Samples were run in freshly-prepared 1% agarose, 1% formaldehyde denaturing gels in fresh 1x MOPS buffer (20 mM MOPS pH 7.0, 5 mM NaOAc, 1 mM EDTA) at 4–10 V/cm for 2.5–4 h.
Denaturing gels were imaged using a Typhoon FLA 9500 imaging system (GE) set to visualize the appropriate fluorescence signal from fluorescently-labeled RNAs. Crosslinking efficacies were quantified in ImageJ by measuring the integrated intensities of the monomer bands and higher-weight bands, subtracting the average of 10–15 background measurements, and dividing the sum of the corrected higher-weight integrated intensities by the sum of all band integrated intensities. In symbols:
where I n is the (background-corrected) integrated intensity of the band corresponding to the RNA _n_-mer, N is the highest order band observed in a given sample, and CE is the crosslinking efficacy.
Total RNA isolation and in vitro condensation inhibition assays
Saccharomyces cerevisiae strain BY4741 was grown to an OD600 of 0.8 AU and was lysed with 425–600 μm glass beads (Sigma-Aldrich) and vortexing for 10 min in a 50 mL conical tube in TRIzol (Ambion). RNA was extracted by using TRIzol reagent according to manufacturer’s instructions. Purified RNA concentration was determined via absorbance at 260 nm using a Nanodrop 2000 device (Thermo Scientific).
To assess the ability of eIF4A1 to decondense RNA in vitro, 10 μM recombinant eIF4A1 or protein storage buffer (200 mM NaCl, 25 mM Tris [pH 7.4], 10% glycerol, 1 mM DTT) was mixed together in with 150 μg/mL yeast total RNA extracts, SYTO RNASelect green fluorescent dye, and either 1 mM ATP or ADPNP (Sigma-Aldrich) in 150 mM KOAc, 1 mM MgCl2, 0.223 mM spermine tetrahydrochloride, 1.34 mM spermidine trihydrochloride, and 8% w/v PEG 3350 in a glass bottom 96-well plate at 37°C. These conditions yield an approximate pH of ~7.4. Polyamine levels recapitulate physiological concentrations (Van Treeck et al., 2018). Drug treatments were performed with DMSO, 10 μM hippuristanol, or 1 μM pateamine A. For each reaction, RNA condensate formation was observed after initial RNA addition by DIC and FITC channels over 30 min with images taken every 5 min. %Droplet area was analyzed using “Particle finder” in ImageJ.
Cell drug treatments and transfections
For drug treatments or transfections, cells were seeded at ~105 cells/mL and were allowed to adhere overnight. For siRNA experiments, cells were transfected with 50 nM of scrambled or eIF4A1 siRNA (see Table S1) using Interferin (polyPLUS) according to the manufacturer’s instructions for 48 h before SG induction. For overexpression experiments, cells were transfected with control (pcDNA3.1, Addgene) or pcDNA3.1-Myc-eIF4A1 (Addgene) plasmids for 48 h prior to SG induction or pmCherry (control) or pmCherry-DDX19A plasmids for DDX19A overexpression.
To induce SGs, arsenite (500 μM in H2O, Sigma-Aldrich), hippuristanol (Hipp, 300 nM or 1 μM in DMSO), or Pateamine A (PatA, 100 nM in DMSO) were added and cells were incubated at 37°C for the allotted times indicated in each assay. To deplete ATP, cells were incubated with 2-deoxy-D-glucose (2DG, 200 mM in H2O, Sigma-Aldrich) and carbonyl cyanide _m_-chlorophenyl hydrazine (CCCP, 100 μM in DMSO, Sigma-Aldrich). For ribopuromycinylation assays, cells were incubated at 37°C with puromycin (10 μg/mL in H2O, Sigma-Aldrich) 5 min prior to fixation or lysis.
Immunoblotting
Following drug treatment or transfection, cells were washed with 37°C phosphate buffered saline (PBS) and lysed with NP-40 lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40 and protease inhibitor cocktail (Thermo Scientific)). Cell lysates were rocked at 4°C for 30 min, and then clarified by centrifugation (13K RPM for 60 s). 4x Nu-PAGE sample buffer was added to lysates to a final concentration of 1x, samples were boiled for 5 min at 95°C, and then loaded into 4%–12% Bis-Tris Nu-PAGE gel and transferred to a nitrocellulose membrane. Membranes were blocked with 5% BSA in Tris-buffered saline with 0.1% Tween-20 (TBST) for an h and then incubated with primary antibody overnight at 4°C. Membranes were washed 3x with TBST, then incubated at room temperature for 2 h in 5% BSA in TBST. Membranes were washed 3x again in TBST and antibody detection was achieved by rocking membranes in Pierce ECL western blotting substrate for 5 min.
Chemiluminescence was visualized on an Image Quant LAS 4000 (GE). Protein band densities were quantified in ImageJ.
smFISH probe design, synthesis, and labeling
DNA smFISH probes targeting human MCM2 mRNA were prepared as follows. First, probe sequences were designed using the Stellaris RNA FISH Probe Designer software (Biosearch Technologies). Probe DNA sequences were synthesized by IDT, and the oligos were pooled. Pooled smFISH probes were then 3′ labeled by 5-Propargylamino-dideoxy-UTP-Atto633 (ddUTP-Atto633) as in Gaspar et al. (2017). Briefly, pooled probes were mixed with a threefold excess of ddUTP-Atto633, with the labeling reaction performed by terminal deoxynucleotidyl transferase (TdT).
Immunofluorescence (IF) and fluorescence in situ hybridization (FISH)
Cells were prepared as described as stated above except grown on No. 1.5 glass coverslips (Thermo). Sequential IF/FISH or IF/smFISH was performed as previously described (Khong et al., 2017, 2018). When IF was performed without FISH, fixed U2OS cells were instead blocked with 5% BSA in PBS for 1 h prior to IF and the antibodies were diluted in 5% BSA in PBS.
Alternatively, fixed cells were simultaneously blocked and permeabilized with 5% BSA in PBS-T (0.1% Triton X-100) for 1 h at room temperature. Thereafter, coverslips were incubated with primary antibody (1:500) overnight at 4°C in 1% BSA in PBS-T. Coverslips were then washed 3X with PBS and incubated with secondary antibody (1:1000) at room temperature for 2 h in 1% BSA in PBS-T.
The primary antibodies used for immunofluorescence (IF) include mouse anti-G3BP1 (5 μg/mL, ab56574(Abcam)), rabbit anti-DDX1 (1:100, 11357–1-AP (Proteintech)), and rabbit anti-eIF4A1 (100 μg/mL, ab31217(Abcam)) and the appropriate secondary antibodies used were goat anti-mouse FITC antibody (1:1000, Abcam (ab6785)), and donkey anti-rabbit Alexa Fluor 555 conjugate antibody (1:500, Abcam (ab150062).
NORAD, PEG3, and DYNH1C1 smFISH probes were custom made using Stellaris® RNA FISH probe Designer (Biosearch Technologies, Inc., Petaluma, CA). The probe sequences are listed in Table S1. _AHNAK_-Quasar670, _TFRC_-Quasar570, and _POLR2A_-Quasar570 probes were purchased premade directly from Biosearch Technologies and resuspended according to the manufacturer’s recommendations. AHNAK smFISH probe sequences are in Khong et al. (2017). Oligo(dT)-Cy3 probes were purchased from IDT.
Microscopy
Fixed U2OS cells stained by IF and/or smFISH, purified SGs, CUG repeat RNA foci, and homopolymer condensates were imaged using a widefield DeltaVision Elite microscope with a 100x NA 1.5 oil objective using a PCO Edge sCMOS camera and SoftWoRx software (GE).
Condensate dilution and helicase assays were imaged using an inverted Nikon Ti Eclipse spinning disk confocal microscope equipped with an environmental chamber and Nikon elements software. Helicase assay imaging was performed at 37°C. All spinning disk confocal images were acquired with a 100x NA 1.5 oil objective and a 2x Andor Ultra 888 EMCCD camera.
Fluorescence recovery after photobleaching (FRAP)
FRAP assays were performed using an inverted Nikon A1R laser scanning confocal microscope equipped with an environmental chamber, a 100x NA 1.5 oil objective, and Nikon Elements software. At least three images were acquired prior to photobleaching followed imaging over the course of recovery. PolyA/pgc FRAP was performed at room temperature.
To analyze recovery, the mean intensity of each bleached region was quantified in ImageJ, and recovery intensities were normalized to the mean of three pre-bleach measurements. Mobile fractions φ M were computed by subtracting the minimum normalized mean intensity _I_0 from the normalized endpoint intensity I F: φ M = I F − _I_0. OligoU recovery in polyA droplets was modeled by a single association exponential curve using Prism Graphpad software.
Partial FRAP was performed similarly, but by only photobleaching approximately half the condensate area.
QUANTIFICATION AND STATISTICAL ANALYSIS
Center, dispersion, and precision measures are defined and found in the figure legends.
Values for n and p, and the meanings of n values are found in the figure legends. Statistical analyses were performed using the Student’s t test in Prism 8.0 (Graphpad) or Excel (Microsoft).
Image analysis
Single molecule FISH analysis was performed using Imaris (Bitplane) as in (Khong et al., 2017, 2018; Khong and Parker, 2018). Briefly, processed Z stacks are opened and the nuclei masked using the DAPI channel. Thereafter, smFISH spots are identified in 3D using the “Spots” tool and the automatic parameters. SGs are similarly identified in 3D with the “Cells” tool, with 3D masks rendered. The number of smFISH spots in SGs is quantified as the number of spots in the 3D rendered SGs. Thus, the fraction of RNA molecules in SGs, the data presented, is this value divided by the total number of smFISH spots identified.
G3BP1, eIF4A1, and oligo(dT) partitioning were quantified using Imaris Imaging Analysis software (Bitplane). In order to measure partitioning, first, the nuclei were masked using the DAPI source channel. Second, individual cells were segmented by creating a region of interest (manually determined) around a single cell using the surface creation wizard. Mean cellular fluorescence intensities for G3BP1, eIF4A1, and oligo(dT) were then obtained under the results tab. Third, SGs were identified using the G3BP1 fluorescent source channel with Imaris Cell Creation wizard with the following parameters (0.0406 μm filter width, manually-determined thresholding, and ≥ 1 voxel). Once SGs were identified by the cell creation wizard, they are then converted to surfaces in order to extract mean fluorescence intensities for G3BP1, eIF4A1 and oligo(dT) inside stress granules. Partitioning was then calculated by taking the ratio of the mean fluorescence intensity in SG to the mean fluorescence intensity in the cell. 20 cells were counted for each sample. For more detailed information, please see Khong et al. (2018).
Line analysis was performed using ImageJ (FIJI). A straight line was drawn across a stress granule using G3BP1 as a source channel. The line is then saved as an ROI. The ROI was then pasted on the corresponding eIF4A1 or DDX1 source channel. Line intensities were then extracted in both channels using the “Plot Profile” tool. Stress granule boundaries (in each source channel) were determined by an intensity threshold halfway between the maximum and minimum intensity values for each source channel. Fifty linescan analyses were performed for each experiment.
Measurements of SG area or PB area per cell area were quantified using the “Particle Finder” tool in ImageJ. The percent of cells with SGs was quantified using the “Count” tool in ImageJ. The fluorescence intensity of puromycin-labeled nascent peptides in individual cells was quantified as mean gray values extracted from ImageJ.
SG and PB interface calculations were quantified by identifying regions of overlap between each mRNP granule and recording these events by using the “count” tool in ImageJ. Once total interfaces were counted, these values were normalized either to PB counts/area (determined using “analyze particles” in ImageJ) or SG counts/area (using “analyze particles” in ImageJ).
The index of dispersion (DI) is a statistical measure of inhomogeneity in a population and is defined to be
where σ2 is the variance of the population distribution (with σ being the population standard deviation) and μ is the mean value. For example, particles in an ideal solution exhibit a dispersion index of 1, as they are Poisson distributed, and substantially higher values indicate particle-particle interactions in that context. Here, dispersion indices were used to quantify inhomogeneity in fluorescence signal and infer the relative extent to which an RNA is assembled, similarly to Jain and Vale (2017).
Indices of dispersion were computed by measuring the mean fluorescence intensity and standard deviation for maximum projections of 1 μm Z stacks or single slices in the case of the dilution experiment. An index was computed for each image using ImageJ, and the displayed index was taken to be the average of the indices for all the separate images. The standard deviations for the distribution of index values for a given set of conditions were used to calculate 95% confidence intervals.
Estimates of eIF4A1 levels per mRNA
For HeLa and U2OS cells we used published values for eIF4A1 from quantitative mass spectroscopy, 1.7 × 107 and 2.2 × 106, respectively (Itzhak et al., 2016; Beck et al., 2011), and mRNA levels for U2OS cells, ~330,000 (Khong et al., 2017). Although previous estimates suggest HeLa cells might contain 200,000 mRNAs per cell (Shapiro et al., 2013) we utilized an estimate of 330,000 mRNAs per cell to be conservative in our estimations. These calculations indicate a ratio of 6.7 eIF4A1 molecules/mRNA to 51 eIF4A molecules/mRNA in U2OS and HeLa cells respectively.
Statistical analysis
Statistical analyses were performed using the Student’s t test in Prism 8.0 (Graphpad) or Excel (Microsoft).
DATA AND CODE AVAILABILITY
All data produced by this study are included in the manuscript or available from the authors upon request.
Supplementary Material
1
2
3
4
5
6
7
8
9
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-G3BP1 antibody | abcam | Cat# ab56574; RRID:AB_941699 |
| Rabbit polyclonal anti-PABPC1 antibody | abcam | Cat# ab21060; RRID:AB_777008 |
| Mouse monoclonal anti-PABPC1 antibody | abcam | Cat# ab6125; RRID:AB_2156878 |
| Rabbit polyclonal anti-eIF4A1 antibody | abcam | Cat# ab31217; RRID:AB_732122 |
| Rabbit polyclonal anti-eIF4G antibody | Santa Cruz | Cat# sc-11373; RRID:AB_2095750 |
| Mouse monoclonal anti-eIF4B antibody | Santa Cruz | Cat# sc-376062; RRID:AB_10988946 |
| Rabbit polyclonal anti-Tia1 antibody | abcam | Cat# ab40693; RRID:AB_2201438 |
| Rabbit polyclonal anti-EDC3 antibody | abcam | Cat#ab169817 |
| Mouse monoclonal anti-Myc antibody | Cell Signaling | Cat# 2278; RRID:AB_490778 |
| Mouse monoclonal anti-puromycin antibody | Millipore Sigma | Cat# MABE343; RRID:AB_2566826 |
| Rabbit polyclonal anti-DDX1 antibody | Proteintech | Cat# 11357-1-AP; RRID:AB_2092222 |
| Rabbit monoclonal anti-GAPDH antibody HRP conjugate | Cell Signaling | Cat# 3683; RRID:AB_1642205 |
| Goat anti-rabbit IgG 647 conjugate secondary antibody | abcam | Cat# ab150079; RRID:AB_2722623 |
| Goat anti-mouse IgG 488 conjugate secondary antibody | abcam | Cat# ab6785; RRID:AB_955241 |
| Donkey anti-rabbit IgG Alexa Fluor 555 conjugate secondary antibody | abcam | Cat# ab150062; RRID:AB_2801638 |
| Goat anti-mouse HRP conjugate antibody | Cell Signaling | Cat# 7076; RRID:AB_330924 |
| Goat anti-rabbit HRP conjugate antibody | Cell Signaling | Cat# 7074; RRID:AB_2099233 |
| Bacterial and Virus Strains | ||
| Quick-change XL1-Blue Supercompentent cells | Agilent technologies | Cat# 200519-4 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium (meta)arsenite | Sigma-Aldrich | Cat# S7400, CAS: 7784-46-5 |
| Pateamine A (PatA) | Low et al., 2005 | N/A |
| Hippuristanol (Hipp.) | Bordeleau et al., 2006a | N/A |
| Carbonyl cyanide m-chlorophenyl hydrazine (CCCP) | Sigma-Aldrich | Cat# C2759, CAS: 555-60-2 |
| RNasin Plus RNase inhibitor | Promega | Cat# N2615 |
| Complete mini EDTA-free protease inhibitor | Sigma-Aldrich | Cat# 11836170001 |
| Adenosine triphosphate (ATP), lithium salt | Millipore Sigma | Cat# 11140965001 |
| Adenylyl-imidodiphosphate (ADPNP), lithium salt | Millipore Sigma | Cat# 10102547001, CAS: 25612-73-1 (free acid) |
| Puromycin (Puro) | Sigma-Aldrich | Cat# P8833, CAS: 58-58-2 |
| Cycloheximide (CHX) | Sigma-Aldrich | Cat# C7698, CAS: 66-81-9 |
| 2-deoxy-D-glucose (2DG) | Sigma-Aldrich | Cat# D8375, CAS: 154-17-6 |
| Polyplus jetPRIME DNA and RNA transfection reagent | VWR | Cat# 89129-922 |
| Recombinant eIF4A1 | Bordeleau et al., 2006a | N/A |
| ZymoPURE plasmid mini-prep kit | ZYMO Research | Cat# D4210 |
| TRIzol Reagent | Ambion | Cat# 15596018 |
| 5-Propargylamino-ddUTP-Atto633 | Axxora | Cat# JBS-NU-1619-633 |
| Terminal Deoxynucleotidyl Transferase (TdT) | Thermo-Fisher | Cat# EP0161 |
| Critical Commercial Assays | ||
| MEGAscript T7 Transcription Kit | Thermo Fisher | Cat# AM1334 |
| Quick-change Site Directed Mutagenesis Kit | Agilent Technologies | Cat# 200523-5 |
| Experimental Models: Cell Lines | ||
| Human U-2 OS cells | Anderson Lab; Kedersha et al., 2016 | N/A |
| Human U-2 OS G3BP1-GFP cells | Taylor Lab; Figley et al., 2014 | N/A |
| Human U-2 OS ΔΔG3BP1/2 cells | Anderson Lab; Kedersha et al., 2016 | N/A |
| Experimental Models: Organisms/Strains | ||
| Saccharomyces cerevisiae strain BY4741 | Brachmann et al., 1998 | Dharmacon, Cat# YSC1049 |
| Oligonucleotides | ||
| Fluorescent RNA oligonucleotides, see Table S1 | IDT | N/A |
| Fluorescence in situ hybridization probes, see Tables S2 and S5 | IDT and Biosearch Technologies | N/A |
| si(eIF4A1) Sense: 5′-GCG AGC CAU UCU ACC UUG Utt-3′ | Ambion | AM16708 |
| si(eIF4A1) Antisense: 5′-ACA AGG UAG AAU GGC UCG Ctg-3′ | Ambion | AM16708 |
| Scrambled siRNA | Ambion | AM4611 |
| Homopolymer RNA potassium salts, see Table S3 | Van Treeck et al., 2018; Amersham Pharmacia Biotech and Sigma-Aldrich | N/A |
| Primers for eIF4A1 mutations, see Table S4 | IDT | N/A |
| Recombinant DNA | ||
| pcDNA3.1 | Gift of Ranum Lab | N/A |
| pcDNA3.1-Myc-eIF4A1 | Gift of Gideon Dreyfuss | Addgene plasmid# 71657 |
| pcDNA3.1-Myc-eIF4A1-E183Q | This paper | N/A |
| pcDNA3.1-Myc-eIF4A1-R362Q | This paper | N/A |
| pmCherry | Gift of Yaron Shav-Tal, Hochberg-Laufer et al., 2019 | N/A |
| pmCherry-DDX19A | Gift of Yaron Shav-Tal Lab Hochberg-Laufer et al., 2019 | N/A |
| pmCherry-DDX19A-DQAD | Gift of Yaron Shav-Tal, Hochberg-Laufer et al., 2019 | N/A |
| pmCherry-DDX19A-DN | Gift of Yaron Shav-Tal, Hochberg-Laufer et al., 2019 | N/A |
| pBS (SK-)-ccr4 | Drosophila Genomics Research Center | clone LD18435, stock # 4406 |
| pBS (SK-)-cycB | Drosophila Genomics Research Center | clone LD07875, stock # 1295561 |
| pFlc-1-nos | Drosophila Genomics Research Center | clone RE53469, stock # 1124533 |
| pFlc-1-pgc | Drosophila Genomics Research Center | clone RE14873, stock # 8512 |
| pcDNA3.1-CUG960 | Gift of Matt Disney | N/A |
| pm_Firre_3.1 | Gfit of John Rinn | N/A |
| Software and Algorithms | ||
| ImageJ (FIJI) | Schindelin et al., 2012; Rueden et al., 2017 | https://imagej.net/Fiji |
| Imaris image analysis software version 9.31.4 | Bitplane | https://imaris.oxinst.com/packages |
| Prism 8 for macOS | Graphpad | https://www.graphpad.com |
Highlights.
- RNA condensates promote intermolecular RNA-RNA interactions
- eIF4A limits the recruitment of RNAs to stress granules in cells
- eIF4A reduces the formation of stress granules in cells
- Recombinant eIF4A1 inhibits RNA condensation in vitro in an ATP-dependent manner
ACKNOWLEDGMENTS
We thank the Parker lab, Amy Buck, and Olke Uhlenbeck for discussions. We thank the University of Colorado BioFrontiers Institute Cell Culture Facility and the BioFrontiers Institute Advanced Light Microscopy Core Facility (NIST-CU cooperative agreement 70NANB15H226 and NIH 1S10RR026680-01A1), John Rinn, and Matt Disney for providing reagents. mCherry-DDX19A plasmids were a kind gift from Yaron Shav-Tal. This work was funded by NSF SCR training grant T32GM08759 (to D.T.), a Banting Postdoctoral Fellowship (to A.K.), NIH GM045443 (to R.P.), and the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Al-Husini N, Tomares DT, Bitar O, Childers WS, and Schrader JM (2018). α-Proteobacterial RNA Degradosomes Assemble Liquid-Liquid Phase-Separated RNP Bodies. Mol. Cell 71, 1027–1039.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N (2006). RNA granules. J. Cell Biol 172, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou AZ, and Klostermeier D (2013). The DEAD-box helicase eIF4A: paradigm or the odd one out? RNA Biol 10, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, and Ivanov P (2017). Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci 130, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumiller WM Jr., Pir Cakmak F, Davis BW, and Keating CD (2016). RNA-Based Coacervates as a Model for Membraneless Organelles: Formation, Properties, and Interfacial Liposome Assembly. Langmuir 32, 10042–10053. [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, and Aebersold R (2011). The quantitative proteome of a human cell line. Mol. Syst. Biol 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, and Pelletier J (2005). Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. USA 102, 10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, and Pelletier J (2006a). Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol 2, 213–220. [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, and Pelletier J (2006b). RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol 13, 1287–1295. [DOI] [PubMed] [Google Scholar]
- Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, Hamon L, Curmi PA, Guerquin-Kern JL, Piétrement O, and Pastré D (2014). Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42, 8678–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, and Boeke JD (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Burke JM, Moon SL, Matheny T, and Parker R (2019). RNase L Reprograms Translation by Widespread mRNA Turnover Escaped by Antiviral mRNAs. Mol. Cell 75, 1203–1217.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, and Wysocka J (2015). RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, and Pelletier J (2016). Hippuristanol - A potent steroid inhibitor of eukaryotic initiation factor 4A. Translation (Austin) 4, e1137381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupníková K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, and Nagamine Y (2008). Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem 283, 35186–35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, and Dreyfuss G (1999). Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol 147, 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MC, Tippana R, Demeshkina NA, Murat P, Balasubramanian S, Myong S, and Ferré-D’Amaré AR (2018). Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 558, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Brittingham GP, Pfeffer S, Surovtsev IV, Pinglay S, Kennedy KJ, Schaffer M, Gutierrez JI, Sang D, Poterewicz G, et al. (2018). mTORC1 Controls Phase Separation and the Biophysical Properties of the Cytoplasm by Tuning Crowding. Cell 174, 338–349.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Dufu K, Lei H, and Reed R (2010). A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun 1, 97 10.1038/ncomms1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ (2001). Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci 26, 597–604. [DOI] [PubMed] [Google Scholar]
- Fay MM, Anderson PJ, and Ivanov P (2017). ALS/FTD-associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 21, 3573–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, and Brangwynne CP (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Koch I, Westhof E, and Nüsslein-Volhard C (1997). RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley MD, Bieri G, Kolaitis RM, Taylor JP, and Gitler AD (2014). Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J Neurosci 34, 8083–8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, and Lamond AI (2002). Paraspeckles: a novel nuclear domain. Curr. Biol 12, 13–25. [DOI] [PubMed] [Google Scholar]
- Fox AH, and Lamond AI (2010). Paraspeckles. Cold Spring Harbor Perspect Biol 2, a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiksen S, and Hearst JE (1979). Binding of 4′-aminomethyl 4,5′,8-tri-methyl psoralen to DNA, RNA and protein in HeLa cells and Drosophila cells. Biochim. Biophys. Acta 563, 343–355. [DOI] [PubMed] [Google Scholar]
- Gaspar I, Wippich F, and Ephrussi A (2017). Enzymatic production of single-molecule FISH and RNA capture probes. RNA 23, 1582–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, and Parker R (2011). The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 43, 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg-Laufer H, Schwed-Gross A, Neugebauer KM, and Shav-Tal Y (2019). Uncoupling of nucleo-cytoplasmic RNA export and localization during stress. Nucleic Acids Res. 47, 4778–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CA, Tran EJ, Noble KN, Alcázar-Román AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shay-Tal Y, Wente SR, and Cole CN (2011). The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev 25, 1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, and Weis K (2019). DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148. [DOI] [PubMed] [Google Scholar]
- Hubstenberger A, Noble SL, Cameron C, and Evans TC (2013). Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev. Cell 27, 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, et al. (2017). P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell 68, 144–157.e5. [DOI] [PubMed] [Google Scholar]
- Itzhak DN, Tyanova S, Cox J, and Borner GHH (2016). Global, quantitative and dynamic mapping of protein subcellular localization. eLife 5, e16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, and Anderson P (2019). Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb. Perspect. Biol 11, a032813 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, and Vale RD (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Brunel C, and Ephrussi A (2011). Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 17, 2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, and Russell R (2014). RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem 83, 697–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, and Pyle AM (2001). Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 29, 121–125. [DOI] [PubMed] [Google Scholar]
- Jarmoskaite I, Denny SK, Vaidyanathan PP, Becker WR, Andreasson JOL, Layton CJ, Kappel K, Shivashankar V, Sreenivasan R, Das R, et al. (2019). A Quantitative and Predictive Model for RNA Binding by Human Pumilio Proteins. Mol. Cell 74, 966–981.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Lei W, Yacono PW, Chen S, Gilks N, Golan DE, and Anderson P (2000). Dynamic shuttling of Tia-1 accompanies the recruitment of mRNA to stress granules. J. Cell Biol 151, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, and Anderson P (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol 169, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, and Anderson P (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol 212, 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, and Parker R (2018). mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol 217, 4124–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, and Parker R (2017). The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 68, 808–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Jain S, Matheny T, Wheeler JR, and Parker R (2018). Isolation of mammalian stress granule cores for RNA-Seq analysis. Methods 137, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, et al. (2018). mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Schmidt BF, Bruchez MP, and McManus CJ (2018). Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res 46, 3742–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, and Fuller-Pace FV (2013). Looking back on the birth of DEAD-box RNA helicases. Biochim. Biophys. Acta 1829, 750–755. [DOI] [PubMed] [Google Scholar]
- Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, Vogt E, Marintchev A, Tanaka J, Fagotto F, Altmann M, et al. (2008). Selective pharmacological targeting of a DEAD box RNA helicase. PLoS ONE 3, e1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Putnam A, and Jankowsky E (2008). ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. USA 105, 20209–20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, and Herschlag D (1998a). The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 37, 2180–2193. [DOI] [PubMed] [Google Scholar]
- Lorsch JR, and Herschlag D (1998b). The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry 37, 2194–2206. [DOI] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, and Liu JO (2005). Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20, 709–722. [DOI] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. (2016). RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 165, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan K, Zhang H, Akef A, Cui XA, Gueroussov S, Cenik C, Roth FP, and Palazzo AF (2013). RanBP2/Nup358 potentiates the translation of a subset of mRNAs encoding secretory proteins. PLoS Biol. 11, e1001545 10.1371/journal.pbio.1001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Bukau B, and Kampinga HH (2018). Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 69, 214–226. [DOI] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Khong A, Lyon K, Parker R, and Stasevich TJ (2019). Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol 21, 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugler CF, Hondele M, Heinrich S, Sachdev R, Vallotton P, Koek AY, Chan LY, and Weis K (2016). ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. eLife 5, e18746 10.7554/eLife.18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat P, Marsico G, Herdy B, Ghanbarian AT, Portella G, and Balasubramanian S (2018). RNA G-quadruplexes at upstream open reading frames cause DHX36- and DHX9-dependent translation of human mRNAs. Genome Biol. 19, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble KN, Tran EJ, Alcázar-Román AR, Hodge CA, Cole CN, and Wente SR (2011). The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev 25, 1065–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro A, Ohtsu T, Svitkin YV, Sonenberg N, and Nakamura Y (2003). RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA 9, 394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Méthot N, Svitkin Y, Merrick WC, and Sonenberg N (1994). Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13, 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016). Principles and Properties of Stress Granules. Trends Cell Biol. 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam AA, and Jankowsky E (2013). DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim. Biophys. Acta 1829, 884–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao BS, and Parker R (2017). Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 114, E9569–E9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Dhir S, Dhir A, Moghaddam AE, Sattentau Q, Meinhart A, and Proudfoot NJ (2018). RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol. Cell 70, 650–662.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr., Richter NJ, and Merrick WC (1999). Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem 274, 12236–12244. [DOI] [PubMed] [Google Scholar]
- Rogers GW Jr., Lima WF, and Merrick WC (2001). Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem 276, 12598–12608. [DOI] [PubMed] [Google Scholar]
- Rowlinson JS, and Widom B (1982). Molecular Theory of Capillarity (New York: Oxford University Press; ). [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific data. BMC Bioinformatics 18, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev R, Hondele M, Linsenmeier M, Vallotton P, Mugler CF, Arosio P, and Weis K (2019). Pat1 promotes processing body assembly by enhancing the phase separation of the DEAD-box ATPase Dhh1 and RNA. eLife 8, e41415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, and Spector DL (2004). Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15, 3876–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Juranek SA, Marks J, De Magis A, Kazemier HG, Hilbig D, Benhalevy D, Wang X, Hafner M, and Paeschke K (2019). DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nat. Commun 10, 2421 10.1038/s41467-019-10432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, and Linnarsson S (2013). Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet 14, 618–630. [DOI] [PubMed] [Google Scholar]
- Sheth U, and Parker R (2006). Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125, 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- Singh AK, Choudhury SR, De S, Zhang J, Kissane S, Dwivedi V, Ramanathan P, Petric M, Orsini L, Hebenstreit D, and Brogna S (2019). The RNA helicase UPF1 associates with mRNAs co-transcriptionally and is required for the release of mRNAs from gene loci. eLife 8, e41444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Atas E, Lindqvist LM, Sonenberg N, Pelletier J, and Meller A (2014). Single-molecule kinetics of the eukaryotic initiation factor 4AI upon RNA unwinding. Structure 22, 941–948. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, and Sonenberg N (2001). The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ohashi S, and Kobayashi S (2014). Roles of YB-1 under arsenite-induced stress: translational activation of HSP70 mRNA and control of the number of stress granules. Biochim. Biophys. Acta 1840, 985–992. [DOI] [PubMed] [Google Scholar]
- Tauber D, and Parker R (2019). 15-Deoxy-Δ12,14-prostaglandin J2 promotes phosphorylation of eukaryotic initiation factor 2a and activates the integrated stress response. J. Biol. Chem 294, 6344–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, and Tazi J (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol 160, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Trcek T, Grosch M, York A, Shroff H, Lionnet T, and Lehmann R (2015). Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun 6, 7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu YT, and Barrientos A (2015). The Human Mitochondrial DEAD-Box Protein DDX28 Resides in RNA Granules and Functions in Mitoribosome Assembly. Cell Rep. 10, 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, and Parker R (2018). Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell 174, 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, and Parker R (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. USA 115, 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Mito M, Kurosaka S, Takumi T, Tanegashima C, Chujo T, Yanaka K, Kingston RE, Hirose T, Bond C, et al. (2016). Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol 214, 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e11. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Burkhardt DH, Rouskin S, Li GW, Weissman JS, and Gross CA (2018). A Stress Response that Monitors and Regulates mRNA Structure Is Central to Cold Shock Adaptation. Mol. Cell 70, 274–286.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SB (1993). Macromolecular crowding effects on macromolecular interactions: some implications for genome structure and function. Biochim. Biophys. Acta 1216, 175–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7
8
9
Data Availability Statement
All data produced by this study are included in the manuscript or available from the authors upon request.