The prion-like domain of Fused in Sarcoma is phosphorylated by multiple kinases affecting liquid- and solid-phase transitions (original) (raw)
Abstract
Fused in Sarcoma (FUS) is a ubiquitously expressed protein that can phase-separate from nucleoplasm and cytoplasm into distinct liquid-droplet structures. It is predominantly nuclear and most of its functions are related to RNA and DNA metabolism. Excessive persistence of FUS within cytoplasmic phase-separated assemblies is implicated in the diseases amyotrophic lateral sclerosis and frontotemporal dementia. Phosphorylation of FUS’s prion-like domain (PrLD) by nuclear phosphatidylinositol 3-kinase-related kinase (PIKK)-family kinases following DNA damage was previously shown to alter FUS’s liquid-phase and solid-phase transitions in cell models and in vitro. However, proteomic data suggest that FUS’s PrLD is phosphorylated at numerous additional sites, and it is unknown if other non-PIKK and nonnuclear kinases might be influencing FUS’s phase transitions. Here we evaluate disease mutations and stress conditions that increase FUS accumulation into cytoplasmic phase-separated structures. We observed that cytoplasmic liquid-phase structures contain FUS phosphorylated at novel sites, which occurred independent of PIKK-family kinases. We engineered phosphomimetic substitutions within FUS’s PrLD and observed that mimicking a few phosphorylation sites strongly inhibited FUS solid-phase aggregation, while minimally altering liquid-phase condensation. These effects occurred independent of the exact location of the phosphomimetic substitutions, suggesting that modulation of PrLD phosphorylation may offer therapeutic strategies that are specific for solid-phase aggregation observed in disease.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are progressive neurodegenerative diseases with overlapping histopathological features (Ferrari et al., 2011; Karch et al., 2018). Subtypes of both diseases can be categorized by the specific proteins that accumulate in neuronal proteinaceous inclusions (Irwin et al., 2015; Saberi et al., 2015). A small percentage of ALS and FTD subtypes feature neuronal inclusions enriched for the fused in sarcoma (FUS) protein (Kwiatkowski et al., 2009; Vance et al., 2009; Snowden et al., 2011). The pathological causes and consequences of FUS aggregation are incompletely known, and there are no drugs that prevent FUS-linked neurodegeneration.
FUS is a ubiquitously expressed 526–amino acid predominantly nuclear protein that supports numerous DNA/RNA-related functions, including transcription, RNA transport, RNA splicing, and the DNA damage response (Zinszner et al., 1997; Yang et al., 1998, 2014; Tan et al., 2012; Mastrocola et al., 2013). FUS consists of an N-terminal low-complexity prion-like domain (PrLD), three RGG repeat motifs, a zinc finger, an RNA recognition motif, and a C-terminal nuclear localization signal (NLS). The PrLD is ∼160 amino acids and is named for its sequence similarity to yeast prion domains (Gitler and Shorter, 2011), which are typically intrinsically disordered and lack sequence complexity (i.e., they are abundant in a few polar residues, with very few charged or hydrophobic residues; Ross and Toombs, 2010). These domains (and similar so-called “prion-like” domains) can facilitate proteins to self-associate and undergo liquid- and/or solid-phase transitions (Franzmann and Alberti, 2019). Conversion into solid aggregates is usually considered a stochastic, pathological event (Wickner et al., 2011), whereas the liquid-phase transitions are considered integral to function (Shin and Brangwynne, 2017). FUS’s PrLD enables the protein to condense into liquid-droplet structures that are distinct from the bulk solvent in a process frequently described as liquid–liquid phase separation (LLPS; Burke et al., 2015; Lin et al., 2017). However, FUS’s PrLD can also form solid amyloid-like aggregates in vitro (Murray et al., 2017).
The condensation of macromolecules such as FUS into distinct liquid phases allows temporal and spatial control of specific cellular functions (Owen and Shewmaker, 2019). Examples of these liquid-phase structures include Cajal bodies, stress granules, and the nucleolus. These diverse condensates are thought to be partially stabilized by proteins with intrinsically disordered domains—such as FUS’s PrLD—that have the capacity to form numerous nonspecific, transient, multivalent interactions (Banani et al., 2017). However, an emerging hypothesis is that the high concentrations of these unstructured domains within liquid condensates can potentiate the formation of intractable pathological solid aggregates (March et al., 2016). In the case of FUS, persistent condensation and/or mislocalization may initiate its solid-phase aggregation along neuroanatomical pathways (Armstrong, 2017). Once in the solid phase, both gain-of-function and loss-of-function mechanisms may contribute to neuronal degeneration (Sharma et al., 2016; Ishigaki and Sobue, 2018). Disrupting the formation of FUS-enriched neuronal inclusions is a therapeutic strategy for FUS-specific subtypes of ALS and FTD.
Post-translational modifications (PTMs) can regulate LLPS (Owen and Shewmaker, 2019; Soding et al., 2020). Low-complexity sequences, such as FUS’s PrLD, are common to phase-separating proteins and are especially susceptible to enzymatic modification due to their relatively open conformational ensembles (Bah and Forman-Kay, 2016). We previously reported that FUS’s PrLD is highly posttranslationally modified under DNA-damaging conditions. Based on our and others’ work, 32 putative phosphorylation sites have been identified (Rhoads et al., 2018b). Twelve of these sites are serine or threonine followed by a glutamine (S/TQ), the consensus sequence for the phosphatidylinositol 3-kinase-related kinases (PIKKs). PIKKs, which include the kinases DNA-PK, ATM, and ATR, are activated during the DNA damage response and have been confirmed to phosphorylate FUS’s PrLD (Gardiner et al., 2008; Deng et al., 2014; Monahan et al., 2017; Rhoads et al., 2018a). Our findings demonstrated that phosphorylation or phosphomimetic substitutions of PIKK consensus sites inhibited FUS’s propensity to undergo solid-phase transitions into amyloid-like aggregates (Monahan et al., 2017); in vitro suppression of LLPS was also observed. However, most of FUS’s putative phosphorylation sites are not PIKK consensus sites, and it is unknown if these sites are actually phosphorylated in cells and if they alter FUS’s state transitions.
In addition to DNA-damaging conditions, FUS has also been shown to respond to oxidative and osmotic stress (Sama et al., 2013). With hyperosmolar stress, wild-type FUS extensively accumulates into cytoplasmic granules, whereas with oxidative stress, ALS-mutant FUS accumulates into stress granules (Vance et al., 2013). However, specific phosphorylation of FUS’s PrLD has not been characterized under these conditions. In this study, we produced phosphospecific antibodies to evaluate phosphorylation of PIKK and non-PIKK consensus sites within FUS’s PrLD following different types of stress in human cell models. We also evaluated ALS-mutant FUS to determine if there was an association between pathological cytoplasmic aggregation and FUS PrLD phosphorylation. We then determined if phosphomimetic substitutions within FUS’s PrLD could alter FUS phase transitions in a non-site-specific manner. Our results suggest that non-PIKK kinases act upon FUS’s PrLD and are capable of modifying FUS’s aggregation propensity. This suggests that pathological FUS aggregation in FTD and ALS could be ameliorated by altering posttranslational modifications.
RESULTS
The FUS prion-like domain is phosphorylated at multiple non-PIKK sites
We previously confirmed and evaluated phosphorylation at PIKK sites (S/TQ) within FUS’s PrLD following DNA damaging stress (Rhoads et al., 2018a). Our preliminary mass spectrometry experiments suggested that non-PIKK consensus sites within the PrLD were also being phosphorylated. We selected three potential non-PIKK sites for corroboration with custom polyclonal antibodies: S57, T71, and S96. Sites S57 and s96 were of interest because they are putative ALS-mutation sites (Rhoads et al., 2018b). Custom antibodies (α-pS57, α-pT71, and α-pS96) that are specific to phosphorylated FUS (pFUS) were produced in rabbits (ThermoFisher and Genscript). Specificity for the phosphorylated epitope, relative to the nonphosphorylated epitope, was confirmed by immunoblotting (Supplemental Figure S1A). Also, antibodies were confirmed not to cross-react with other phosphoepitopes (Supplemental Figure S1B).
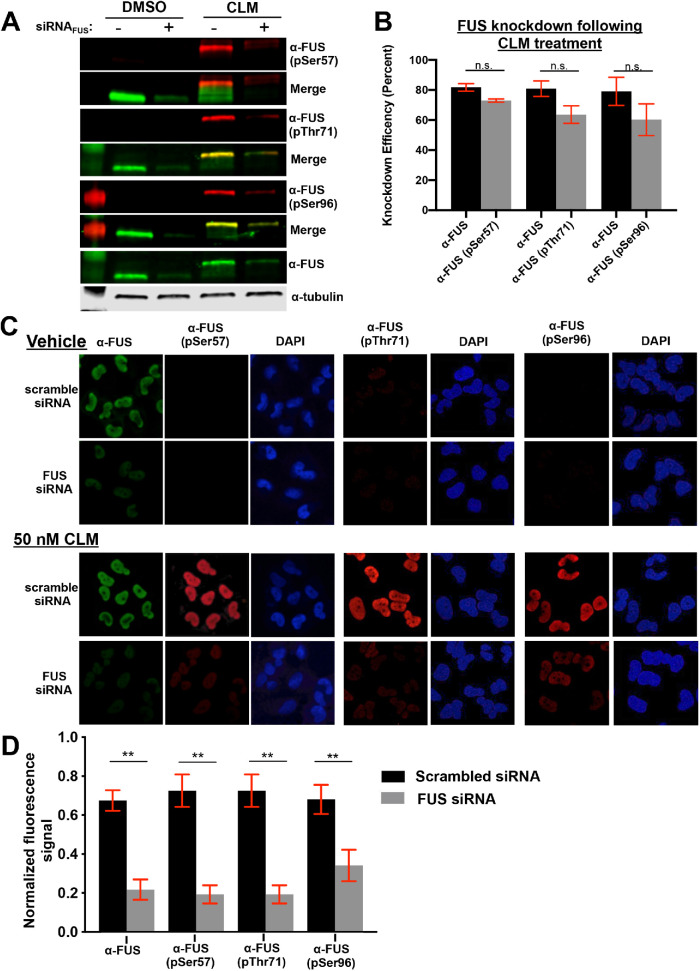
Antibody specificity to full-length FUS from human H4 neuroglioma cells was tested by Western blotting. H4 cells were treated with the DNA-damaging agent calicheamicin, as previously described (Rhoads et al., 2018a). The pFUS antibodies from treated cells, reacted with protein species that migrated similarly to species recognized by commercial FUS antibodies (Figure 1A). The custom antibodies gave no signal at the same position as FUS in the no-treatment control; however, nonspecific bands of different molecular weights were present (Supplemental Figure S1C). To ensure that the phosphospecific antibodies were identifying FUS species, knockdown experiments (siRNA) were performed. Quantification of Western blot signals revealed similar knockdown efficiency of FUS and pFUS signals using the same FUS-specific siRNA (Figure 1B). The custom antibodies were analyzed using immunofluorescence microscopy of H4 cells to verify their specificity to FUS, which is predominantly a nuclear protein. After calicheamicin treatment, the pFUS antibodies yielded strong nuclear signals (Figure 1C) that were eliminated, similarly to FUS signals, by FUS siRNA (Figure 1D).
FIGURE 1:
Phosphospecific antibodies recognize non-PIKK sites within FUS’s prion-like domain. (A) H4 cells with FUS knockdown by siRNA treated with DMSO or 50 nM calicheamicin (CLM) were analyzed by Western blots probed with commercially available FUS and custom phospho-FUS antibodies. (B) Quantification of percent reduction in Western blot band intensity of both FUS and phospho-FUS blots in A (n = 3). Raw data in S1A. (C, D) H4 neuroglioma cells treated with 50 nM CLM after FUS knockdown were fixed and probed with commercially available FUS and custom phosphospecific antibodies. Nuclear fluorescence signal was quantified and normalized to total fluorescence for each experiment. Figure data analyzed using Student’s t test (n = 3).
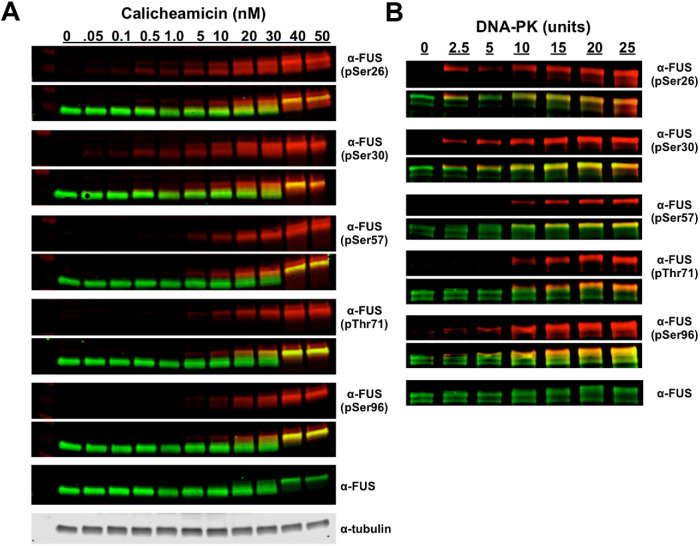
Our previous work found that low concentrations of calicheamicin (∼0.5 nM) induced PIKK-kinase phosphorylation of the FUS PrLD at two PIKK consensus sites: S26 and S30 (Rhoads et al., 2018a). When H4 cells were treated with a calicheamicin dose series, the α-pS57, α-pT71, and α-pS96 antibodies detected FUS species at and above ∼10 nM calicheamicin (Figure 2A). This phosphorylation was completely inhibited with a PIKK-kinase inhibitor (discussed further below; see Figure 4A). The phospho-bands detected by α-pS57, α-pT71, and α-pS96 overlapped with the higher–apparent molecular weight commercial α-FUS bands; this slower electrophoretic migration occurs when FUS is multiphosphorylated (Deng et al., 2014; Monahan et al., 2017; Figure 2A). The antibodies specific to PIKK consensus sites (pS26 and pS30) identify FUS before, during, and after its shift to higher molecular weight, which could indicate that phosphorylation occurs first at PIKK sites. The potential for PIKK kinases to phosphorylate these non-PIKK-consensus sites (S57, T71, and S96) in FUS’s PrLD were corroborated by in vitro assays using recombinant PIKK kinase (DNA-PK) and maltose binding protein–tagged FUS (MBP-FUS; Figure 2B).
FIGURE 2:
DNA damage induces multiphosphorylation of FUS’s prion-like domain at PIKK and non-PIKK sites. (A) H4 cells treated with a dose series of calicheamicin were analyzed by Western blot probed with anti-phospho-FUS (pSer26, pSer30, pSer57, pThr71, and pSer96) and anti-FUS antibodies. (B) Recombinant DNA-PK was used to phosphorylate MBP-FUS in vitro_._ Reaction was analyzed by Western blot and probed with commercial FUS and phosphospecific antibodies.
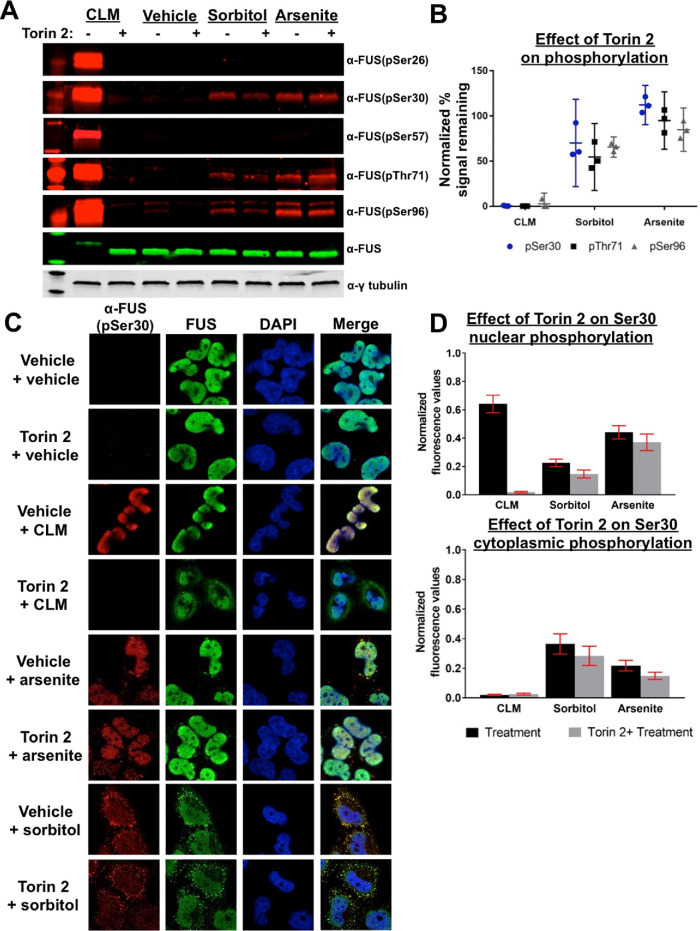
FIGURE 4:
Inhibition of PIKK-family kinases does not prevent phosphorylation of the FUS prion-like domain following osmotic or oxidative stress. (A) Phosphorylation status of FUS from H4 cells treated with or without torin 2 under varying stress conditions were analyzed by Western blot. (B) Quantification of band fluorescence normalized to total FUS; error bars represent 95% CI (n = 3). (C) Phosphorylation of FUS in H4 cells treated with or without Torin 2 under varying stress conditions. Fixed cells imaged using confocal microscopy. Cells were probed with FUS and phospho-FUS(pS30) antibodies. (D) Quantification of nuclear and cytoplasmic phospho-FUS(pS30); fluorescence error bars represent 95% CI (n = 3).
Phosphorylation of FUS occurs independent of PIKK-family kinases following osmotic and oxidative stress
Because each custom antibody was specific to its unique epitope in cross-reactivity assays (Supplemental Figure S1B), we concluded that the phosphorylation of FUS’s PrLD is not limited to the 12 S/TQ consensus sites, and other non-PIKK kinases may also act on this domain. However, because of the low sequence complexity of FUS’s PrLD, it could not be ruled out that phosphospecific antibodies cross-react to other PIKK-site phosphoepitopes nonspecifically. We therefore asked if other stress conditions that affect FUS cell biology would reveal distinct phosphorylation patterns that would be independent of PIKK kinase activity.
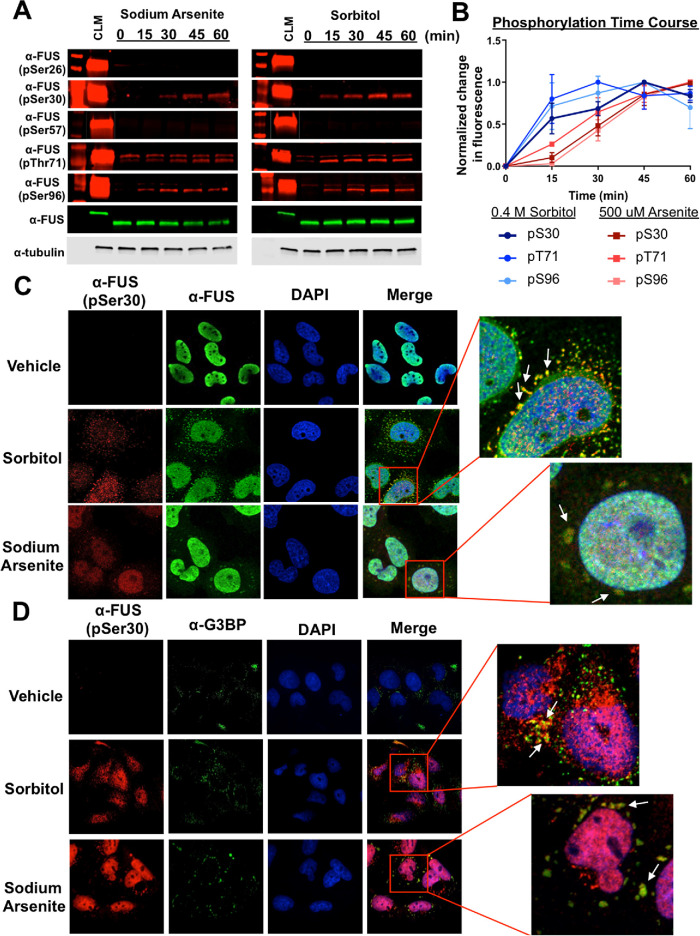
Previous work demonstrated that both sorbitol (osmotic stress) and sodium arsenite (oxidative stress) affect mutant or wild-type FUS subcellular localization (Andersson et al., 2008; Sama et al., 2013). We analyzed the phosphorylation status of FUS’s PrLD at PIKK and non-PIKK sites following treatment of H4 cells with these stressors (Figure 3A). Western blotting indicated that S30, T71, and S96 are phosphorylated following both treatments, whereas S26 and S57 are unchanged. For the α-pS30, α-pT71, and α-pS96 antibodies, higher– and lower–molecular weight bands were also evident following treatment, but FUS siRNA knockdown confirmed that these bands were not specific to FUS (Supplemental Figure S1D). Quantification of the Western blot signals showed an immediate increase in phosphorylation at S30, T71, and S96 within 15 min following sorbitol treatment (Figure 3B). In contrast, there was a gradual increase in phosphorylation following sodium arsenite treatment that did not plateau until ∼ 45 min (Figure 3B).
FIGURE 3:
FUS is phosphorylated at both PIKK and non-PIKK sites following non–DNA damaging stress. (A) H4 cells treated with 500 µM sodium arsenite or 0.4 M sorbitol at various time points were analyzed by Western blot using anti-phospho-FUS (pSer26, pSer30, pSer57, pThr71, and pSer96) and anti-FUS antibodies. The time courses show that only three of the five sites analyzed are phosphorylated by non–DNA damaging stress. (B) The normalized band signal intensities from A; 95% CI error bars (n = 3). (C) H4 cells treated with either sodium arsenite or sorbitol for 1 h were analyzed using confocal microscopy. Both FUS and phospho-FUS (pSer30—representative images) are found in cytoplasmic granules. (D) H4 cells treated with either sodium arsenite or sorbitol for 1 h were analyzed using confocal microscopy. Phospho-FUS (pSer30—representative images) colocalizes with stress granule marker G3BP.
FUS remains nuclear following PIKK-kinase phosphorylation (Rhoads et al., 2018a). However, because osmotic and oxidative stress can cause FUS to accumulate in cytoplasmic granules (Andersson et al., 2008; Sama et al., 2013), we asked if phospho-FUS species could be seen in these structures by immunofluorescence microscopy. H4 cells were treated with sodium arsenite or sorbitol as before and then immuno-stained with the custom FUS antibodies. Figure 3C shows representative images using commercial and α-pS30 FUS antibodies. Both pFUS and FUS colocalize in cytoplasmic inclusions. The α-pT71 and α-pS96 antibodies also stain cytoplasmic structures (Supplemental Figure S3) but are inconclusive due to cross-reactivity with other species (Supplemental Figure S1B). To determine if the pFUS-positive cytoplasmic granules were stress granules, immunostaining was performed with α-G3BP and α-TIA1 (Figure 3D; Supplemental Figure S4); both markers colocalized with phospho-FUS antibodies, suggesting that stress granules contain phosphorylated FUS.
In previous work, we showed that PIKKs phosphorylate the FUS PrLD following DNA damage, but phosphorylation could be eliminated with the addition of PIKK-specific inhibitors (Rhoads et al., 2018a). To determine if phosphorylation following osmotic and oxidative stress is facilitated by PIKK kinases, here we used the PIKK-specific inhibitor torin 2 (Udayakumar et al., 2016; Blackford and Jackson, 2017), which we confirmed to be a broadly acting PIKK kinase inhibitor (Supplemental Figure S2A). Torin 2 treatment completely abrogated DNA damage-induced (calicheamicin) phosphorylation at all PIKK and non-PIKK sites, but had only a small effect on osmotic stress-induced phosphorylation and essentially no effect on oxidative stress-induced phosphorylation (Figure 4, A and B).
When we pre-treated cells with torin 2, the morphology of FUS-positive granules did not change and they still reacted with the phosphoantibodies (Figure 4C). At the three sites analyzed, we see that torin 2 dramatically reduced the nuclear phospho-FUS signal when cells were treated with calicheamicin, but not with sorbitol or sodium arsenite (Figure 4D; Supplemental Figure S2B). Cytoplasmic phospho-FUS levels under all conditions are not significantly reduced by torin 2 (Figure 4D; Supplemental Figure S2B). The above data suggest that phosphorylated FUS is present in cytoplasmic granules and may be modified by an unidentified nuclear or cytoplasmic kinase(s).
The phosphorylated ALS mutant FUS is present in cytoplasmic inclusions
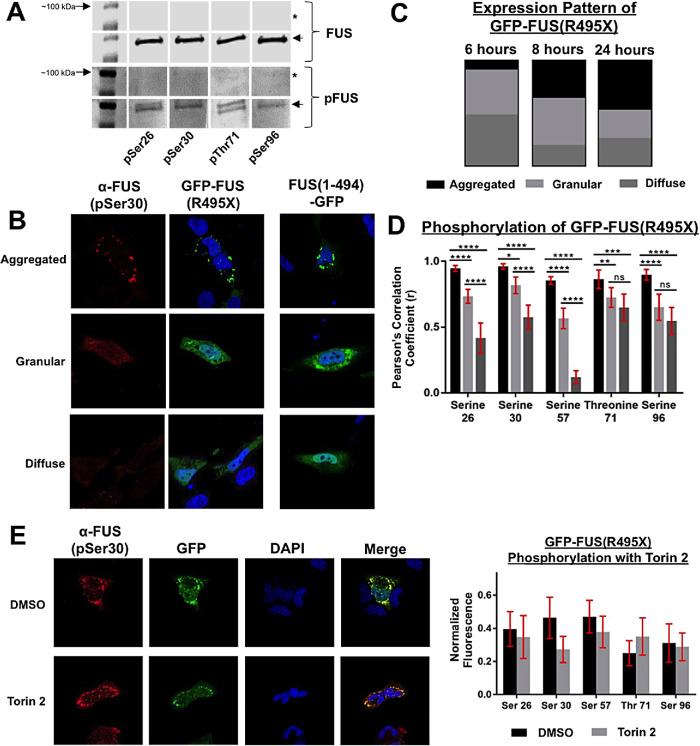
ALS-associated mutations that disrupt FUS’s C-terminal NLS cause FUS to accumulate in the cytoplasm, which is hypothesized to be part of the pathological mechanism causing FUS-linked ALS (Dormann et al., 2010). Because we observed that FUS’s PrLD is capable of being phosphorylated at non-PIKK sites and by non-PIKK kinases, we asked if mutant cytoplasm-confined FUS could be phosphorylated. We expressed an N-terminal GFP-tagged FUS containing an ALS-causing nonsense mutation that eliminated FUS’s NLS (FUS(R495X)) in H4 cells. Expression of FUS(R495X) in H4 cells, followed by immunoprecipitation and Western blotting with phosphospecific antibodies, indicated that mutant FUS’s PrLD was being phosphorylated in the cytoplasm (Figure 5A; Supplemental Figure S5A).
FIGURE 5:
Phosphorylated ALS-mutant FUS is present in cytoplasmic granules. (A) H4 cell transfected with GFP-FUS(R495X) for 24 h or untransfected control (*). GFP-FUS(R495X) was pulled down from cell lysates using GFP immunoprecipitation (IP). IP products were analyzed by Western blot and probed with anti-FUS (Santa Cruz) and phospho-FUS (pSer26, pSer30, pThr71, and pSer96). GFP-FUS(495X) is denoted with an arrow at roughly 100 kDa. (B) Ectopic expression of mutant FUS(R495X) in H4 cells with N- or C-terminal GFP. Anti-phospho-FUS(pSer30) antibody was used to probe for phosphorylated FUS. (C) Quantification of number of cells expressing diffuse, granular, or aggregated FUS(R495X) at 6, 8, or 24 h posttransfection (n = 3). (D) Quantification using Pearson’s coefficient of correlation of phospho-FUS (pSer26, pSer30, pSer57, pThr71, and pSer96) to the GFP-FUS(R495X) signal; error bars represent 95% CI (n = 30). (E) H4 cells transfected with GFP-FUS(495X) treated with torin 2, 6 h posttransfection. Cells were analyzed 8 h posttransfection and probed with phospho-FUS antibodies. Error bars represent 95% CI (n = 30).
Using immunofluorescence microscopy, we characterized cytoplasmic mutant FUS expression patterns. Expression of GFP-FUS(495X) for 6, 8, or 24 h yielded diffuse, granular, or aggregated cytoplasmic patterns (Figure 5, B and C). To confirm these results, we also used a C-terminal GFP-tagged FUS (FUS(1-494-GFP)). Both N-terminal and C-terminal GFP-FUS constructs showed similar cytoplasmic accumulation (Figure 5B). At 6 h posttransfection, the majority of mutant FUS was in a diffuse or granular state. By 24 h, the aggregated pattern was more prevalent. We assessed the phosphorylation of diffuse, granular, and aggregated FUS(R495X) at 24 h posttransfection and quantified phosphorylation at both PIKK and non-PIKK consensus sites ((Figure 5, A and D; Supplemental Figure S4). The phosphosignal had the highest correlation coefficient with the GFP-signal in the aggregated inclusion state, and decreased as the expression became more diffuse.
To determine if phosphorylation of mutant FUS’s PrLD is dependent on PIKK-family kinases, H4 cells were treated with torin 2 after 6 h of transfection—before aggregation was observed. The inhibition of PIKK-family kinases did not significantly affect mutant FUS phosphorylation at any residues analyzed (Figure 5E; Supplemental Figure S5). These data indicate that ALS-mutant FUS can be phosphorylated by cytoplasmic kinases and phospho-FUS is enriched within cytoplasmic foci.
Mutant FUS—with or without phosphomimetic substitutions at non-PIKK sites—forms cytoplasmic inclusions with liquid-like properties
In our previous work, we found that phosphomimetic substitutions (S/T→E) of all 12 PIKK consensus sites within the PrLD dramatically decreased cytoplasmic aggregation of ALS-mutant FUS(495X) in cell culture (Monahan et al., 2017). We asked if phosphomimetic substitution (S/T→E) at four non-PIKK consensus sites (4Ev3 and 4Ev4; diagrammed in FUS toxicity and aggregation can be altered by non-PIKK phosphomimetic substitution in Figure 7A) would alter cytoplasmic inclusion formation of mutant FUS(1-494-GFP) in H4 cells. For controls, we used constructs with alanine or guanine substituted at all the PIKK sites (12A or 12E), as well as a construct with no substitutions (0E). We analyzed the expression patterns 24 h posttransfection using live-cell imaging (Figure 6A).
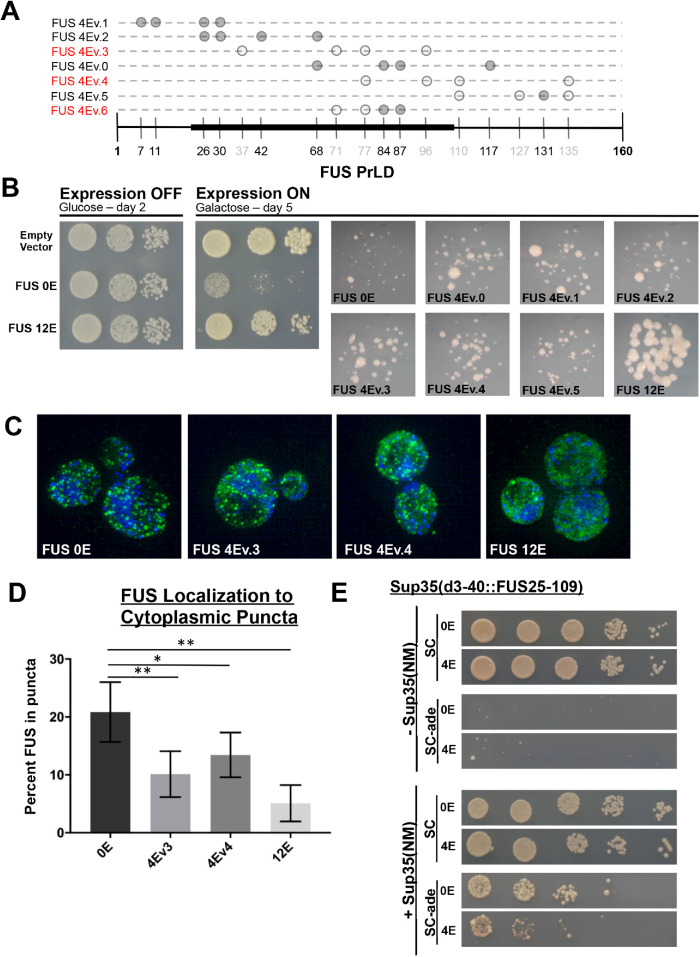
FIGURE 7:
Non-PIKK phosphomimetic substitution decreases FUS toxicity and prion-like aggregation in yeast. (A) Schematic of the various phosphomimetic constructs used in the lab. Solid gray circles indicate PIKK consensus sites and light gray circles indicate non-PIKK sites. The constructs are in red to indicate their use in subsequent experiments. The black-highlighted axis indicates the FUS fragment inserted into Sup35 and used in Panel E. (B) Phosphomimetic substitution in the prion-like domain rescues FUS toxicity in yeast. (C) Ectopic expression of FUS 4Ev3 and 4Ev4 analyzed by structured illumination microscopy. Cells were probed with anti-FUS. (D) Quantification of FUS signal in punctate structures compared with total FUS expression; error bars represent 95% CI. Figure data analyzed using a Student _t_-test (n = 9) (E) Sup35-FUS or Sup35-FUS 4E were expressed in yeast on SC or SC-ade media. Sup35NM was added to promote prion formation under both conditions.
FIGURE 6:
ALS-mutant FUS forms cytoplasmic droplets; phosphomimetic substitutions in the prion-like domain do not alter droplet dynamics. (A) Representative images of FUS(494)-GFP phosphomimetic constructs 24 h posttransfection. (B) Average number of large (>1 m2) or small (<1 m2) FUS(494)-GFP cytoplasmic granules per cell 24 h posttransfection; error bars represent SEM (n = 17). (C) FRAP half-times of FUS(494)-GFP 24 h posttransfection; error bars represent 95% CI (n = 30). Student’s t test was used for statistical analysis.
Expression of FUS(1-494)-GFP (0E) resulted in the formation of numerous cytoplasmic inclusions with different morphologies, from smooth to amorphous (Figure 6A). The 12A construct produced amorphous, relatively small puncta throughout the cytoplasm of cells. The expression of the 12E variant had the most profound effect, with noticeably more soluble protein. The 4E phosphomimetic constructs resulted in patterns like those in the 0E control. We counted the number of granules in cells transfected with each construct (Figure 6B). There was a significant decrease in the number of granules present in cells containing the 12E construct from all other constructs, as previously observed (Monahan et al., 2017). However, the 4E constructs appeared much like 0E.
Different variants of mutant FUS have previously been shown to form condensates with liquid-like characteristics (Niaki et al., 2020). We asked if the FUS(1-494) inclusions would have liquid-like behavior and if phosphomimetic substitutions would alter their dynamics. We used fluorescence recovery after photobleaching (FRAP) to assess the dynamics of mutant FUS cytoplasmic inclusions (Alberti et al., 2019). All FUS constructs had similar mobile fractions and half-times of recovery, suggesting similar dynamics (Figure 6C). Each construct also displayed liquid-like characteristics with recovery on the time scale of seconds. We also performed FRAP with cells treated with sodium arsenite, since it is reported to drive cytoplasmic FUS into granules; the fluorescence recovery times were similar for all variants (Supplemental Figure S6) These data suggest that a few phosphomimetic substitutions in the PrLD do not dramatically alter FUS’s LLPS in cells, which could be because a percentage of FUS is typically phosphorylated at sites within the PrLD when contained within liquid-like cytoplasmic inclusions.
FUS toxicity and aggregation can be altered by non-PIKK phosphomimetic substitution
Our previous work in yeast models revealed a link between human FUS’s cytoplasmic aggregation and toxicity when ectopically expressed (Kryndushkin et al., 2011; Monahan et al., 2017, 2018). FUS expressed in yeast models displays detergent resistance and dye-binding properties more typical of solid-phase aggregates (Fushimi et al., 2011; Kryndushkin et al., 2011). In yeast, we previously found that phosphomimetic substitutions (S/T→E) of the PrLD’s 12 PIKK consensus sites reduced FUS’s aggregation propensity and caused a concomitant reduction in its toxicity (Monahan et al., 2017). However, it was not known if the PIKK consensus sites were special, or if other potential phosphorylation sites within the PrLD could likewise inhibit aggregation and toxicity.
We compared the effect of PIKK versus non-PIKK consensus site phosphomimetic substitution using five full-length FUS variants that each had four unique S/T→E substitutions (Figure 7A). The constructs contained substitutions at PIKK consensus sites (4Ev0, v1, v2), non-PIKK consensus sites (4Ev3, v4), or both (4Ev5). The different combinations of 4E substitutions were expressed at similar levels and significantly decreased toxicity regardless of the specific substitutions being PIKK or non-PIKK consensus sites (Figure 7B; Supplemental Figure S7, A and B). Substitutions that overlapped a core region (aa 39–95), previously defined by solid-state NMR as being important for solid-phase aggregation (Murray et al., 2017), appeared to have a slightly greater suppression of toxicity, although not statistically significant (Supplemental Figure S7C). We used structured illumination microscopy of yeast cells expressing GFP-tagged FUS (4Ev3 and 4Ev4) and found that phosphomimetic substitutions at non-PIKK sites could cause FUS to have a more diffuse expression pattern than WT FUS (Figure 7, C and D), which is consistent with previous results in which substitutions were made at all the PIKK sites (Monahan et al., 2017).
Phosphomimetic substitutions in the core of FUS’s PrLD inhibit prion-like behavior in a yeast model
FUS’s PrLD is compositionally similar to the naturally occurring prion proteins that form self-propagating toxic amyloid in yeast (Kryndushkin et al., 2011; McGlinchey et al., 2011). Human disease proteins that resemble yeast prion proteins have previously been evaluated using quantitative prion scoring methods that exploit well-characterized prion assays. With these methods, human prion-like sequences are substituted for segments within the yeast prion protein Sup35 (Kim et al., 2013). The sequences can then be scored for how well they support prion-like aggregation, which is phenotypically reported by growth on media lacking adenine (essentially, the prion-like aggregation of the fusion protein results in its loss of function; Tuite et al., 2015).
We replaced the nucleating portion of the Sup35 prion domain (aa 3–40) with a section of the FUS PrLD (aa 25–109). We also engineered a variant (4Ev6; Figure 7A) with four phosphomimetic substitutions within the region that appeared to protect best against FUS toxicity. The WT and 4Ev6 fusions with Sup35 were both able to complement a sup35Δ strain (Figure 7E, top two rows). Both fusions were able to perform normal translational termination by not permitting read-through of the ade-1 gene with an internal nonsense codon (nongrowth on media lacking adenine; Figure 7E, rows 3 and 4). Transient expression of Sup35NM, which is a nonfunctional amino-terminal fragment of Sup35, can promote prion formation of the full-length protein (Ter-Avanesyan et al., 1994). After Sup35NM was transiently overexpressed, the FUS-Sup35 fusions gained the ability to grow on media lacking adenine. However, induction of growth for 4Ev6 was reduced by ∼1 log relative to the nonmimetic fusion. This suggests that phosphorylation within a few sites (either PIKK or non-PIKK) of FUS’s PrLD may be able to inhibit prion-like aggregation.
Non-PIKK phosphomimetic substitutions inhibit FUS solid-phase aggregation in vitro
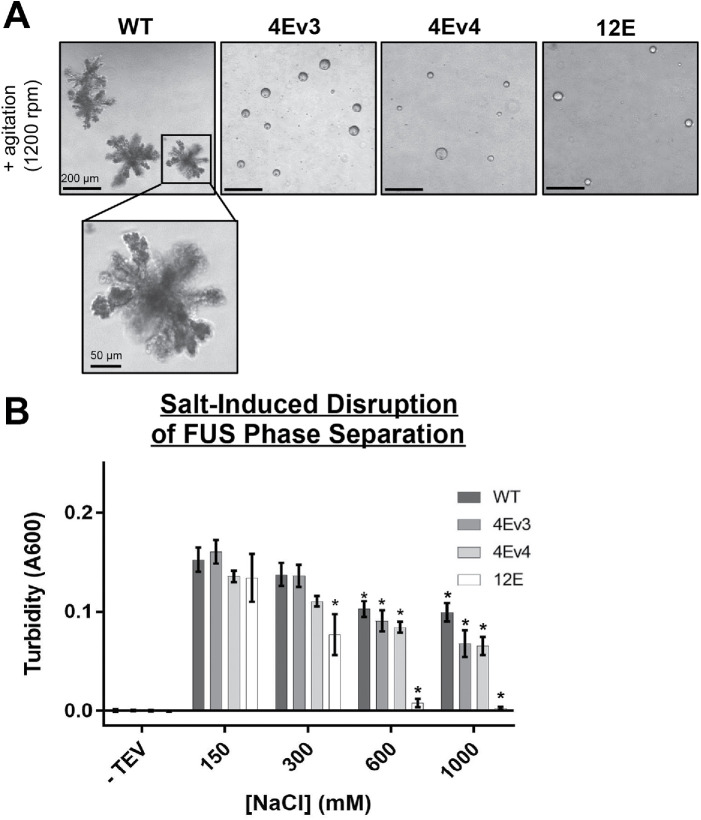
We and others previously showed that recombinant FUS can undergo liquid- and solid-phase transitions into dynamic droplet or solid aggregate structures in vitro (Patel et al., 2015; Monahan et al., 2017). Wild-type full-length FUS will undergo LLPS and form liquid droplets observable by differential interference contrast (DIC) microscopy, but after prolonged , solid amorphous aggregates form (Monahan et al., 2017). We created two FUS-MBP phosphomimetic constructs with the same 4 non-PIKK S/T→E substitutions described above (Figure 7A; 4Ev3 and 4Ev4). We examined phase separation of these two non-PIKK phosphomimetic constructs using DIC. We also included a control with phosphomimetic substitutions at all 12 PIKK sites (12E), which were previously found to be sufficient to inhibit the formation of solid aggregates when FUS was subjected to overnight agitation. We found that the FUS constructs that contained four non-PIKK phosphomimetic substitutions in the PrLD were able to form droplet structures, but failed to form solid aggregates with prolonged agitation (Figure 8A; Supplemental Figure S8A). Amorphous aggregates of FUS start to form within 6 h of agitation, while the droplet structures formed by the phosphomimetic variants persist up to 48 h with agitation (Supplemental Figure S8A). To ensure that these droplets were in a liquid-phase separated state, we treated the agitated proteins with 1,6-hexanediol, a chemical probe used to disrupt weak hydrophobic interactions (Kroschwald et al., 2017). Wild-type full-length FUS solid aggregates were unaffected by 1,6-hexanediol treatment, while the phosphomimetic FUS droplets dissolved upon exposure (Supplemental Figure S8B).Using turbidity as a reporter for LLPS, we also found that high salt could significantly suppress phase separation for both FUS-4Ev3 and FUS-4Ev4 when compared with the baseline condition (150 mM NaCl; Figure 8B), which is similar to results observed for phosphomimetic substitutions at 6 or 12 PIKK consensus sites (Monahan et al., 2017). These results suggest that non-PIKK kinases that act upon a few sites within the PrLD could have dramatic effects on FUS’s phase separation, especially in preventing irreversible solid aggregate formation.
FIGURE 8:
Phosphomimetic substitution reduces FUS solid-phase aggregation in vitro. (A) Differential interference microscopy of full-length and phosphomimetic variants of FUS (4Ev3, 4Ev4, or 12E). Maltose binding protein (MBP)–tagged FUS proteins were agitated for 1 day at 25°C after the addition of TEV protease. (B) Turbidity assay of full-length FUS in the presence of varying salt concentrations. Turbidity was assessed 45 min following TEV addition (n = 10). Two-way ANOVA was used for statistical analysis (*indicates significance relative to 150 mM NaCl).
DISCUSSION
Post-translational modifications of FUS have been proposed to regulate its inclusion and function in membraneless organelles (Owen and Shewmaker, 2019). Aberrant formation of membraneless organelles has also been linked to FUS-associated neurodegenerative disease (Shin and Brangwynne, 2017). To better understand the link between PTMs and FUS phase transitions, we analyzed both PIKK and non-PIKK consensus site phosphorylation of wild-type FUS’s PrLD under cellular stress conditions and ectopically expressed mutant FUS. Our data suggest that the FUS PrLD is differentially phosphorylated depending on the stress conditions within the cell. We evaluated how phosphomimetic substitutions at PIKK and non-PIKK sites affected FUS’s phase separation and aggregation. Our data suggest that phosphorylation at non-PIKK sites can have similar inhibitory effects on phase separation and aggregation as previously observed with PIKK sites (Monahan et al., 2017); however, phosphomimetic substitutions in FUS’s PrLD appear to be particularly detrimental to solid-phase transitions.
Previous mass spectrometry analysis of FUS indicated 32 putative phosphorylation sites in the PrLD of FUS (Rhoads et al., 2018a). We and others previously confirmed phosphorylation of three PIKK consensus sites by PIKK-family kinases following DNA damage (Gardiner et al., 2008; Deng et al., 2014; Rhoads et al., 2018a). In this study, we provide data showing that three non-PIKK consensus sites (S57, T71, and S96) are phosphorylated following different types of cellular stress. The inhibition of PIKK-family kinases did not prevent phosphorylation of FUS’s PrLD following osmotic or oxidative stress, suggesting that other kinases may regulate this domain. The NetPhos algorithm predicts that protein kinase C (PKC) is the most likely candidate kinase for sites S57 and S96, and cyclin-dependent kinase 5 (cdk5) is the most likely for T71. Our preliminary experiments using pharmacological inhibitors of these kinases were inconclusive. We also evaluated ALS-mutant FUS that was confined to the cytoplasm due to defects in its NLS. Mutant cytoplasmic FUS was phosphorylated by one or more presumably cytoplasmic kinase(s) that were not affected by PIKK inhibition.
We also observed that PIKK-family kinases have the potential to phosphorylate non-S/TQ consensus sites within the PrLD following DNA damage, which is consistent with bioinformatic data on PIKK substrates that indicate that noncanonical phosphorylation is not uncommon (kinaseNET). However, non-PIKK sites appeared to be phosphorylated subsequent to PIKK sites. In conclusion, we observed differential phosphorylation of PIKK and non-PIKK consensus sites depending on the type and extent of stress response elicited from mutations, DNA damage, oxidative stress, or osmotic stress. This suggests that FUS’s phosphoproteoform can change according to the specific function it is performing or the stress it responding to.
Previous work shows that the cytoplasmic FUS phase separates into stress granules. Here we found that under stress conditions that caused FUS accumulation into cytoplasmic granules, phospho-FUS was detectable in G3BP-positive stress granules. The rates of phosphorylation observed by Western blotting were stress-dependent and correlated with the dynamics previously reported for stress granule formation under different stress conditions (Wheeler et al., 2016). These data suggest a role for site-specific phosphorylation of FUS that could be important for its function or localization within stress granules (both into or out of). However, the temporal relationship between phosphorylation and granule dynamics requires further characterization.
A recent study showed that numerous mutant FUS constructs can separate into droplets in vitro (Niaki et al., 2020). The liquid-like dynamics of FUS varied depending on the type or location of the mutation. Here, we corroborated these findings and observed that NLS-mutant FUS (FUS(R495X)) can liquid-phase separate in cells. We biophysically characterized mutant FUS and determined that it is in a liquid-like state in cytoplasmic granules. When four phosphomimetic substitutions were introduced into the PrLD, no differences in FUS dynamics within granules were observed by FRAP. The dynamics was similar regardless of where the phosphomimetic substitutions occurred. When 12 substitutions were introduced, the effect was largely observed as more diffuse FUS, but FRAP dynamics of cytoplasmic granules remained largely the same. Likewise, mutant FUS-positive cytoplasmic granules did not have altered FRAP dynamics even when cells were treated with sodium arsenite to induce PrLD phosphorylation. Thus, FUS dynamics within these structures appears not to be greatly altered by PrLD phosphorylation. This may be because cytoplasmic liquid-state granules are complex heterogeneous structures, in which FUS might be a minority species and subject to many other overriding interactions.
Phosphorylation of FUS’s PrLD may be more critical for preventing pathological solid-state transitions. A current hypothesis suggests a link between membraneless organelle dynamics and formation of toxic cytoplasmic inclusion in neurodegenerative disorders like ALS (Wolozin and Ivanov, 2019). The high concentrations of proteins like FUS within condensates may potentiate molecular interactions that lead to solid/irreversible aggregate formation (Shin and Brangwynne, 2017). Our in vitro data with recombinant FUS suggest that phosphorylation of the PrLD can have profound inhibiting effects on solid-phase transition while minimally affecting LLPS. Also, these effects are nonspecific; phosphomimetic substitutions had similar effects regardless of their exact locations within the PrLD. In yeast, we observed similarly that four phosphomimetic substitutions were enough to suppress prion/amyloid-like aggregation and proteotoxicity. Toombs et al. (2010) previously discovered that the amino acid composition of prion domains can be more important for amyloid-like solid-phase aggregation than the specific order of the amino acids; charged groups are especially unfavorable. Likewise, the parallel in-register amyloid model of FUS PrLD proposed by Murray et al. (2017) would be strongly disfavored by the introduction of charged groups. Our phosphomimetic data are consistent with these findings, suggesting that substitution at specific PIKK sites is not required to have a general anti-aggregation effect on the solid phase.
In the case of NLS-deficient mutant FUS (FUS(R495X)), which localizes to the cytoplasm in either a diffuse, granular, or aggregated state, we observed that punctate FUS appears to have greater PrLD phosphorylation. A possibility is that the environment of some liquid phase–separated environments favors more phosphorylation (Rai et al., 2018), which could have protective effects against solid-phase aggregation. Hyperphosphorylation of some neurodegenerative-associated proteins within pathological inclusions could be the marks of failed solubilization mechanisms, rather than promoters of aggregation (Li et al., 2011; Hergesheimer et al., 2019).
While FUS’s PrLD has 12 PIKK consensus and numerous other putative phosphorylation sites, in our observations, the FUS proteoform generally consists of protein, with only a few phosphorylation events following most stress. This is evident because when FUS is highly phosphorylated it visibly migrates more slowly by Western blot (Deng et al., 2014; Monahan et al., 2017). We observed that a few phosphomimetic substitutions could have dramatic effects on solid-state aggregation, regardless of their exact position in the PrLD. Phosphomimetic substitutions are not perfect substitutes for phosphoserine and phosphothreonine, and are more subtle than the addition of phosphate groups to amino acids, so possibly the phosphomimetic effects understate the inhibitory effects of phosphorylation. Tilting the balance toward slightly greater phosphorylation of the FUS proteoform in vivo could be a therapeutic strategy for ALS and FTD subtypes. Further research into the kinase and phosphatase regulation of FUS is required.
METHODS
Cell culture/transfections/FUS knockdowns
H4 neuroglioma (ATCC HTB-148) cells were cultured in DMEM (Sigma D6429) supplemented with 10% fetal bovine serum (Sigma F6178) and 1% penicillin–streptomycin (Corning 30-002-Cl). Cells were lysed with a modified RIPA buffer (200 mM NaCl, 100 mM Tris-HCl pH 8, 0.5% sodium deoxycholate, 1% Triton X-100, 670 mM phenylmethylsulfonyl fluoride, 1250 units of benzonase nuclease [Sigma E8263], 150 μl protease inhibitor cocktail [Thermo 1861278], and 100 μl phosphatase inhibitor [Thermo 78426]) for 30 min on ice.
DNA was transfected into H4 cells at ∼70–80% confluency using Lipofectamine 2000 (Thermo 11668027) and OptiMEM (Life Technologies 31985070) at a ratio of 3 μg DNA to 2.5 μl Lipofectamine 2000 and incubated at 37°C for 24 h unless otherwise stated. FUS knockdowns were done with Lipofectamine RNAiMAX (Thermo 13778075) and 30 pmol siRNA (FUS [Thermo 4392420] and negative control [Thermo 4390843]) for 48 h at 37°C.
H4 cells were treated with the following reagents for 1 h at 37°C unless otherwise stated: 15 nM or 50 nM calicheamicin-γ (generous gift from Pfizer), 0.4 M sorbitol (Sigma S1876), 500 μM sodium arsenite (Chem Cruz 301816), and 200 nM torin 2 (Selleckchem S2817).
Cloning/Plasmids
WT FUS, FUS 12E, and GFP-FUS 495X plasmids were obtained from Monahan et al. (2017). Plasmids were generated either through PCR cloning (Thermo F531S) of genes into appropriate parent vectors at multiple cloning sites or through site-directed mutagenesis (New England Biolabs E0552S), with the exception of FUS 12A, which was synthesized by GenScript (Piscataway, NJ). Plasmids used for protein purification were derived from 6xHis-MBP-FUS (source Addgene 98651); plasmids used for immunofluorescence and live-cell imaging were derived from C1-eGFP (source Addgene 54759); plasmids used for yeast toxicity were derived from pH317 (2μ LEU2 PGAL; source Shewmaker lab); plasmids used for yeast SIM imaging were derived from pH316 (CEN LEU2 PGAL; source Shewmaker lab).
Custom antibodies and peptide specificity
Production of Serine 26 and Serine 30 phosphoantibodies was previously described in Rhoads et al. (2018a) by GenScript (Piscataway, NJ). Serine 57, Threonine 71, and Serine 96 were produced similarly using synthetic peptides FUS 51-63 GQSSYS(p-S)YGQSQN, FUS 65-79 GYGTQS{p-T}PQGYGSTC, and FUS 91-105 YGQQS{p-S}YPGYGQQPC as immunogens for antibody production in rabbits, respectively. T71 and S96 peptide synthesis and antibody production was performed by Genscript, while S57 production was performed by ThermoFisher (Lafayette, CO).
Specificity of phosphoantibodies was verified through FUS knockdowns and peptide dilutions. FUS knockdowns were analyzed through Western blot and immunocytochemistry procedures. Nitrocellulose membranes (BioRad 1620112) were saturated with 8 M urea before being loaded on the Hybri-Slot Manifold blotting apparatus (BRL 1052MM). Unmodified and phosphorylated FUS peptides were blotted and probed with respective antibodies following the Western blotting protocol.
Immunocytochemistry and FRAP
For fixed cell imaging, cells were grown on glass coverslips for 24 h before any treatments and fixation with 4% paraformaldehyde (Sigma P6148). The cells were permeabilized with -20°C methanol and blocked with 5% normal goat serum (Abcam ab7481) with 0.05% sodium azide (Life Technologies 50062Z). The following antibodies were used to probe the fixed cells: FUS antibodies (Abcam ab154141, custom rabbit phospho-FUS antibodies), G3BP (BD 61112), and TIA1 (Santa Cruz 166247). Secondaries used to detect primary antibodies were AlexaFluors AF488 and AF568 (ThermoFisher A-11001, A-11011). Nuclei were stained using the Mounting media with DAPI (ThermoFisher P36931). Slides were imaged using Zeiss 700 and the Nikon A1R. Immunofluorescence quantification was done using the raw integral density in Fiji (Schindelin et al., 2012). Values were normalized to the highest intensity in the data set. Mutant FUS(495X) phosphorylation was quantified using Pearson’s correlation coefficient to the GFP signal. FUS(494)-GFP granule area and number was quantified using particle analysis on ImageJ.
For live-cell imaging, cells were grown in glass-bottomed microwell dishes 24 h before transfection. The cells were incubated at 37°C for either 12 or 24 h posttransfection. After either time point, the medium was changed to dye-free DMEM (Thermo #21063029). The FRAP data were collected on the Nikon A1R. The center of a granule, marked by a 0.3-μm region of interest, was bleached at 100% power for 1.9 s. The recovery was analyzed for 98 s (∼1.6 min). The recovery was quantified using the time series analyzer V3 plugin on Fiji. The bleached pixel intensity was subtracted from each data point and then data points were normalized to the pixel intensity before the bleaching occurred.
Western blotting
Lysates were mixed with 4× NuPAGE LDS Sample Buffer (Thermo NP0008) and run through AnyKD precast gels (BioRad 4569034) at 80 V for 2 h. Gels were transferred through either a Trans-Blot Turbo Transfer System (Bio-Rad 1704150) or eBlot L1 (GenScript L00686) onto nitrocellulose membranes (BioRad 1620112). Membranes were blocked with 6% milk (BioRad 1706404) in Tris-buffered saline (TBS). Primary and secondary antibodies were diluted in TBS with 0.1% Tween-20 (Sigma P7949). The following primary antibodies were used to probe the blots: FUS antibodies (Santa Cruz 373698, Abcam ab154141, Bethyl A300-293A, custom rabbit phospho-FUS antibodies), gamma tubulin (Sigma T6557), and GFP (Roche 11814460001). Primary antibodies were detected with secondaries conjugated to IRDye fluorescent probes (LI-COR 926-68021, 926-32210). Blots were imaged with the Odyssey CLx imaging system (LI-COR). Band densitometry quantification was done using Image Studio software (Li-COR). Phosphobands were normalized to endogenous FUS band intensity.
In vitro DNA-PK phosphorylation (2B)
A plasmid encoding for 6xHis-MBP-FUS (Addgene 98651) was transformed into NiCo(DE3)-competent Escherichia coli (New England BioLabs C2529H). Cell pellets were collected from 1-l cultures induced with 0.5 mM IPTG after continued growth at 37°C for 4 h. Pellets were sonicated in 20 mM NaPi pH 7, 1 M NaCl, and 10 mM imidazole with a protease inhibitor tablet (Roche 11836170001) and spun down at 20,000 × g for 1 h at 4°C. The supernatant was filtered through 0.8 μm surfactant-free cellulose acetate syringe filters (Corning 431221) before being run through a nickel column (GE 17-5286-01). The column was washed with 20 mM NaPi pH 7, 1 M NaCl, and 30 mM imidazole before elution with 20 mM NaPi pH 7, 1 M NaCl, and 300 mM imidazole. The eluate was collected and spun down with 50-kDa centrifuge filters (Millipore UFC505024) and stored in 75% 20 mM NaPi pH 7, 1 M NaCl, and 25% glycerol.
In vitro DNA-PK reactions were carried out using a DNA-PK kinase enzyme system (Promega V4106) in a reaction following the manufacturer’s protocol containing 5 μg recombinant MBP-FUS, 200 μM ATP, and varying doses of DNA-PK. The reactions were incubated at room temperature for 1 h before being prepped for Western blotting.
Phase separation and turbidity
A plasmid encoding for 6xHis-MBP-FUS (Addgene 98651) and phosphomimetic derivatives were transformed into BL21(DE3)-competent E. coli (New England BioLabs C2529H). Cell pellets were collected from 1-l cultures induced with 0.5 mM IPTG after continued growth at 37°C for 4 h. Pellets were sonicated in 20 mM Tris-HCl, 1 M NaCl, and 1 mM EDTA, pH 7.4, with a protease inhibitor tablet (Roche 11836170001) and spun down at 20,000 × g for 1 h at 4°C. The supernatant was filtered through 0.8 μm surfactant-free cellulose acetate syringe filters (Corning 431221) before being run through an MBPTrap column (GE 17-5286-01). The column was washed with 20 mM Tris-HCl, 1 M NaCl, and 1 mM EDTA, pH 7.4 before elution with 10 mM maltose in the buffer. The eluate was collected and spun down with 50-kDa centrifuge filters (Millipore UFC505024) and stored with 25% glycerol.
Phase separation samples were prepared in 20 mM Tris HCl and 150 mM NaCl, pH 7.4 with 5 µM of MBP-FUS and 5 units of ProTEV Plus (Promega 20200703) and agitated overnight at 1200 rpm at 25°C. Samples were aliquoted onto glass slides and imaged through differential interference microscopy (Olympus IX73). Phase-separated, agitated protein samples were subjected to 10% 1,6-hexanediol (Sigma 629118). Protein samples were aliquoted into a glass microscopy dish and 10% 1,6-hexanediol (final concentration) was added. Images were taken using differential interference microscopy (Leica DMi1) before treatment and at 5, 10, and 30 min during treatment. Turbidity measurements were made with 5 µM MBP-FUS in 20 mM Tris HCl and varying concentrations of NaCl ranging from 150 mM to 1 M. Samples were incubated with 10 units of ProTEV Plus (Promega) for 45 min at 30°C, and absorbance at 600 nm was measured using a BioTek Cytation 5 imaging reader.
Yeast
The yeast strain BY4741 (his3D1 leu2D0 met15D0 ura3D0 PIN+) was used for toxicity and aggregation assays. For the toxicity assay, single colonies were grown overnight at 30°C and dilution spotted on glucose and galactose plates. For structured illumination microscopy, overnight cultures were induced with galactose for 8 h at 30°C before being fixed with 4% PFA and washed with 0.1 M KPO4 and 0.1 M KPO4/1.2 M sorbitol buffer. The cells were treated with 100T zymolyase (Zymo Research E1005) to form spheroplasts. Glass slides were treated with 0.1% poly-D-lysine and the spheroplasts settled on the slide before being permeabilized with -20°C methanol. Slides were blocked with PBS-BSA and probed with rabbit anti-FUS polyclonal antibody (Bethyl 300-293A). Alexa Fluor 488 Conjugate #4412 secondary was used to detect the FUS antibody before being mounted with Prolong mounting media with DAPI (ThermoFisher P36931) and covered with a glass coverslip. Samples were imaged using the Zeiss Elyra PS.1 with three rotated gratings.
Yeast Sup35 knockout strain (kar1-1, SWQ5, ade2-1, his3, leu2, trp1, ura3, sup35::KanMx) was complemented by Sup35 or Sup35-FUS fusion plasmids. A galactose-inducible Sup35NM plasmid was transformed into the yeast strains and induced for 16 h at 30 C before being plated on synthetic complete media and media lacking adenine.
Supplementary Material
Acknowledgments
We thank Zoey Barnette for assistance with Western blotting, Matt Stinson for help with purifying protein, Jeremy Smyth for sharing his FRAP expertise, and Dennis McDaniel for assistance with confocal microscopy. We are grateful to Nick Fawzi for many helpful discussions. This work was possible due to support from NIH awards R35GM119790 and R01GM118530.
Abbreviations used:
FUS
Fused in Sarcoma
LLPS
liquid–liquid phase separation
PIKK
phosphatidylinositol 3-kinase-related kinase
PrLD
prion-like domain
PTM
posttranslational modification.
Footnotes
REFERENCES
- Alberti S, Gladfelter A, Mittag T (2019). Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell , 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P (2008). The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol , 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong R (2017). Neuronal cytoplasmic inclusions in tau, TDP-43, and FUS molecular subtypes of frontotemporal lobar degeneration share similar spatial patterns. Folia Neuropathol , 185–192. [DOI] [PubMed] [Google Scholar]
- Ashley AK, Kemp CJ (2018). DNA-PK, ATM, and ATR: PIKKing on p53. Cell Cycle , 275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A, Forman-Kay JD (2016). Modulation of intrinsically disordered protein function by post-translational modifications. J Biol Chem , 6696–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol , 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, Jackson SP (2017). ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell , 801–817. [DOI] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL (2015). Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell , 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, Ito D, Murray ME, Dickson DW, Seyfried NT, Kukar T (2014). FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J Neurosci , 7802–7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, et al. (2010). ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J , 2841–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Kapogiannis D, Huey ED, Momeni P (2011). FTD and ALS: a tale of two diseases. Curr Alzheimer Res , 273–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann TM, Alberti S (2019). Prion-like low-complexity sequences: key regulators of protein solubility and phase behavior. J Biol Chem , 7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Long C, Jayaram N, Chen X, Li L, Wu JY (2011). Expression of human FUS/TLS in yeast leads to protein aggregation and cytotoxicity, recapitulating key features of FUS proteinopathy. Protein Cell , 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner M, Toth R, Vandermoere F, Morrice NA, Rouse J (2008). Identification and characterization of FUS/TLS as a new target of ATM. Biochem J , 297–307. [DOI] [PubMed] [Google Scholar]
- Gitler AD, Shorter J (2011). RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion , 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergesheimer RC, Chami AA, de Assis DR, Vourc’h P, Andres CR, Corcia P, Lanznaster D, Blasco H (2019). The debated toxic role of aggregated TDP-43 in amyotrophic lateral sclerosis: a resolution in sight? Brain , 1176–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DJ, Cairns NJ, Grossman M, McMillan CT, Lee EB, Van Deerlin VM, Lee VM, Trojanowski JQ (2015). Frontotemporal lobar degeneration: defining phenotypic diversity through personalized medicine. Acta Neuropathol , 469–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki S, Sobue G (2018). Importance of functional loss of FUS in FTLD/ALS. Front Mol Biosci , 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Wen N, Fan CC, Yokoyama JS, Kouri N, Ross OA, Hoglinger G, Muller U, Ferrari R, Hardy J, _et al._International Frontotemporal Dementia -Genomics Consortium, I.C.f.F.D.P.S.P.G.C., International Parkinson’s Disease Genomics, C. (2018). Selective genetic overlap between amyotrophic lateral sclerosis and diseases of the frontotemporal dementia spectrum. J Am Med Assoc Neurol , 860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature , 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Alberti S (2017). Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. _Matters_1–7. [Google Scholar]
- Kryndushkin D, Wickner RB, Shewmaker F (2011). FUS/TLS forms cytoplasmic aggregates, inhibits cell growth and interacts with TDP-43 in a yeast model of amyotrophic lateral sclerosis. Protein Cell , 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. (2009). Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science , 1205–1208. [DOI] [PubMed] [Google Scholar]
- Li HY, Yeh PA, Chiu HC, Tang CY, Tu BP (2011). Hyperphosphorylation as a defense mechanism to reduce TDP-43 aggregation. PLoS One , e23075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Currie SL, Rosen MK (2017). Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem , 19110–19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March ZM, King OD, Shorter J (2016). Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res , 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS (2013). The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem , 24731–24741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, Kryndushkin D, Wickner RB (2011). Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci USA , 5337–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan ZT, Rhoads SN, Yee DS, Shewmaker FP (2018). Yeast models of prion-like proteins that cause amyotrophic lateral sclerosis reveal pathogenic mechanisms. Front Mol Neurosci , 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng W, et al. (2017). Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J , 2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R (2017). Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell , 615–627 e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaki AG, Sarkar J, Cai X, Rhine K, Vidaurre V, Guy B, Hurst M, Lee JC, Koh HR, Guo L, et al. (2020). Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-linked FUS mutations. Mol Cell , 82–94 e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen I, Shewmaker F (2019). The Role of post-translational modifications in the phase transitions of intrinsically disordered proteins. Int J Mol Sci . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell , 1066–1077. [DOI] [PubMed] [Google Scholar]
- Rai AK, Chen JX, Selbach M, Pelkmans L (2018). Kinase-controlled phase transition of membraneless organelles in mitosis. Nature , 211–216. [DOI] [PubMed] [Google Scholar]
- Rhoads SN, Monahan ZT, Yee DS, Leung AY, Newcombe CG, O’Meally RN, Cole RN, Shewmaker FP (2018a). The prionlike domain of FUS is multiphosphorylated following DNA damage without altering nuclear localization. Mol Biol Cell , 1786–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads SN, Monahan ZT, Yee DS, Shewmaker FP (2018b). The role of post-translational modifications on prion-like aggregation and liquid-phase separation of FUS. Int J Mol Sci . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Toombs JA (2010). The effects of amino acid composition on yeast prion formation and prion domain interactions. Prion , 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi S, Stauffer JE, Schulte DJ, Ravits J (2015). Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol Clin , 855–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sama RR, Ward CL, Kaushansky LJ, Lemay N, Ishigaki S, Urano F, Bosco DA (2013). FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J Cell Physiol , 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Lyashchenko AK, Lu L, Nasrabady SE, Elmaleh M, Mendelsohn M, Nemes A, Tapia JC, Mentis GZ, Shneider NA (2016). ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat Commun , 10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science . [DOI] [PubMed] [Google Scholar]
- Snowden JS, Hu Q, Rollinson S, Halliwell N, Robinson A, Davidson YS, Momeni P, Baborie A, Griffiths TD, Jaros E, et al. (2011). The most common type of FTLD-FUS (aFTLD-U) is associated with a distinct clinical form of frontotemporal dementia but is not related to mutations in the FUS gene. Acta Neuropathol , 99–110. [DOI] [PubMed] [Google Scholar]
- Soding J, Zwicker D, Sohrabi-Jahromi S, Boehning M, Kirschbaum J (2020). Mechanisms for active regulation of biomolecular condensates. Trends Cell Biol , 4–14. [DOI] [PubMed] [Google Scholar]
- Tan AY, Riley TR, Coady T, Bussemaker HJ, Manley JL (2012). TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc Natl Acad Sci USA , 6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN (1994). The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics , 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, McCarty BR, Ross ED (2010). Compositional determinants of prion formation in yeast. Mol Cell Biol , 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite MF, Staniforth GL, Cox BS (2015). [PSI(+)] turns 50. Prion , 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayakumar D, Pandita RK, Horikoshi N, Liu Y, Liu Q, Wong KK, Hunt CR, Gray NS, Minna JD, Pandita TK, Westover KD (2016). Torin2 suppresses ionizing radiation-induced DNA damage repair. Radiat Res , 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. (2009). Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science , 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Scotter EL, Nishimura AL, Troakes C, Mitchell JC, Kathe C, Urwin H, Manser C, Miller CC, Hortobagyi T, et al. (2013). ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum Mol Genet , 2676–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R (2016). Distinct stages in stress granule assembly and disassembly. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman D, Kelly AC, Gorkovskiy A (2011). The yeast prions [PSI+] and [URE3] are molecular degenerative diseases. Prion , 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B, Ivanov P (2019). Stress granules and neurodegeneration. Nat Rev Neurosci , 649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Embree LJ, Tsai S, Hickstein DD (1998). Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem , 27761–27764. [DOI] [PubMed] [Google Scholar]
- Yang L, Gal J, Chen J, Zhu H (2014). Self-assembled FUS binds active chromatin and regulates gene transcription. Proc Natl Acad Sci USA , 17809–17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Sok J, Immanuel D, Yin Y, Ron D (1997). TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci (Pt 15), 1741–1750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.