The RecA Protein as a Model Molecule for Molecular Systematic Studies of Bacteria: Comparison of Trees of RecAs and 16S rRNAs from the Same Species (original) (raw)
. Author manuscript; available in PMC: 2011 Oct 6.
Published in final edited form as: J Mol Evol. 1995 Dec;41(6):1105–1123. doi: 10.1007/BF00173192
Abstract
The evolution of the RecA protein was analyzed using molecular phylogenetic techniques. Phylogenetic trees of all currently available complete RecA proteins were inferred using multiple maximum parsimony and distance matrix methods. Comparison and analysis of the trees reveal that the inferred relationships among these proteins are highly robust. The RecA trees show consistent subdivisions corresponding to many of the major bacterial groups found in trees of other molecules including the α, β, γ, δ, and ε Proteobacteria, cyanobacteria, high-GC gram-positives, and the Deinococcus-Thermus group. However, there are interesting differences between the RecA trees and these other trees. For example, in all the RecA trees the proteins from gram-positives species are not monophyletic. In addition, the RecAs of the cyanobacteria consistently group with the RecAs of the high-GC gram-positives. To evaluate possible causes and implications of these and other differences, phylogenetic trees were generated for small-subunit rRNA sequences from the same (or closely related) species as represented in the RecA analysis. The trees of the two molecules using these equivalent species-sets are highly congruent and have similar resolving power for close, medium, and deep branches in the history of bacteria. The implications of the particular similarities and differences between the trees are discussed. Some of the features that make RecA useful for molecular systematics and for studies of protein evolution are also discussed.
INTRODUCTION
Molecular systematics has become the primary way to determine evolutionary relationships among microorganisms because morphological and other phenotypic characters are either absent or change too rapidly to be useful for phylogenetic inference (Woese 1987). Not all molecules are equally useful for molecular systematic studies and the molecule of choice for most such studies of microorganisms has been the small-subunit of the rRNA (SS-rRNA). Comparisons of SS-rRNA sequences have revolutionized the understanding of the diversity and phylogenetic relationships of all organisms, and in particular those of microorganisms (Fox et al. 1980, Olsen 1988, Olsen et al. 1994, Pace et al. 1986, Sogin 1989, Woese 1991, Woese 1987). Some of the reasons that SS-rRNA sequence comparisons have been so useful include: SS-rRNAs are present in, and have conserved sequence, structure, and function among, all known species of free-living organisms as well as mitochondria and chloroplasts (Pace et al. 1986, Woese 1987); genes encoding SS-rRNAs are relatively easy to clone and sequence even from uncharacterized or unculturable species (Eisen et al. 1992, Lane et al. 1985, Medlin et al. 1988, Olsen et al. 1986, Weisburg et al. 1991); the conservation of some regions of primary structure and large sections of secondary structure aids alignment of SS-rRNA sequences between species (Woese 1987); the evolutionary substitution rate between species varies greatly within the molecule allowing for this one molecule to be used to infer relationships among both close and distant relatives (Pace et al. 1986, Woese 1987); and it is generally considered unlikely that SS-rRNA genes have undergone lateral transfers between species (Pace et al. 1986), thus the history of SS-rRNA genes should correspond to the history of the species from which they come. The accumulating database of SS-rRNA sequences, which now includes over 3000 complete or nearly complete sequences (Maidak et al. 1994), provides an extra incentive to focus on this molecule.
Despite the advantages and successes of using SS-rRNA sequences to determine microbial phylogenetic relationships, there are potential problems with relying on only SS-rRNA-based phylogenetic trees (e.g., Hasegawa and Hashimoto 1993, Rothschild et al. 1986). First, there are some characteristics of SS-rRNA genes that may lead to trees based on them being inaccurate including: over-estimation of the relatedness of species with similar nucleotide frequencies (such as could occur in unrelated thermophiles) (Embley et al. 1993, Vawter and Brown 1993, Viale et al. 1994, Weisburg et al. 1989b, Woese et al. 1991), non-independence of substitution patterns at different sites (Gutell et al. 1994, Schoeniger and Von Haeseler 1994), variation in substitution rates between lineages (e.g., Wolfe et al. 1992, Bruns and Szaro 1992, Nickrent and Starr 1994), and ambiguities in alignments between distantly related taxa. Even if the trees inferred from SS-rRNA genes accurately reflect the evolutionary history of these genes, they might not accurately reflect the history of the species as a whole. For example, lateral transfers between species might cause the genomes of some species to have mosaic evolutionary histories. Although it is unlikely that SS-rRNAs have been stably transferred between species (see above), other genes may have been. Therefore, to understand the history of entire genomes, and to better understand the extent of mosaicism within species, it is important to compare and contrast the histories of different genes from the same species. Finally, since SS-rRNA genes are present in multiple copies in many bacteria (Jinks-Robertson and Nomura 1987, Nomura et al. 1977), it is possible that the genes being compared between species are paralogous not orthologous. This could cause the gene trees to be different from the species trees. For these and other reasons, researchers interested in microbial systematics have begun to compare and contrast the relationships of other molecules with those of the SS-rRNA. The choice of which additional molecule to use is a difficult one. Many potential candidates have arisen and each has its advantages. Examples include HSP70 (Boorstein et al. 1994, Gupta et al. 1994, Rensing and Maier 1994), GroEL (Viale et al. 1994), EF-TU (Ludwig et al. 1994; Delwiche et al. 1995), ATPase-β-subunit (Ludwig et al. 1994), 23S rRNA (Ludwig et al. 1992), and RNA polymerases (Klenk and Zillig 1994). Another potential choice is RecA.
The RecA protein of Escherichia coli is a small (352 aa) yet versatile protein with roles in at least three distinct cellular processes: homologous DNA recombination, SOS induction, and DNA damage induced mutagenesis (Kowalczykowski et al. 1994). This diversity of genetic functions is paralleled by multiple biochemical activities including DNA binding (double and single-stranded), pairing and exchange of homologous DNA, ATP hydrolysis, and coproteolytic cleavage of the LexA, λcI, and UmuD proteins (Kowalczykowski et al. 1994). It has been 30 years since the isolation of the first recA mutants in E. coli (Clark and Margulies 1965) and 15 years since the sequencing of the corresponding recA gene (Sancar et al. 1980; Horii et al. 1980). In that time, studies of the wild type and mutant RecA proteins and genes have yielded a great deal of information about the structure-function relationships of the protein, as well as about the general mechanisms of homologous recombination (Clark and Sandler 1994, Kowalczykowski 1991, Roca and Cox 1990). Such studies have been facilitated greatly by the publication of the crystal structure of the E. coli RecA protein alone, and bound to ADP (Story and Steitz 1992, Story et al. 1992).
Genes encoding proteins with extensive amino-acid sequence similarity to the E. coli RecA have been cloned and sequenced from many other bacterial species. Included among these are complete open reading frames from many of the major bacterial phyla as well as an open reading frame from the nucleus of Arabidopsis thaliana that encodes a protein that functions in the chloroplast (Table 1). Partial open reading frames are available from many additional bacterial species. The high levels of sequence similarity, even between proteins from distantly related taxa, and the demonstration that many of the functions and activities of the E. coli RecA are conserved in many of these other proteins (Angov and Camerini-Otero 1994, Gutman et al. 1994, Roca and Cox 1990, Wetmur et al. 1994), suggest that these proteins are homologs of the E. coli RecA.
Table 1.
RecA and SS-rRNA sequences.
| Species (by Phylum) | Abbr. | RecA. | #aa | SS-rRNA1,2 | RecA Refs. |
|---|---|---|---|---|---|
| Proteobacteria | |||||
| Acetobacter polyoxogenes | Act.po | D13183 | 348 | ABA.PASTER* | (Tayama et al. 1993) |
| Acidiphilium facilis | Acd.f | D16538 | 354 | ACDP.FACI2 | (Inagaki et al. 1993) |
| Acinetobacter calcoaceticus | Acn.c | L26100 | 349 | ACN.CALCOA | (Gregg-Jolly and Ornston 1994) |
| Agrobacterium tumefaciens | Ag.t | L07902 | 363 | AG.TUMEFAC | (Wardhan et al. 1992) |
| Azotobacter vinelandii | Az.v | S96898 | 349 | F.LUTESCEN* | (Venkatesh and Das 1992) |
| Bordetella pertussis | Bd.p | X53457 | 352 | BRD.PERTUS | (Favre et al. 1991, Favre and Viret 1990) |
| Brucella abortus | Br.a | L00679 | 360 | BRU.ABORTS | (Tatum et al. 1993) |
| Burkholderia cepacia3 | Bu.c | D90120 | 347 | BUR.CEPACI | (Nakazawa et al. 1990) |
| Campylobacter jejuni | Ca.j | U03121 | 343 | CAM.JEJUNI | (Guerry et al. 1994) |
| Enterobacter agglomerans 4 | En.a | P33037 | 354 | ER.HERBICO | (Rappold and Klingmueller 1993) |
| Erwinia carotovara | Er.c | X55554 | 342 | ER.CAROTOV | (Zhao and McEntee 1990) |
| Escherichia coli | Es.c | V00328 | 353 | E.COLI | (Horii et al. 1980, Sancar et al. 1980) |
| Haemophilus influenzae | Ha.i | L07529 | 354 | H.INFLUENZ | (Zulty and Barcak 1993) |
| Helicobacter pylori | He.p | Z35478 | 347 | HLB.PYLOR3 | (Haas 1994) |
| Legionella pneumophila | Le.p | X55453 | 348 | LEG.PNEUMO | (Zhao and Dreyfus 1990) |
| Magnetospirillum magnetotacticum5 | Ma.m | X17371 | 344 | MAG.MAGNE2 | (Berson et al. 1990) |
| Methylobacillus flagellatum | Mb.f | M35325 | 344 | MBS.FLAGEL | (Gomelsky et al. 1990) |
| Methylomonas clara | Mm.c | X59514 | 342 | MLM.METHYL* | (Ridder et al. 1991) |
| Methylophilus methylotrophus | Mp.m | unpub. | 342 | MLP.METHY1 | (Emmerson 1995, pers. commun) |
| Myxococcus xanthus 1 | Mx.x1 | L40367 | 342 | MYX.XANTHU | (Inouye 1995, pers. commun.) |
| Myxococcus xanthus 2 | Mx.x2 | L40368 | 358 | n/a6 | (Inouye 1995, pers. commun.) |
| Neisseria gonorrhoeae | Ne.g | X17374 | 348 | NIS.GONORR | (Fyfe and Davies 1990) |
| Proteus mirabilis | Pr.m | X14870 | 355 | ARS.NASONI* | (Akaboshi et al. 1989) |
| Proteus vulgaris | Pr.v | X55555 | 325 | P.VULGARIS | (Zhao and McEntee 1990) |
| Pseudomonas aeruginosa | Ps.a | X52261 | 346 | PS.AERUGIN | (Sano and Kageyama 1987) |
| Pseudomonas fluorescens | Ps.f | M96558 | 352 | PS.FLAVESC* | (De Mot et al. 1993) |
| Pseudomonas putida | Ps.p | L12684 | 355 | PS.PUTIDA | (Luo et al. 1993) |
| Rhizobium leg. phaseoli | Rz.p | X62479 | 360 | RHB.LEGUM6* | (Michiels et al. 1991) |
| Rhizobium leg. viciae | Rz.l | X59956 | 351 | RHB.LEGUM8 | (Selbitschka et al. 1991) |
| Rhizobium meliloti | Rz.m | X59957 | 348 | RHB.MELIL2 | (Selbitschka et al. 1991) |
| Rhodobacter capsulatus | Rh.c | X82183 | 355 | RB.CAPSUL2 | (Fernandez de Henestrosa 1994) |
| Rhodobacter sphaeroides | Rh.s | X72705 | 343 | RB.SPHAER2 | (Calero et al. 1994) |
| Rickettsia prowazekii | Ri.p | U01959 | 340 | RIC.PROWAZ | (Dunkin and Wood 1994) |
| Serratia marcescens | Se.m | M22935 | 354 | SER.MARCES | (Ball et al. 1990) |
| Shigella flexneri | Sh.f | X55553 | 353 | n/a | (Zhao and McEntee 1990) |
| Thiobacillus ferrooxidans | Tb.f | M26933 | 346 | THB.CALDUS* | (Ramesar et al. 1989) |
| Vibrio anguillarum | Vi.a | M80525 | 348 | V.ANGUILLA | (Gammie and Crosa 1991, Tolmasky et al. 1992) |
| Vibrio cholerae | Vi.c | U10162 | 354 | V.CHOLERAE | (Margraf et al. 1995, Stroeher et al. 1994) |
| Xanthomonas oryzae | Xa.o | unpub. | 355 | XAN.ORYZAE | (Mongkolsuk 1995, pers. commun.) |
| Yersinia pestis | Ye.p | X75336 | 356 | YER.PESTIS | (Kryukov et al. 1993) |
| Gram Positives | |||||
| Acholeplasma laidlawii | Acp.l | M81465 | 331 | ACP.LAIDLA | (Dybvig and Woodard 1992) |
| Bacillus subtilis | Ba.s | X52132 | 347 | B.SUBTILIS | (Stranathan et al. 1990) |
| Corynebacterium glutamicum | Co.g | X77384 | 376 | Z46753 | (Billman-Jacobe 1994, Kerins et al. 1994) |
| Lactococcus lactis | La.l | M88106 | 365 | LCC.LACTIS | (Duwat et al. 1992a) |
| Mycobacterium leprae | Myb.l | X73822 | 711 | MYB.LEPRAE | (Davis et al. 1994) |
| Mycobacterium tuberculosis | Myb.t | X58485 | 790 | MYB.TUBER2 | (Davis et al. 1991) |
| Mycoplasma mycoides | Myp.m | L22073 | 345 | M.MYCOIDES | (King et al. 1994) |
| Mycoplasma pulmonis | Myp.p | L22074 | 339 | M.PULMONIS | (King et al. 1994) |
| Staphylococcus aureus | Sta.a | L25893 | 347 | STP.AUREUS | (Bayles et al. 1994) |
| Streptococcus pneumoniae | Stc.p | Z17307 | 388 | STC.SALIVA* | (Martin et al. 1992) |
| Streptomyces ambofaciens | Stm.a | Z30324 | 372 | STM.AMBOFA | (Aigle et al. 1994) |
| Streptomyces lividans | Stm.l | X76076 | 374 | STM.LIVIDA | (Nussbaumer and Wohlleben 1994) |
| Streptomyces violaceus7 | Stm.v | U04837 | 377 | STM.COELI3* | (Yao and Vining 1994) |
| Cyanobacteria/Chloroplasts | |||||
| Arabidopsis thaliana | Ar.t | M98039 | 439 | NICO.TAB_C* | (Binet et al. 1993, Cerutti et al. 1992) |
| Anabaena variabilis | An.v | M29680 | 358 | X59559* | (Owttrim and Coleman 1989) |
| Synechococcus sp. PCC7942 | Sy.79 | unpub. | 361 | PHRM.MINUT* | (Coleman 1995) |
| Synechococcus sp. PCC7002 | Sy.70 | M29495 | 348 | SYN.6301* | (Murphy et al. 1987, Murphy et al. 1990) |
| Deinococcus-Thermus Group | |||||
| Deinococcus radiodurans8 | De.r | U01876 | 363 | D.RADIODUR | (Gutman et al. 1994) |
| Thermus aquaticus | Th.a | L20095 | 340 | T.AQUATICU | (Angov and Camerini-Otero 1994, Wetmur et al. 1994) |
| Thermus thermophilus | Th.t | D13792 | 340 | T.THMOPHL | (Kato and Kuramitsu 1993, Wetmur et al. 1994) |
| Chlamydia/Planctomyces | |||||
| Chlamydia trachomatis | Ch.t | U16739 | 352 | CLM.TRACHO | (Larsen 1994, Zhang et al. 1994) |
| Spirochaetes | |||||
| Borrelia burgdorferi | Bo.b | unpub. | 365 | BOR.BURGDO | (Huang 1995, pers. commun.) |
| Bacteroides | |||||
| Bacteroides fragilis | Bct.f | M63029 | 318 | BAC.FRAGIL | (Goodman and Woods 1990) |
| Thermophilic O2 Reducers | |||||
| Aquifex pyrophilus | Aq.p | L23135 | 348 | AQU.PYROPH | (Wetmur et al. 1994) |
| Thermotogales | |||||
| Thermotoga maritima | Tg.m | L23425 | 356 | TT.MARITIM | (Wetmur et al. 1994) |
The diversity and number of species from which sequences are available makes RecA a potentially useful tool for molecular systematic studies of bacteria. Previously, Lloyd and Sharp (1993) tested the utility of RecA comparisons for phylogenetic studies. They concluded that RecA comparisons were probably only useful for determining relationships among closely related bacterial species. However, they were limited by the number and diversity of RecA sequences that were available at the time. I have reanalyzed the evolution of RecA using 40 additional sequences. In this paper, analysis is presented that shows that the RecA protein is a good alternative or supplement to SS-rRNA for molecular systematic studies of all bacteria, not just of closely related species. Phylogenetic trees of the 65 complete RecA protein sequences were inferred using a variety of phylogenetic methods. Statistical analysis and comparisons of trees generated by the different phylogenetic methods suggests that the RecA phylogeny is highly consistent and robust. The RecA trees are compared to trees of SS-rRNA sequences from the same or very closely related species as represented in the RecA trees. Overall, the trees of the two molecules are highly congruent. The implications of the particular similarities and differences between the RecA-based and SS-rRNA-based trees are discussed. Some of the features of RecA that make it a potentially useful molecular chronometer are also discussed.
METHODS
Sequences and alignment
All RecA sequences used in this paper were obtained from the National Center for Biotechnology Information (NCBI) databases by electronic mail (Henikoff 1993) except for those from Methylophilius methylotrophus (Emmerson 1995), Xanthomonas oryzae (Mongkolsuk 1995), Synechococcus sp. PCC7942 (Coleman 1995), and Borrelia burgdorferi (Huang 1995) which were kindly provided prior to submission. Accession numbers for those in databases are given in Table 1. The amino-acid sequences of the RecA proteins were aligned both manually and with the clustalw multiple sequence alignment program (Thompson et al. 1994). The RecA alignment was used as a block and aligned with the sequences of the RadA protein from an Archaea (Clark and Sandler 1994, Clark 1995) and RecA-like proteins from eukaryotes (Ogawa et al. 1993), also using clustalw.
For the comparison of RecA and SS-rRNA trees, a complete or nearly complete SS-rRNA sequence was chosen to represent each species for which a complete RecA protein was available. For most of the RecA proteins, a complete SS-rRNA sequence was available from the same species. The remaining species (those for which a RecA sequence was available but a complete or nearly complete SS-rRNA was not) were represented by a “replacement” SS-rRNA from a different species. The choice of which replacement sequence to use was determined in one of two ways. For those RecAs for which a partial SS-rRNA was available from the same species, the complete or nearly complete SS-rRNA that was most similar to the partial sequence was used. Similarity was determined by comparisons using the Ribosomal Database Project (RDP) computer server (Maidak et al. 1994) and blastn searches (Altschul et al. 1990) of the NCBI databases by electronic mail (Henikoff 1993). For those RecAs for which even a partial SS-rRNA sequence was not available from the same species, a replacement SS-rRNA was chosen from a species considered to be a close relative. A SS-rRNA was not used to represent the Shigella flexneri RecA because this protein was identical to the E. coli RecA. For the majority of the SS-rRNA phylogenetic analysis, only one SS-rRNA sequence was used to represent the two RecAs from Myxococcus xanthus. For some of the analysis an additional SS-RNA from a close relative of M. xanthus was also included. The SS-rRNA sequences used and the species from which they come are listed in Table 1. The SS-rRNA sequences were obtained already aligned from the RDP (Maidak et al. 1994), with the exception of those from Corynebacterium glutamicum and Anabaena sp. PCC7120, which were obtained from the NCBI and were aligned to the other sequences manually. Entry names and numbers are listed in Table 1.
Phylogenetic trees
Phylogenetic trees were generated from the sequence alignments using computer algorithms implemented in the PHYLIP (Felsenstein 1993), PAUP (Swofford 1991), and GDE (Smith 1994, Smith et al. 1994) computer software packages. Trees of the RecA sequences were generated using two parsimony methods (the protpars program in PHYLIP and the heuristic search algorithm of PAUP) and three distance methods (the least-squares method of De Soete (De Soete 1983) as implemented in GDE, and the Fitch-Margoliash (Fitch and Margoliash 1967) and neighbor-joining methods (Saitou and Nei 1987) as implemented in PHYLIP). Trees of the SS-rRNA sequences were generated using one parsimony method (the dnapars algorithm of PHYLIP) and the same three distance methods as used for the RecA trees. For the trees generated by the protpars, dnapars, Fitch-Margoliash, and neighbor-joining methods, 100 bootstrap replicates were conducted by the method of Felsenstein (1985) as implemented in PHYLIP.
For the distance-based phylogenetic methods listed above, estimated evolutionary distances between each pair of sequences were calculated for input into the tree-reconstruction algorithms. Pairwise distances between RecA proteins were calculated using the protdist program of PHYLIP and the PAM matrix-based distance correction (Felsenstein 1993). Pairwise distances between SS-rRNA sequences were calculated in two ways: the method of Olsen (1988) (as implemented by the count program of GDE) was used for the trees generated by the De Soete method; and the two-parameter model of Kimura (1980) (as implemented by the dnadist program of PHYLIP) was used for the Fitch-Margoliash and neighbor-joining trees.
Regions of the alignments for which homology of residues could not be reasonably assumed were excluded from the phylogenetic analysis. For the SS-rRNA trees, the alignment of SS-rRNA sequences was extracted from an alignment of thousands of sequences in the RDP database (Maidak et al. 1994). This RDP alignment was generated using both primary and secondary structures as a guide to assist in the assignment of homology (Maidak et al. 1994). Therefore it was assumed that the aligned regions were likely homologous. Nevertheless, regions of high sequence variation (as determined by a 50% consensus mask using the consensus program of GDE) were excluded from the phylogenetic analysis since these regions are perhaps most likely to contain non-homologous residues. The SS-rRNA alignment and a list of the 1061 alignment positions used for phylogenetic analysis are available on request. For the RecA analysis, the assignment of homology in the alignment was based only on similarity of primary structure (as determined by the clustalw program). Regions of ambiguity in the alignment were considered to potentially include non-homologous residues and thus were excluded from the phylogenetic analysis. Such regions were identified by comparing alignments generated by the clustalw program using a variety of alignment parameters. Parameters varied included scoring matrices (PAM, BLOSUM, and identity matrices were used) and gap opening and extension penalties. Alignments were compared by eye to detect differences and those regions that contained different residues in the different alignments were considered ambiguous.
Character states and changes
Analysis of character states and changes over evolutionary history was done using the MacClade 3.04 program (Maddison and Maddison 1992). For each alignment position, all unambiguous substitutions as well as all unambiguous non-conservative substitutions were counted. Non-conservative substitutions were defined as amino-acid changes that were not within the following groups: (V-I-L-M), (F-W-Y), (D-E-N-Q), (K-R), (G-A), and (S-T).
Computer programs
GDE, PHYLIP, and clustalw were obtained by anonymous FTP from the archive of the Biology Department at the University of Indiana (ftp.bio.indiana.edu). PAUP was obtained from David Swofford and is now available from Sinauer Associates, Inc., Sunderland, MA. GDE, PHYLIP, and clustalw were run on a Sparc10 workstation and MacClade and PAUP on a Power Macintosh 7100/66. Unless otherwise mentioned, all programs were run with default settings.
RESULTS AND DISCUSSION
The potential of using RecA for phylogenetic studies of bacteria was first addressed by Lloyd and Sharp (1993). In a detailed analysis of the evolution of recA genes from 25 species of bacteria, they showed that phylogenetic trees of RecA proteins appeared to be reliable for determining relationships among closely related bacterial species. Specifically, for the Proteobacteria, the branching patterns of RecA proteins were highly congruent to branching patterns of SS-rRNA genes from the same or similar species. However, the RecA and SS-rRNA trees were not highly congruent for relationships between sequences from more distantly related species. Lloyd and Sharp concluded that this was due to a low resolution of the deep branches in the RecA tree. However, this low resolution of deep branches could have been due to poor representation of certain taxa in their sample set. Of the recA sequences available at the time, only six were from species outside the Proteobacteria. The diversity as well as the number of recA sequences available has increased greatly since Lloyd and Sharp’s study (see Table 1). Therefore, I have re-analyzed the evolution of recA including these additional sequences with a specific focus on determining whether recA comparisons can provide reasonable resolution of moderate to deep branches in the phylogeny of bacteria. The analysis presented here focuses on amino-acid comparisons for two reasons. First, for highly conserved proteins such as RecA, it is likely that amino-acid trees will be less biased by multiple substitutions at particular sites and base-composition variation between species than trees of the corresponding nucleotide sequences (Hasegawa and Hashimoto 1993; Viale et al. 1994, Lloyd and Sharp 1993). In addition, Lloyd and Sharp (1993) presented specific evidence suggesting that DNA-level comparisons of the recA genes between distantly related taxa might be misleading.
Alignment of RecA sequences
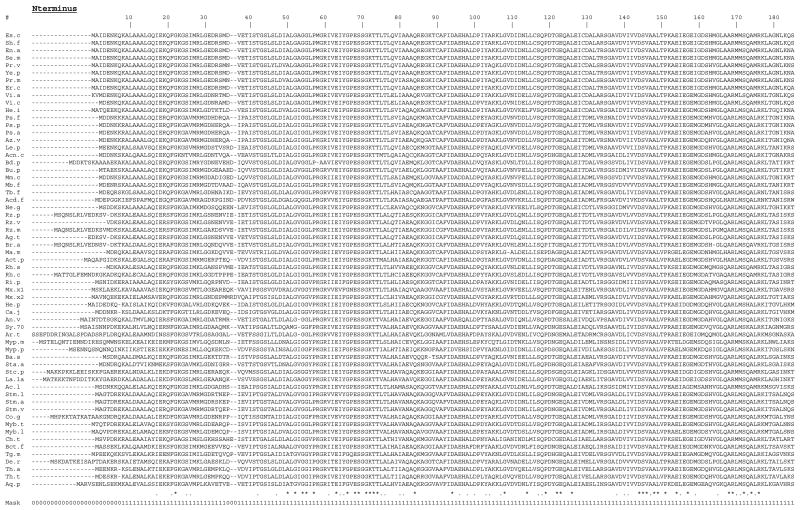
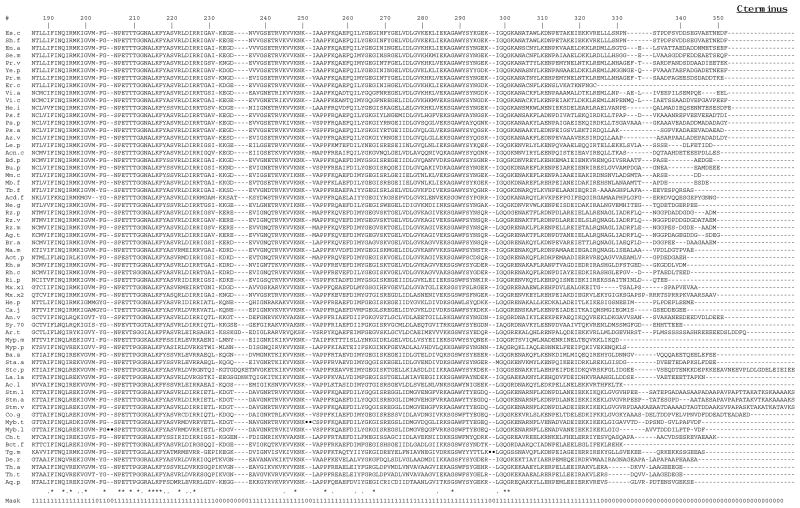
An alignment of the sequences of the complete RecA proteins is shown in Figure 1. Aligning sequences is an integral part of any molecular systematic study because each aligned position is assumed to include only homologous residues from the different molecules. Assignment of homology, as represented by the sequence alignments, can be highly controversial, and differences in alignments can cause significant differences in phylogenetic conclusions (Gatesy et al. 1993, Lake 1991). To limit such problems, regions for which homology of residues cannot be unambiguously assigned should be excluded from phylogenetic analysis. Thus for a molecule to be useful for molecular systematic studies, alignments between species should be relatively free of ambiguities. This is one of the main advantages of using SS-rRNA genes over other genes for phylogenetic analysis. Assignment of homology for SS-rRNA sequences can be aided by alignment of both primary and secondary structures (Woese 1987). In addition, regions of high primary structural conservation that are interspersed throughout the molecule help align less conserved regions. Since RecA is a highly conserved protein, it has the potential to be useful for phylogenetics because the assignment of homology should be relatively unambiguous (Lloyd and Sharp 1993). Regions of ambiguity in the RecA alignment shown in Fig. 1 were determined by comparing this alignment to those generated using different alignment parameters (see Methods). Regions of the alignment were considered to be ambiguous if they contained different residues in the different alignments, as suggested by Gatesy et al. (1993). Overall, the majority of the alignment was determined to be free of ambiguities and thus can be used with confidence for the phylogenetic analysis. The four regions of ambiguity (the C- and N-termini (corresponding to E. coli amino-acids 1–7 and 320–352) and two short regions corresponding to E. coli amino-acids 36–37 and 231–236)) were excluded from the phylogenetic analysis. The 313 alignment positions used are indicated by the sequence mask shown in Fig. 1.
Figure 1.
Alignment of complete RecA sequences.
The alignment was generated using the clustalw multiple sequence alignment program. Dashes (-) represent alignment gaps. Three insertions that are present in only one sequence each (Myb.t, Myb.l, and Tg.m) and the first 80 aa of the A. thaliana protein are left out for space reasons and are indicated by a ••. Conservation of alignment positions as determined by the clustalw program is indicated by * (identical aa in all) and. (similar aa in all). The alignment positions used in phylogenetic analysis are indicated by the sequence mask (1=used, 0=not used). Sequence abbreviations are described in Table 1.
Another potential source of variation and error in phylogenetic reconstruction from sequences lies in assigning a weight to give insertion or deletion differences (indels) between species. Other than in the C- and N-terminal regions, there are few indels in the RecA alignment (see Fig. 1). Most of the indels are in regions of ambiguous alignment as identified above, and thus were not included in the phylogenetic analysis. The phylogenetic results were not affected whether the few remaining indels were included or not (data not shown). Of the indels in regions of unambiguous alignment most are isolated (in only one species) and only one amino acid in length. There are two very large indels - one in each of the Mycobacterium RecAs. These are protein introns that are removed by post-translational processes (Davis et al. 1991, Davis et al. 1994). There is a 4 aa indel in the Thermotoga maritima RecA (see Fig. 1). There only indels that have obvious phylogenetic relevance are the single amino acid gaps found in the cyanobacterial and the A. thaliana RecAs all at the same position --E. coli position 53 (see below for discussion of this).
Another aspect of the RecA alignment that is relevant to molecular systematics is the degree of conservation of different alignment positions. I have used the RecA phylogeny and parsimony character-state analysis to characterize the patterns of aminoacid substitutions at different sites of the molecule (see Methods). The number of inferred substitutions varies a great deal across the molecule. The number of total substitutions ranges from 0 (at 58 positions) to 38 (at one position) with a mean of 9.4. The number of non-conservative substitutions varies from 0 (at 111 positions) to 27 (at one position) with a mean of 4.8. The variation in the substitution patterns across the molecule suggests that RecA comparisons may have phylogenetically useful information at multiple evolutionary distances.
Generation of phylogenetic trees
To examine the utility of the RecA comparisons for molecular systematics, the RecA trees were compared to trees of the same species based on studies of other molecules. Such a comparison is useful for a few reasons. First, congruence among trees of different molecules indicates both that the genomes of the species are not completely mosaic and that the molecular systematic techniques being used are reliable (Miyamoto and Fitch 1995). Differences in the branching patterns between trees of different molecules can help identify genetic mosaicism, unusual evolutionary processes, or inaccuracies in one or both of the trees. Differences in resolution and significance of particular branches can help identify which molecules are useful for specific types of phylogenetic comparisons. Since differences in species sampled have profound effects on tree generation (e.g., (Lecointre et al. 1993)), to best compare the phylogenetic resolution of trees of different molecules the analysis should include sequences from the same species. Fortunately, SS-rRNA sequences were available for most of the species represented in the RecA data set. Therefore it was possible to generate SS-rRNA trees for essentially the same species-set as represented in the RecA trees. For those species for which RecA sequences were available but SS-rRNA sequences were not, SS-rRNA sequences were used from close relatives (see Methods). A list of the sequences used is in Table 1.
Phylogenetic trees of the RecAs and SS-rRNAs were generated from the sequence alignments using multiple phylogenetic techniques (see Methods). The trees were generated without an outgroup and thus can be considered unrooted. However, since rooting of trees is helpful for a variety of reasons, a root was determined for both the RecA and SS-rRNA trees. In both cases, the root was determined to be the sequence from Aquifex pyrophilus. For the SS-rRNA trees, this rooting was chosen because analyses of sequences from all three kingdoms of organisms indicate that the deepest branching bacterial SS-rRNA is that of A. pyrophilus (Burggraf et al. 1992; Pitulle et al. 1994). Although it seems reasonable to assume that the deepest branching bacterial RecA would also be that of A. pyrophilus, if there have been lateral transfers or other unusual evolutionary processes, the RecA trees could be rooted differently than the SS-rRNA trees. Therefore the rooting of the RecA sequences was tested by constructing trees using likely RecA homologs from Archaea and eukaryotes as outgroups (see Methods). In both neighbor-joining and protpars trees, the deepest branching bacterial protein was that of A. pyrophilus (not shown). However, the alignments of the RecAs with the Archaeal and eukaryotic RecA-like proteins include many regions of ambiguity. Therefore, only 140 alignment positions were used in this analysis and the trees showed little resolution within the bacteria. In addition, the bootstrap values for the deep branching of the A. pyrophilus RecA were low (<30 in all cases). Thus although the rooting of the RecA trees to the A. pyrophilus protein is reasonable it should be considered tentative. The rooting will likely be better resolved as more sequences become available from eukaryotes and Archaea.
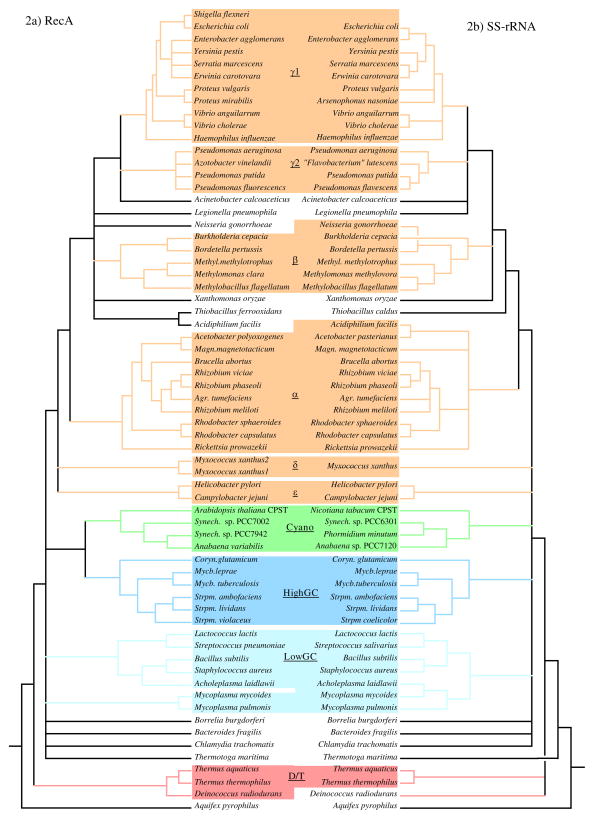
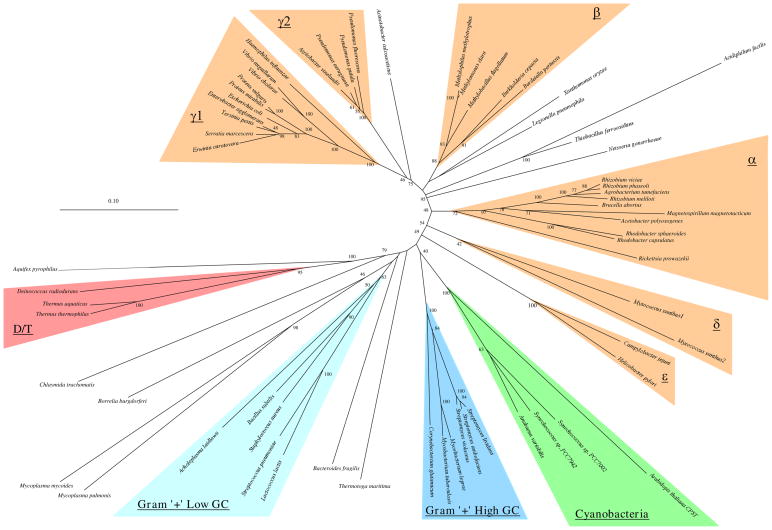
The analysis and comparison of the phylogenetic trees focused on a few specific areas. First, bootstrap values were used to get an estimate of the degree that the inferred branching patterns reflect the characteristics of the entire molecule. In addition, since phylogenetic methods differ in the range of evolutionary scenarios in which they accurately reconstruct phylogenetic relationships (Hillis 1995), comparison of the trees generated by the different methods was used to identify the phylogenetic patterns that were most robust for that particular molecule. To summarize the differences and similarities among the trees inferred by the different methods, strict-consensus trees of all the trees of each molecule were generated (Figure 2). Since consensus trees lose some of the information of single trees and since they only show the areas of agreement among trees (and not the phylogenetic patterns in the areas of difference), it is also useful to examine individual trees. A comparison of the Fitch-Margoliash trees for the two molecules is shown in Figure 3. The other trees are available from the author on request. Finally, the SS-rRNA trees determined here were compared to those determined with more sequences to help identify patterns that might be due to poor sampling of the species here.
Figure 2.
Comparison of consensus trees for RecA and SS-rRNA.
Strict-rule consensus trees representing the phylogenetic patterns found in all trees generated by multiple methods for each molecule are shown. The RecA consensus (A) was generated from the PAUP, protpars, Fitch-Margoliash, De Soete and neighbor-joining trees (see Methods). The SS-rRNA consensus (B) was generated from the dnapars, Fitch-Margoliash, De Soete and neighbor-joining trees. Comparable species are aligned in the middle and species are ordered to minimize branch crossing (note two crossed branches in SS-rRNA tree). Consensus clades are shaded for each molecule.
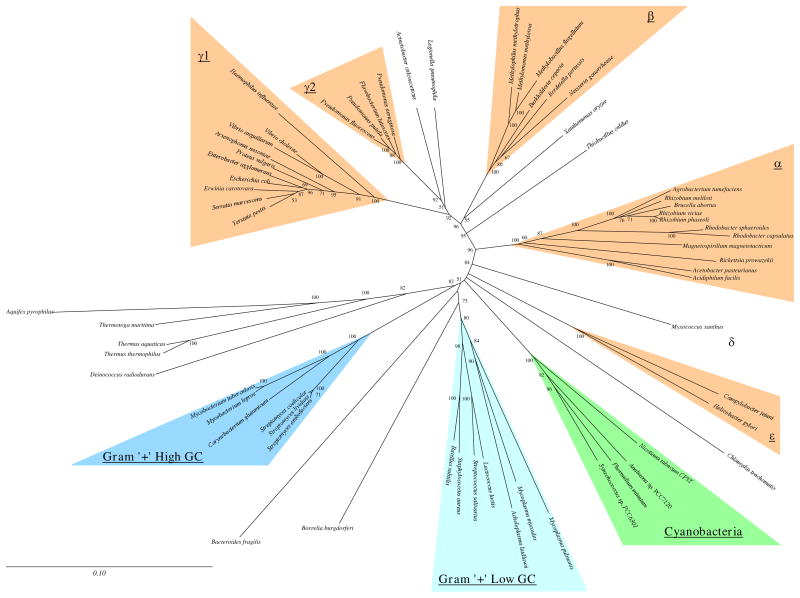
Figure 3.
Fitch-Margoliash trees for RecA (A) and SS-rRNA (B).
Trees were generated from the multiple sequence alignments by the method of Fitch and Margoliash. Regions of ambiguous alignment and indels were excluded from the analysis (see Methods). For the RecA tree, distances were calculated using the protdist program of PHYLIP with a PAM-matrix based distance correction. For the SS-rRNA tree, distances were calculated using the dnadist program of PHYLIP and the Kimura-2-parameter distance correction. Consensus clades representing groups found in all phylogenetic methods are highlighted. Branch lengths and scale bars correspond to estimated evolutionary distance. Bootstrap values when over 40 are indicated.
A quick glance at the trees in Fig. 2 and 3 shows that the patterns for each molecule are highly robust (there is high resolution in the consensus trees) and that the patterns are similar between the two molecules. To aid comparison of the trees of the two molecules, sequences have been grouped into consensus clades based on the patterns found in the consensus trees (Fig. 2, Table 2). Clades of RecA sequences were chosen to represent previously characterized bacterial groups as well as possible. Comparable clades were determined for the SS-rRNA sequences (Table 2). The clades are named after the rRNA-based classification of most of the members of the clade (Maidak et al. 1994). These clades are highlighted in the trees in Fig. 2 and 3. Sequences from the same or similar species are aligned in the middle in Fig. 2 to ease comparison of the two consensus trees. Besides being found in trees generated by all the phylogenetic methods used, the consensus clades have high bootstrap values for the methods in which bootstrapping was performed (Table 2). Thus we believe that the clades are consistent and reliable groupings of the RecA and SS-rRNA sequences. In the following sections, some of the implications of the similarities and differences within and between the RecA and SS-rRNA trees are discussed. The discussion has been organized by phylogenetic groups.
Table 2.
Consensus Phylogenetic Groups
| Clade | Species in RecA Consensus Clade6 | Comprable SS-RNA Consensus ?1,2,3 | RecA Bootstrap4 | sRNA Bootstrap5 | ||||
|---|---|---|---|---|---|---|---|---|
| PP | NJ | FM | DP | NJ | FM | |||
| Proteobacteria - γ17 | Escherichia coli, Shigella flexneri, Yersinia pestis, Erwinia carotovara, Serratia marcescens, Enterobacter agglomerans, Proteus vulgaris, Pr. mirabilis, Vibrio cholerae, V. anguillarum, Haemophilus influenzae | YES | 78 | 91 | 100 | 100 | 100 | 100 |
| Proteobacteria - γ2 | Azotobacter vinelandii, Pseudomonas aeruginosa, Ps. putida, Ps. fluorescens | YES | 100 | 100 | 100 | 100 | 100 | 100 |
| Proteobacteria - γ | γ1, γ2 Acinetobacter calcoaceticus | YES (+ Legpn) | 33 | 63 | 75 | 48 | 85 | 92 |
| Proteobacteria - β1 | Methylobacillus flagellatum, Methylomonas clara, Methylophilus methylotrophus, Burkholderia cepacia, Bordetella pertussis | YES (+ Neigo) | 74 | 84 | 88 | 100 | 100 | 100 |
| Proteobacteria - β2 | Thiobacillus ferrooxidans, Acidiphilium facilis | No | 100 | 100 | 100 | * | * | * |
| Proteobacteria - βγ | γ, β1, β2, Xanthomonas oryzae, Neisseria gonorrhoeae, Legionella pneumophila | YES (−Acifa) | 53 | 86 | 95 | 90 | 94 | 95 |
| Proteobacteria - α | Rhodobacter capsulatus, Rho. sphaeroides, Rhizobium meliloti, Rhi. viciae, Rhi. phaseoli, Acetobacter polyoxogenes, Magnetospirillum magnetotacticum, Brucella abortus, Agrobacterium tumefaciens, Rickettsia prowazekii | YES (+Acifa) | 14 | 68 | 72 | 100 | 100 | 100 |
| Proteobacteria - αβγ | αβγ | YES | 10 | 57 | 58 | 93 | 96 | 96 |
| Proteobacteria - δ | Myxococcus xanthus 1, M. xanthus 2 | YES | 43 | 71 | 42 | *8 | * | * |
| Proteobacteria - ε | Campylobacter jejuni, Helicobacter pylori | YES | 100 | 100 | 100 | 100 | 100 | 100 |
| Proteobacteria | γ, β, α, δ, ε | NO | 14 | 38 | 49 | * | * | 36 |
| Gram”+” High GC | Corynebacterium glutamicum, Streptomyces ambofaciens, S. violaceus, S. lividans, Mycobacterium tuberculosis, Myb. leprae | YES | 97 | 100 | 100 | 100 | 100 | 100 |
| Gram”+” Low GC | Bacillus subtilis, Lactococcus lactis, Streptococcus pneumoniae, Staphylococcus aureus, Acholeplasma laidlawii | YES (+ Mycpn, Mycge) | 27 | 59 | 63 | 50 | 56 | 80 |
| Mycoplasmas | Mycoplasma mycoides, Myp. pulmonis | YES (+ Achla) | 88 | 100 | 98 | 71 | 88 | 84 |
| Cyanobacteria | Arabidopsis thaliana, Anabaena variabilis, Synechococcus sp. PCC7942, Syn.sp. PCC7002 | YES | 100 | 96 | 91 | 100 | 100 | 100 |
| Deinococcus-Thermus | Deinococcus radiodurans, Thermus aquaticus, T. thermophilus | NO | 95 | 96 | 95 | * | * | * |
Proteobacteria
The Proteobacteria phylum includes most but not all the traditional gram-negative bacterial species (Stackebrandt et al. 1988). This phylum has been divided into five phylogenetically distinct groups (α, β, γ, δ, and ε) mostly based on SS-rRNA comparisons (Olsen et al. 1994, Rainey et al. 1993, Stackebrandt et al. 1988, Woese 1987). The available RecA sequences are heavily biased towards the Proteobacteria (Table 1) and thus much of the discussion will focus on this phylum. With the species represented in this analysis, the Proteobacterial RecA sequences form a monophyletic clade in all phylogenetic methods (Fig. 2). In contrast, with essentially the same species-set, the Proteobacterial SS-rRNA sequences do not consistently form a clade (Fig. 2, positions of Campylobacter jejuni, Helicobacter pylori, and Myxococcus xanthus), although they do in some of the phylogenetic methods (e.g., Fig. 3). This was surprising since the Proteobacterial group was defined based on SS-rRNA comparisons (Stackebrandt et al. 1988). When additional SS-rRNA sequences are included in phylogenetic analysis, M. xanthus, C. jejuni, and H. pylori consistently branch with the other Proteobacteria (Maidak et al. 1994; Olsen et al. 1994). The lack of resolution of the position of these species in the SS-rRNA versus RecA trees was not due to using only one SS-rRNA sequence to represent the two M. xanthus RecAs -- the same pattern was seen when the SS-rRNA sequence from another δ species was also included. Thus in this case the RecA trees can be considered to have higher resolution than the SS-rRNA trees since the RecA trees show a relationship between species that is only consistently detected in SS-rRNA trees with more sequences.
Subdivisions corresponding to the α, β, γ, δ, and ε groups are detected in both the RecA and SS-rRNA trees and the placement of species into these subdivisions is nearly the same for the two molecules (Fig. 2, Table 2). Thus the RecA comparisons support the division of the Proteobacteria into these groups as well as the classification of particular species into the groups here. There are other phylogenetic patterns that are the same in the RecA and SS-rRNA trees here. Examples include the separation of the _Pseudomonas_-Azotobacter γs (γ2 here) from the Haemophilus, Proteus, and enteric γs (γ1 here); the monophyly of the enteric bacteria (represented here by E. coli, S. flexneri, Erwinia carotovara, Enterobacter agglomerans and Yersinia pestis); the relatedness of the Rhizobium species, Agrobacterium tumefaciens and Brucella abortus; the placement of Acinetobacter calcoaceticus into the γ supergroup; an affiliation between the γ’s and the β’s into what can be called a β-γ supergroup; and the grouping of Legionella pneumophilia, Neisseria gonorrhoeae, Xanthomonas oryzae, and the Thiobacillus species somewhere in the γ- β supergroup. In all these cases, the relationships have been suggested by other studies of SS-rRNA sequences (see (Maidak et al. 1994; Olsen et al. 1994; Woese 1987)). The finding of the same patterns in the RecA trees serves to confirm the previous suggestions of the phylogenetic associations indicated between these species. Thus even though the RecA trees are based on analysis of highly conserved protein sequences, they do appear to have resolution for even close relatives as suggested by Lloyd and Sharp (1993).
Most of the differences between the RecA and SS-rRNA trees for the Proteobacteria are in areas of low resolution (differences among the trees generated by the different methods) or low bootstrap values for one or both of the molecules and thus are probably not biologically significant. For example, the differences in the grouping of the δ and ε clades within the Proteobacteria discussed above appears to be due to a lack of resolution of the SS-rRNA trees with the species represented here. In addition, the branching order between Haemophilus influenzae, the Proteus species, the Vibrios, and the enteric species is ambiguous in the SS-rRNA trees yet it is consistent in the RecA trees. In other cases, the SS-rRNA trees appear to have more resolution than the RecA trees. For example, the specific position of the RecA from L. pneumophilia is ambiguous (Fig. 2a) yet the SS-rRNA of this species consistently groups with the γ1 and γ2 groups, and thus can be considered part of the γ clade (Fig. 2b, Table 2). Analysis of other SS-rRNA sequences suggests that the position of the Legionellaceae in the γ subgroup is robust (Fry et al. 1991; Weisburg et al. 1989a). Similarly, the exact position of the N. gonorrhoeae RecA is ambiguous, yet the N. gonorrhoeae SS-rRNA groups consistently with the β clade.
There are branching patterns within the Proteobacteria that have high resolution and robustness for each molecule but are different between the two. The most striking example of this is the phylogenetic position of the sequences from Acidiphilium facilis. The A. facilis RecA branches with the Thiobacillus ferrooxidans RecA in the β– γ supergroup in all trees (Fig. 2) and the node joining these two species has very high bootstrap values (Table 2). However, the corresponding A. facilis SS-rRNA consistently branches with species in the α clade also with high bootstrap values. Thus either the SS-rRNA and RecA genes of A. facilis have different phylogenetic histories, or one of the trees is inaccurate. The grouping of Acidiphilium species within the α subgroup appears to be a reliable representation of the SS-rRNA relationships (Lane et al. 1992; Sievers et al. 1994), so it is unlikely that the SS-rRNA tree here is biased by species sampling. It has been suggested that the A. facilis RecA sequence contains many sequencing errors and it is currently being resequenced (Roca 1995). Errors in the sequence would explain the unusual amino acids found in the A. facilis RecA in otherwise highly conserved regions (Fig. 1) and the extremely long branch length for this sequence in all phylogenetic methods (Fig. 3). Thus the position of the A. facilis RecA in the trees may not represent the actual evolutionary history of this gene.
M. xanthus, the only δ Proteobacteria represented in this analysis, is the only species known to encode two RecA proteins. There are at least two plausible explanations for this: lateral transfer from another species or gene duplication. The phylogenetic analysis of the two proteins helps limit the possibilities for when and how a duplication or lateral transfer could have occurred. In all the RecA trees, the two M. xanthus proteins branch together, showing that they are more related to each other than to any other known RecAs. However, the node joining them is quite deep indicating that the degree of evolutionary separation between them is quite high. Thus if a duplication event was what led to these two genes in the same species, it apparently happened reasonably early in the history of the δ clade. If one of these sequences was obtained by a lateral transfer from another species, most likely, the donor was another δ species. It is interesting that the bootstrap values for the node joining the two M. xanthus RecAs are relatively low in all methods (Table 2). This indicates that the branching together is not very stable and is affected by the choice of alignment positions used in the phylogenetic analysis. Perhaps there was a gene conversion event after a lateral transfer or duplication and only certain regions of the recA genes underwent the conversion. Alternatively, the low bootstraps could also be explained if a duplication occurred right at or near the time of separation of the δ clade from the other Proteobacterial groups. The specific history of these two genes will probably be best resolved by studies of RecAs in other δ species.
Gram-positive bacteria
Previous studies have shown that gram-positive species are divided into multiple phylogenetically distinct groups (Woese 1987). Whether these distinct groups are monophyletic has been the subject of a great deal of research and debate (e.g., (Gupta et al. 1994; Van De Peer et al. 1994; Weisburg et al. 1989c; Woese 1987)). For example, studies of HSP70 genes (Viale et al. 1994) and some studies of rRNA genes (Woese 1987) suggest the gram-positives are monophyletic while studies of EF-TUs (Ludwig et al. 1994), ATPaseβ (Ludwig et al. 1994) and different studies of rRNA genes (Van De Peer et al. 1994) suggest they are not.
Species from two of the gram-positive groups, the low-GCs and the high-GCs, are represented in the analysis here (Table 1). In all the RecA and SS-rRNA trees inferred in this study, the sequences from the high-GC species cluster together (Fig. 2). In addition these species have the same branching patterns within this group in all trees of both molecules. Thus the RecA data support the phylogenetic coherence of as well as the branching topology within the high-GC clade. In contrast, the RecA and SS-rRNA trees are not congruent for the relationships among sequences from low-GC gram-positive species. In all the SS-rRNA trees, the sequences from species considered to be low-GC gram-positives are monophyletic, as might be expected, since the classification of these species was based on SS-rRNA comparisons. However in all the RecA trees the sequences from the low-GCs are not monophyletic (e.g., Fig. 3). This may be due to a combination of poor species sampling and unusual evolutionary patterns. In four of the five RecA trees only one RecA, that of the spirochaete Borrelia burgdorferi, prevents the low-GCs as a whole from being monophyletic (e.g., Fig. 3). The bootstrap values for the position of the B. burgdorferi RecA are relatively low in all of these trees, and since this is the only sample from the spirochaetes, its position may be unreliable. In addition, in three out of four of the SS-rRNA trees, the B. burgdorferi sequence is an outgroup to the low-GCs. Thus with the species sampled here the B. burgdorferi sequences tend to group with the sequences from low-GCs. Yet another factor that could contribute to a biased placement of the B. burgdorferi RecA is the apparent high rate of sequence change in the mycoplasmal RecAs, which can be seen by their long branch lengths (Fig. 3a). A rapid rate of mycoplasmal protein evolution has been thought to complicate trees of other proteins (e.g., (Ludwig et al. 1994)). The inclusion of additional sequences from the spirochetes and other low-GC gram-positives may help resolve whether this difference between the RecA and SS-rRNA trees is biologically significant.
With the species represented here, the branching between the high and low-GCs is unresolved in both the RecA and SS-rRNA trees. Interestingly, in all the RecA trees, the proteins from the high-GCs form a group with the cyanobacterial proteins. Thus the gram-positives are non-monophyletic for RecA proteins. Analysis of other genes has suggested that the cyanobacteria and gram-positives are sister groups (e.g., (Van De Peer et al. 1994; Viale et al. 1994; Woese 1987)). However this is one of the few if not the only case in which the cyanobacterial genes consistently group with genes from high-GCs to the exclusion of those from the low-GCs. Since this relationship is found in all the RecA trees it appears to be robust. However, the bootstrap values for the node linking these two groups are moderate (31–40) indicating that this association is a good, but not great, representation of the relationships of RecA sequences.
Cyanobacteria
The RecA and SS-rRNA trees both show the cyanobacteria forming a coherent clade. The nuclear encoded chloroplast RecA from A. thaliana groups consistently with the cyanobacterial RecAs. This suggests that the A. thaliana recA gene is derived from the recA gene of a cyanobacterial-like ancestor to the A. thaliana chloroplast and that, as has been demonstrated for many other genes, it was transferred to the nucleus after endosymbiosis. Given the high degree of sequence conservation in RecAs, it is possible that studies of chloroplast evolution might be aided by sequencing of additional nuclear encoded chloroplast RecAs. In addition, all the RecAs from this group (including the A. thaliana RecA) contain an alignment gap not found in any other RecAs (see Fig. 1). This could serve as a sequence signature for cyanobacterial and chloroplast RecAs and further serves to demonstrate the relatedness among chloroplasts and cyanobacteria. As discussed above, the cyanobacterial RecAs group with those of the high-GC gram-positives in all trees.
Deinococcus/Thermus group
The RecAs of Deinococcus radiodurans and the two Thermus species form a clade with high bootstrap values in all the trees (see Table 2, Fig. 2). Analysis of other data suggests that these species are part of a clade (Ludwig et al. 1994; Weisburg et al. 1989b). However, these sequences do not consistently form a clade in the SS-rRNA trees here (they form a clade only in the dnapars tree (not shown)). Inclusion of additional SS-rRNA sequences allows for better resolution of this clade, probably because of GC content variation among the species (Embley et al. 1993). Thus with the species used here, the RecA trees show resolution of the _Deinococcus_-Thermus group while the SS-rRNA trees do not. This may be due to less of a GC bias in the RecA sequences this in the SS-rRNA sequences, as suggested by Lloyd and Sharp (1993). The RecA analysis also supports previous assertions that this group is one of the deeper branching bacterial phyla (Weisburg et al. 1989b), and shows that RecA has resolution even for deep branches. However, this conclusion relies on the rooting of the RecA tree to the A. pyrophilus sequence which has low support (see above).
Other taxa
There is little resolution in the RecA trees regarding the position of the Thermotoga maritima, Chlamydia trachomatis, and Bacteroides fragilis proteins. These RecA proteins do not show consistent affiliations with any individual sequences or groups (Fig. 2, Fig. 3) and the bootstrap values for their positions in the individual trees are low (Fig. 3). I believe that this is due to these sequences being the only representatives from large phylogenetic groups (Thermotogales, Chlamydia, and Bacteroides, respectively). Using the same sets of sequences as in the RecA trees, the SS-rRNA trees show a similar lack of resolution for sequences that are individual representatives of large groups (in this case, C. trachomatis, B. fragilis, and Borrelia burgdorferi). It would be useful to have more RecA genes from these phylogenetic groups to better determine if the RecA and SS-rRNA based trees are congruent for these bacterial groups. It is interesting that although the specific positions of the T. maritima RecA is ambiguous, it never branches below the _Deinococcus_-Thermus sequences as the T. maritima SS-rRNA does in all the SS-rRNA trees. Thus even if the rooting of the RecA tree with A. pyrophilus is incorrect, the A. pyrophilus and T. maritima RecAs never branch immediately near each other as they do in the SS-rRNA trees. Since the RecA tree appears to be less biased by GC content variation (as suggested by Lloyd and Sharp (1993)) than SS-rRNA analysis, it seems plausible that the close branching of the T. maritima and A. pyrophilus SS-rRNAs may be caused by GC content convergence.
Conclusions
Comparison of phylogenetic results for particular taxa using different genes can help determine what genes are useful for evolutionary studies as well as whether different genes have different histories (as could be caused by lateral transfers). However, in order to make direct comparisons it is important to remove as many variables in the studies of the different genes. For example, many researchers studying bacterial systematics compare phylogenetic trees of particular genes to standard trees of SS-rRNA sequences. Yet when these trees have differences with the SS-rRNA trees it is not always clear whether the differences are due to use of different techniques (SS-rRNA trees tend to be constructed with maximum likelihood methods while such methods are still difficult to apply to large numbers of protein sequences), the inclusion of different sets of species (there are some 3000 SS-rRNA sequences that can be used), or true differences in branching or resolution power of different molecules. In the analysis presented here I have compared phylogenetic trees of RecA and SS-rRNA sequences using similar techniques from essentially the same sets of species. Overall, the branching patterns and powers of resolution of the two molecules are highly similar. The similar branching patterns lend support to the general pattern of bacterial systematics inferred from SS-rRNA sequences. This indicates either that the potential problems with SS-rRNA trees have little effect on phylogenetic results or that the RecA trees are biased in the same ways by these problems. In some cases, the RecA trees have resolution where the SS-rRNA trees do not (e.g., for the monophyly of the Proteobacteria and the grouping of D. radiodurans and the Thermus species) and in other cases the reverse is true -- the SS-rRNA trees have resolution (e.g., the position of T. maritima; the placement of L. pneumophilia within the γ-Proteobacteria and the monophyly of the low-GC gram-positives). The lack of resolution of some of the deep branches in the RecA trees is likely related to the species sampled -- a similar lack of resolution is seen in SS-rRNA trees when using the same species set. Therefore RecA appears to be as good a model for studies of molecular systematics of bacteria as SS-rRNA. It remains to be seem whether some of the unusual patterns in the RecA trees (such as the grouping of the cyanobacteria with the high-GC gram-positives and the branching of T. maritima above the Deinococci-Thermus group) are supported by future studies.
In conclusion I would like to emphasize some of the features of RecA that make it a good choice for molecular systematic studies. Among protein encoding genes RecA is relatively easy to clone from new species -- either by degenerate PCR (e.g., (Duwat et al. 1992a, Duwat et al. 1992b, Dybvig et al. 1992, Dybvig and Woodard 1992, Quivey and Faustoferri 1992)) or functional complementation of the radiation sensitivity of recA mutants from other species (Calero et al. 1994, De Mot et al. 1993, Favre et al. 1991, Gomelsky et al. 1990, Tatum et al. 1993). RecA protein function appears to be conserved in all bacteria and there are similar proteins in eukaryotes and Archaea (Clark and Sandler 1994), although whether these can be used reliably for phylogenetic analysis of all three kingdoms remains to be seen. Like with SS-rRNAs, some regions of RecA are virtually completely conserved between species and other regions are variable even between close relatives. This allows for resolution of relationships among both close and distant relatives. The high conservation of size and sequence among RecAs makes alignments virtually unambiguous, limiting complications due to incorrect assignment of homology. In addition since RecA sequences can be compared at the protein and the DNA level it may be possible to limit problems due to nucleotide composition convergence between species. However, perhaps most importantly, I have shown here that phylogenetic trees of RecA sequences have similar topologies and similar resolution to trees of SS-rRNA sequences from the same species. This not only demonstrates that the genomes of these species are not completely mosaic (these two genes have similar phylogenies) but also that molecular systematics of bacteria is reliable and that RecA comparisons are useful for such molecular systematic studies.
Finally, I would like to suggest two additional reasons why researchers might want to choose RecA for molecular systematic studies. First, the cloning and sequencing of recA genes from new species facilitates the creation of recA mutants which are useful to have for laboratory studies of bacterial species. Also, with the availability of the crystal structure of the E. coli protein and with information about the phenotypes of 100s of recA mutants, I believe RecA can become a model for studies of protein evolution.
Acknowledgments
I would like to thank P. C. Hanawalt for support and encouragement; M. B. Eisen for help with computer analysis; S. Smith and J. Felsenstein for making their computer programs freely available; M. Huang, J. Coleman, A. J. Clark, S. Sandler, W. Finch, and S. Mongkolsuk for making sequences available prior to publication; M. Feldman for the use of computer equipment; D. Pollock, M. B. Eisen, M-I. Benito, H. Hamilton, D. Distel, and J. D. Palmer for helpful discussions and suggestions. I also want to thank A. I. Roca for help in all aspects of this project including and especially the identification of errors in recA sequences and aid in getting access to unpublished sequences. During the course of this research I have received support from a pre-doctoral fellowship from the N. S. F., an N. I. H. grant to P. C. Hanawalt, and a tuition grant from the Woods Hole Institution to attend the Summer Workshop on Molecular Evolution, 1994.
References
- Aigle B, Schneider D, Decaris B. Genbank entry Z30324. 1994. [Google Scholar]
- Akaboshi E, Yip ML, Howard-Flanders P. Nucleotide sequence of the recA gene of Proteus mirabilis. Nucleic Acids Res. 1989;17:4390. doi: 10.1093/nar/17.11.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Angov E, Camerini-Otero RD. The recA gene from the thermophile Thermus aquaticus YT-1: cloning, expression, and characterization. J Bacteriol. 1994;176:1405–1412. doi: 10.1128/jb.176.5.1405-1412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball TK, Wasmuth CR, Braunagel SC, Benedik MJ. Expression of Serratia marcescens extracellular proteins requires recA. J Bacteriol. 1990;172:342–349. doi: 10.1128/jb.172.1.342-349.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles KW, Brunskill EW, Iandolo JJ, Hruska LL, Huang S, Pattee PA, Smiley BK, Yasbin RE. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Gene. 1994;147:13–20. doi: 10.1016/0378-1119(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Berson AE, Peters MR, Waleh NS. Nucleotide sequence of recA gene of Aquaspirillum magnetotacticum. Nucleic Acids Res. 1990;18:675. doi: 10.1093/nar/18.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billman-Jacobe H. Genbank entry X77384. 1994. [Google Scholar]
- Binet M-N, Osman M, Jagendorf AT. Genomic nucleotide sequence of a gene from Arabidopsis thaliana encoding a protein homolog of Escherichia coli RecA. Plant Physiol. 1993;103:673–674. doi: 10.1104/pp.103.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Bruns TD, Szaro TM. Rate and mode differences between nuclear and mitochondrial small-subunit rRNA genes in mushrooms. Mol Biol Evol. 1992;9:836–855. doi: 10.1093/oxfordjournals.molbev.a040760. [DOI] [PubMed] [Google Scholar]
- Burggraf S, Olsen GJ, Stetter KO, Woese CR. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Calero S, Fernandez de Henestrosa AR, Barbe J. Molecular cloning, sequence and regulation of expression of the recA gene of the phototrophic bacterium Rhodobacter sphaeroides. Mol Gen Genet. 1994;242:116–120. doi: 10.1007/BF00277356. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Osman M, Grandoni P, Jagendorf AT. A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci USA. 1992;89:8068–8072. doi: 10.1073/pnas.89.17.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Margulies AD. Isolation and characterization of recombinant-deficient mutants of Escherichia coli. Proc Natl Acad Sci USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Sandler SJ. Homologous genetic recombination: the pieces begin to fall into place. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- Clark AJ. Personal communication. 1995.
- Coleman J. Personal communication. 1995.
- Davis EO, Sedgwick SG, Colston MJ. Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J Bacteriol. 1991;173:5653–62. doi: 10.1128/jb.173.18.5653-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EO, Thangaraj HS, Brooks PC, Colston MJ. Evidence of selection for protein introns in the recAs of pathogenic mycobacteria. EMBO J. 1994;13:699–703. doi: 10.1002/j.1460-2075.1994.tb06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mot R, Laeremans T, Schoofs G, Vanderleyden J. Characterization of the recA gene from Pseudomonas fluorescens OE 28.3 and construction of a recA mutant. J Gen Microbiol. 1993;139:49–57. doi: 10.1099/00221287-139-1-49. [DOI] [PubMed] [Google Scholar]
- De Soete G. A least squares algorithm for fitting additive trees to proximity data. Psychometrika. 1983;48:621–626. [Google Scholar]
- Delwiche CF, Kuhsel M, Palmer JD. Phylogenetic analysis of tufA sequences indicates a cyanobacterial origin of all plastids. Mol Phylogen Evol. 1995;4:110–128. doi: 10.1006/mpev.1995.1012. [DOI] [PubMed] [Google Scholar]
- Dunkin SM, Wood DO. Isolation and characterization of the Rickettsia prowazekii recA gene. J Bacteriol. 1994;176:1777–1781. doi: 10.1128/jb.176.6.1777-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwat P, Ehrlich SD, Gruss A. A general method for cloning recA genes of gram-positive bacteria by polymerase chain reaction. J Bacteriol. 1992a;174:5171–5175. doi: 10.1128/jb.174.15.5171-5175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwat P, Ehrlich SD, Gruss A. Use of degenerate primers for polymerase chain reaction cloning and sequencing of the Lactococcus lactis subsp. lactis recA gene. Appl Environ Microbiol. 1992b;58:2674–2678. doi: 10.1128/aem.58.8.2674-2678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybvig K, Hollingshead SK, Heath DG, Clewell DB, Sun F, Woodard A. Degenerate oligonucleotide primers for enzymatic amplification of recA sequences from gram-positive bacteria and mycoplasmas. J Bacteriol. 1992;174:2729–2732. doi: 10.1128/jb.174.8.2729-2732.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybvig K, Woodard A. Cloning and DNA sequence of a mycoplasmal recA gene. J Bacteriol. 1992;174:778–784. doi: 10.1128/jb.174.3.778-784.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Smith SW, Cavanaugh CM. Phylogenetic relationships of chemoautotrophic bacterial symbionts of Solemya velum Say (Mollusca: Bivalvia) determined by 16S rRNA sequence analysis. J Bacteriol. 1992;174:3416–3421. doi: 10.1128/jb.174.10.3416-3421.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Thomas RH, Williams RAD. Reduced thermophilic bias in the 16S rDNA sequence from Thermus ruber provides further support for a relationship between Thermus and Deinococcus. Syst Appl Microbiol. 1993;16:25–29. [Google Scholar]
- Emmerson PT. Personal communication. 1995.
- Favre D, Cryz SJ, Jr, Viret JF. Cloning of the recA gene of Bordetella pertussis and characterization of its product. Biochimie. 1991;73:235–44. doi: 10.1016/0300-9084(91)90208-i. [DOI] [PubMed] [Google Scholar]
- Favre D, Viret JF. Nucleotide sequence of the recA gene of Bordetella pertussis. Nucleic Acids Res. 1990;18:4243. doi: 10.1093/nar/18.14.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evol. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP version 3.5c. University of Washington; Seattle, WA: 1993. [Google Scholar]
- Fernandez de Henestrosa AR. Genbank entry X82183. 1994. [Google Scholar]
- Finch WM. Personal communication. 1995.
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Fox GE, Stackebrandt E, Hespell RB, Gibson J, Maniloff J, Dyer TA, Wolfe RS, Balch WE, Tanner RS, Magrum LJ, Zablen LB, Blakemore R, Gupta R, Bonen L, Lewis BJ, Stahl DA, Leuhrsen KH, Chen KN, Woese CR. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Fry NK, Warwick S, Saunders NA, Embley TM. The use of 16S ribosomal RNA analyses to investigate the phylogeny of the family Legionellaceae. J Gen Microbiol. 1991;137:1215–1222. doi: 10.1099/00221287-137-5-1215. [DOI] [PubMed] [Google Scholar]
- Fyfe JA, Davies JK. Nucleotide sequence and expression in Escherichia coli of the recA gene of Neisseria gonorrhoeae. Gene. 1990;93:151–156. doi: 10.1016/0378-1119(90)90151-g. [DOI] [PubMed] [Google Scholar]
- Gammie AE, Crosa JH. Co-operative autoregulation of a replication protein gene. Mol Microbiol. 1991;5:3015–3023. doi: 10.1111/j.1365-2958.1991.tb01861.x. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Desalle R, Wheeler W. Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Mol Phylog Evol. 1993;2:152–157. doi: 10.1006/mpev.1993.1015. [DOI] [PubMed] [Google Scholar]
- Gomelsky M, Gak E, Chistoserdov A, Bolotin A, Tsygankov YD. Cloning, sequence and expression in Escherichia coli of the Methylobacillus flagellatum recA gene. Gene. 1990;94:69–75. doi: 10.1016/0378-1119(90)90469-8. [DOI] [PubMed] [Google Scholar]
- Goodman HJ, Woods DR. Molecular analysis of the Bacteroides fragilis recA gene. Gene. 1990;94:77–82. doi: 10.1016/0378-1119(90)90470-c. [DOI] [PubMed] [Google Scholar]
- Gregg-Jolly LA, Ornston LN. Genbank entry L26100. 1994. [Google Scholar]
- Guerry P, Pope PM, Burr DH, Leifer J, Joseph SW, Bourgeois AL. Development and characterization of recA mutants of Campylobacter jejuni for inclusion in attenuated vaccines. Infect Immun. 1994;62:426–432. doi: 10.1128/iai.62.2.426-432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Golding GB, Singh B. Hsp70 phylogeny and the relationship between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1994;39:537–540. doi: 10.1007/BF00173424. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman PD, Carroll JD, Masters CI, Minton KW. Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene. 1994;141:31–37. doi: 10.1016/0378-1119(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Haas R. Genbank entry Z35478. 1994. [Google Scholar]
- Hasegawa M, Hashimoto T. Ribosomal RNA tree misleading? Nature. 1993;361:23. doi: 10.1038/361023b0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Sequence analysis by electronic mail server. Trends Biochem Sci. 1993;18:267–268. doi: 10.1016/0968-0004(93)90179-q. [DOI] [PubMed] [Google Scholar]
- Hillis DM. Approaches for assessing phylogenetic accuracy. Syst Biol. 1995;44:3–16. [Google Scholar]
- Horii T, Ogawa T, Ogawa H. Organization of the recA gene of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:313–317. doi: 10.1073/pnas.77.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WM. Personal communication. 1995.
- Inagaki K, Tomono J, Kishimoto N, Tano T, Tanaka H. Cloning and sequence of the recA gene of Acidiphilium facilis. Nucleic Acids Res. 1993;21:4149. doi: 10.1093/nar/21.17.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M. Personal communication. 1995.
- Jinks-Robertson S, Nomura M. In: Ribosomes and tRNAEscherichia coli and Salmonella typhimurium cellular and molecular biology. Neidhardt FC, editor. American Society for Microbiology; Washington, D.C: 1987. pp. 1358–1385. [Google Scholar]
- Kato R, Kuramitsu S. RecA protein from an extremely thermophilic bacterium, Thermus thermophilus HB8. J Biochem. 1993;114:926–929. doi: 10.1093/oxfordjournals.jbchem.a124278. [DOI] [PubMed] [Google Scholar]
- Kerins SM, Fitzpatrick R, O’Donohue M, Dunican L. Genbank entry X75085. 1994. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- King KW, Woodard A, Dybvig K. Cloning and characterization of the recA genes from Mycoplasma pulmonis and M. mycoides subsp. mycoides. Gene. 1994;139:111–115. doi: 10.1016/0378-1119(94)90532-0. [DOI] [PubMed] [Google Scholar]
- Klenk H-P, Zillig W. DNA-dependent RNA polymerase subunit b as a tool for phylogenetic reconstructions: branching topology of the archaeal domain. J Mol Evol. 1994;38:420–432. doi: 10.1007/BF00163158. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SS, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC. Biochemical and biological function of Escherichia coli RecA protein: behavior of mutant RecA proteins. Biochimie. 1991;73:289–304. doi: 10.1016/0300-9084(91)90216-n. [DOI] [PubMed] [Google Scholar]
- Kryukov VM, Suchkov IY, Sazykin IS, Mishankin BN. Genbank entry X75336. 1993. [Google Scholar]
- Lake JA. The order of sequence alignment can bias the selection of tree topology. Mol Biol Evol. 1991;8:378–385. doi: 10.1093/oxfordjournals.molbev.a040654. [DOI] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S rRNA sequences for phylogenetic analysis. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ, Harrison AP, Stahl DA, Pace B, Giovannoni SJ, Olsen GJ, Pace NR. Evolutionary relationships among sulfur- and iron-oxidizing eubacteria. J Bacteriol. 1992;174:269–278. doi: 10.1128/jb.174.1.269-278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SH. Genbank entry U16739. 1994. [Google Scholar]
- Lecointre G, Philippe H, Van Le HL, Le Guyader H. Species sampling has a major impact on phylogenetic inference. Mol Phylog Evol. 1993;2:205–224. doi: 10.1006/mpev.1993.1021. [DOI] [PubMed] [Google Scholar]
- Lloyd AT, Sharp PM. Evolution of the recA gene and the molecular phylogeny of bacteria. J Mol Evol. 1993;37:399–407. doi: 10.1007/BF00178869. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Kirchhof G, Klugbauer N, Weizenegger M, Betzl D, Ehrmann M, Hertel C, Jilg S, Tatzel R, Zitzelsberger H, Liebl S, Hochberger M, Shah J, Lane D, Wallnöfer PR, Shleifer KH. Complete 23S ribosomal RNA sequences of gram-positive bacteria with a low DNA G plus C content. Syst Appl Microbiol. 1992;15:487–501. [Google Scholar]
- Ludwig W, Neumaier J, Klugbauer N, Brockmann E, Roller C, Jilg S, Reetz K, Schachtner I, Ludvigsen A, Bachleitner M, Fischer U, Schleifer KH. Phylogenetic relationships of bacteria based on comparative sequence analysis of elongation factor TU and ATP-synthase beta-subunit genes. Antonie Van Leeuwenhoek. 1994;64:285–305. doi: 10.1007/BF00873088. [DOI] [PubMed] [Google Scholar]
- Luo J, Burns G, Sokatch JR. Construction of chromosomal recA mutants of Pseudomonas putida PpG2. Gene. 1993;136:263–266. doi: 10.1016/0378-1119(93)90476-j. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. MacClade Version 3. Sinauer Associates, Inc; Sunderland, MA: 1992. [Google Scholar]
- Maidak BL, Larsen N, McCaughey MJ, Overbeek R, Olsen GJ, Fogel K, Blandy J, Woese CR. The ribosomal database project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margraf RL, Roca AI, Cox MM. The deduced Vibrio cholerae RecA amino acid sequence. Gene. 1995;152:135–136. doi: 10.1016/0378-1119(94)00686-m. [DOI] [PubMed] [Google Scholar]
- Martin B, Ruellan JM, Angulo JF, Devoret R, Claverys JP. Identification of the recA gene of Streptococcus pneumoniae. Nucleic Acids Res. 1992;20:6412. doi: 10.1093/nar/20.23.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like ribosomal RNA-coding regions. Gene. 1988;71:491–500. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Michiels J, Vande Broek A, Vanderleyden J. Molecular cloning and nucleotide sequence of the Rhizobium phaseoli recA gene. Mol Gen Genet. 1991;228:486–490. doi: 10.1007/BF00260644. [DOI] [PubMed] [Google Scholar]
- Miyamoto MM, Fitch WM. Testing species phylogenies and phylogenetic methods with congruence. Syst Biol. 1995;44:64–76. [Google Scholar]
- Mongkolsuk S. Personal communication. 1995.
- Murphy RC, Bryant DA, Porter RD, de Marsac NT. Molecular cloning and characterization of the recA gene from the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 1987;169:2739–2747. doi: 10.1128/jb.169.6.2739-2747.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC, Gasparich GE, Bryant DA, Porter RD. Nucleotide sequence and further characterization of the Synechococcus sp. strain PCC 7002 recA gene: complementation of a cyanobacterial recA mutation by the Escherichia coli recA gene. J Bacteriol. 1990;172:967–976. doi: 10.1128/jb.172.2.967-976.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T, Kimoto M, Abe M. Cloning, sequencing, and transcriptional analysis of the recA gene of Pseudomonas cepacia. Gene. 1990;94:83–88. doi: 10.1016/0378-1119(90)90471-3. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Starr EM. High rates of nucleotide substitution in nuclear small-subunit (18S) rDNA from holoparasitic flowering plants. J Mol Evol. 1994;39:62–70. doi: 10.1007/BF00178250. [DOI] [PubMed] [Google Scholar]
- Nomura M, Morgan EA, Jaskunas S. Genetics of bacterial ribosomes. Ann Rev Genet. 1977;11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- Nussbaumer B, Wohlleben W. Identification, isolation and sequencing of the recA gene of Streptomyces lividans TK24. FEMS Microbiol Lett. 1994;118:57–63. doi: 10.1111/j.1574-6968.1994.tb06803.x. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Olsen GJ. Phylogenetic analysis using ribosomal RNA. Meth Enzymol. 1988;164:793–812. doi: 10.1016/s0076-6879(88)64084-5. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: a rRNA approach. Ann Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owttrim GW, Coleman JR. Regulation of expression and nucleotide sequence of the Anabaena variabilis recA gene. J Bacteriol. 1989;171:5713–5719. doi: 10.1128/jb.171.10.5713-5719.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR, Olsen GJ, Woese CR. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986;45:325–326. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- Pitulle C, Yang Y, Marchiani M, Moore ERB, Siefert JL, Aragno M, Jurtshuk PJ, Fox GE. Phylogenetic position of the genus Hydrogenobacter. Int J Syst Bacteriol. 1994;44:620–626. doi: 10.1099/00207713-44-4-620. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Faustoferri RC. In vivo inactivation of the Streptococcus mutans recA gene mediated by PCR amplification and cloning of a recA DNA fragment. Gene. 1992;116:35–42. doi: 10.1016/0378-1119(92)90626-z. [DOI] [PubMed] [Google Scholar]
- Rainey FA, Toalster R, Stackebrandt E. Desulfurella acetivorans, a thermophilic, acetate-oxidizing and sulfur-reducing organism, represents a distinct lineage within the Proteobacteria. Syst Appl Microbiol. 1993;16:373–379. [Google Scholar]
- Ramesar RS, Abratt V, Woods DR, Rawlings DE. Nucleotide sequence and expression of a cloned Thiobacillus ferrooxidans recA gene in Escherichia coli. Gene. 1989;78:1–8. doi: 10.1016/0378-1119(89)90308-9. [DOI] [PubMed] [Google Scholar]
- Rappold CSJ, Klingmueller W. Genbank entry P33037. 1993. [Google Scholar]
- Rensing SA, Maier UG. Phylogenetic analysis of the stress-70 protein family. J Mol Evol. 1994;39:80–86. doi: 10.1007/BF00178252. [DOI] [PubMed] [Google Scholar]
- Ridder R, Marquardt R, Esser K. Molecular cloning and characterization of the recA gene of Methylomonas clara and construction of recA deficient mutant. Appl Microbiol Biotechnol. 1991;35:23–31. doi: 10.1007/BF00180631. [DOI] [PubMed] [Google Scholar]
- Roca AI. Personal communication. 1995.
- Roca AI, Cox MM. The RecA protein: structure and function. Crit Rev Biochem Mol Biol. 1990;25:415–456. doi: 10.3109/10409239009090617. [DOI] [PubMed] [Google Scholar]
- Rothschild LJ, Ragan MA, Coleman AW, Heywood P, Gerbi SA. Are rRNA sequences the Rosetta stone of phylogenetics. Cell. 1986;47:640. doi: 10.1016/0092-8674(86)90505-2. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sancar A, Stachelek C, Konigsberg W, Rupp WD. Sequences of the recA gene and protein. Proc Natl Acad Sci USA. 1980;77:2611–2615. doi: 10.1073/pnas.77.5.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y, Kageyama M. The sequence and function of the recA gene and its protein in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1987;208:412–419. doi: 10.1007/BF00328132. [DOI] [PubMed] [Google Scholar]
- Schoeniger M, Von Haeseler A. A stochastic model for the evolution of autocorrelated DNA sequences. Mol Phylog Evol. 1994;3:240–247. doi: 10.1006/mpev.1994.1026. [DOI] [PubMed] [Google Scholar]
- Selbitschka W, Arnold W, Priefer UB, Rottschafer T, Schmidt M, Simon R, Puhler A. Characterization of recA genes and recA mutants of Rhizobium meliloti and Rhizobium leguminosarum biovar viciae. Mol Gen Genet. 1991;229:86–95. doi: 10.1007/BF00264217. [DOI] [PubMed] [Google Scholar]
- Sievers M, Ludwig W, Teuber M. Phylogenetic positioning of Acetobacter, Gluconobacter, Rhodopila and Acidiphilium species as a branch of acidophilic bacteria in the alpha-subclass of proteobacteria based on 16S ribosomal DNA sequences. Syst Appl Microbiol. 1994;17:189–196. [Google Scholar]
- Smith SW. Genetic Data Environment. Version 2.2a. Harvard Genome Laboratory; Cambridge, MA: 1994. [Google Scholar]
- Smith SW, Overbeek R, Woese CR, Gilbert W, Gillevet PM. The genetic data environment: an expandable GUI for multiple sequence analysis. CABIOS. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- Sogin ML. Evolution of eukaryotic microorganisms and their small subunit ribosomal RNA. Amer Zool. 1989;29:487–500. [Google Scholar]
- Stackebrandt E, Murray RGE, Trüper HG. Proteobacteria classis nov., a name for the phylogenetic taxon that includes ‘purple bacteria and their relatives’. Int J Syst Bacteriol. 1988;38:321–325. [Google Scholar]
- Story RM, Bishop DK, Kleckner N, Steitz TA. Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the RecA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Story RM, Weber IT, Steitz TA. The structure of the E. coli RecA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Stranathan MC, Bayles KW, Yasbin RE. The nucleotide sequence of the recE+ gene of Bacillus subtilis. Nucleic Acids Res. 1990;18:4249. doi: 10.1093/nar/18.14.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroeher UH, Lech AJ, Manning PA. Gene sequence of recA+ and construction of recA mutants of Vibrio cholerae. Mol Gen Genet. 1994;244:295–302. doi: 10.1007/BF00285457. [DOI] [PubMed] [Google Scholar]
- Swofford D. Phylogenetic Analysis Using Parsimony (PAUP) Version 3.0d. Illinois Natural History Survey; Champaign, Ill: 1991. [Google Scholar]
- Tatum FM, Morfitt DC, Halling SM. Construction of a Brucella abortus recA mutant and its survival in mice. Microb Pathog. 1993;14:177–185. doi: 10.1006/mpat.1993.1018. [DOI] [PubMed] [Google Scholar]
- Tayama K, Fukaya M, Takemura H, Okumura H, Kawamura Y, Horinouchi S, Beppu T. Cloning and sequencing the recA+ genes of Acetobacter polyoxogenes and Acetobacter aceti: construction of recA- mutants of by transformation- mediated gene replacement. Gene. 1993;127:47–52. doi: 10.1016/0378-1119(93)90615-a. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky ME, Gammie AE, Crosa JH. Characterization of the recA gene of Vibrio anguillarum. Gene. 1992;110:41–48. doi: 10.1016/0378-1119(92)90442-r. [DOI] [PubMed] [Google Scholar]
- Van De Peer Y, Neefs JM, De Rijk P, De Vos P, De Wachter R. About the order of divergence of the major bacterial taxa during evolution. Syst Appl Microbiol. 1994;17:32–38. [Google Scholar]
- Vawter L, Brown WM. Rates and patterns of base change in the small subunit ribosomal RNA gene. Genetics. 1993;134:597–608. doi: 10.1093/genetics/134.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh TV, Das HK. The Azotobacter vinelandii recA gene: sequence analysis and regulation of expression. Gene. 1992;113:47–53. doi: 10.1016/0378-1119(92)90668-f. [DOI] [PubMed] [Google Scholar]
- Viale AM, Arakaki AK, Soncini FC, Ferreyra RG. Evolutionary relationships among eubacterial groups as inferred from GroEL (chaperonin) sequence comparisons. Int J Syst Bacteriol. 1994;44:527–533. doi: 10.1099/00207713-44-3-527. [DOI] [PubMed] [Google Scholar]
- Wardhan H, McPherson MJ, Harris CA, Sharma E, Sastry GR. Molecular analysis of the recA gene of Agrobacterium tumefaciens C58. Gene. 1992;121:133–6. doi: 10.1016/0378-1119(92)90171-k. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Dobson ME, Samuel JE, Dasch GA, Mallavia LP, Baca O, Mendelco L, Sechrest JE, Weiss E, Woese CR. Phylogenetic diversity of the Rickettsiae. J Bacteriol. 1989a;171:4202–4206. doi: 10.1128/jb.171.8.4202-4206.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Giovannoni SG, Woese CR. The Deinococcus-Thermus phylum and the effect of rRNA composition on phylogenetic tree construction. Syst Appl Microbiol. 1989b:128–134. doi: 10.1016/s0723-2020(89)80051-7. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Tully JG, Rose DL, Petzel JP, Oyaizu H, Yang D, Mandelco L, Sechrest J, Lawrence TG. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989c;171:6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur JG, Wong DM, Ortiz B, Tong J, Reichert F, Gelfand DH. Cloning, sequencing, and expression of RecA proteins from three distantly related thermophilic eubacteria. J Biol Chem. 1994;269:25928–25935. [PubMed] [Google Scholar]
- Woese CR. The use of ribosomal RNA in reconstructing relationships among bacteria. In: Selander RK, Clark AG, Whittam TS, editors. Evolution at the molecular level. Sinauer Associates, Inc; Sunderland, MA: 1991. pp. 1–24. [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Achenbach L, Rouviere P, Mandelco L. Archaeal phylogeny: reexamination of the phylogenetic position of Archaeoglobus fulgidus in the light of certain coposition-induced artifacts. Syst Appl Microbiol. 1991;14:364–371. doi: 10.1016/s0723-2020(11)80311-5. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Katz-Downie DS, Morden CW, Palmer JD. Evolution of the plastid ribosomal RNA operon in a nongreen parasitic plant: accelerated sequence evolution, altered promoter structure, and tRNA pseudogenes. Plant Mol Biol. 1992;18:1037–1048. doi: 10.1007/BF00047707. [DOI] [PubMed] [Google Scholar]
- Yao W, Vining LC. Cloning and sequence analysis of a recA-like gene from Streptomyces venezuelae ISP5230. FEMS Microbiol Lett. 1994;118:51–56. doi: 10.1111/j.1574-6968.1994.tb06802.x. [DOI] [PubMed] [Google Scholar]
- Zhang D, Fan H, McClarty G, Brunham RC. Genbank entry U15281. 1994. [Google Scholar]
- Zhao X, Dreyfus LA. Expression and nucleotide sequence analysis of the Legionella pneumophila recA gene. FEMS Microbial Lett. 1990;70:227–232. doi: 10.1111/j.1574-6968.1990.tb13983.x. [DOI] [PubMed] [Google Scholar]
- Zhao XJ, McEntee K. DNA sequence analysis of the recA genes from Proteus vulgaris, Erwinia carotovora, Shigella flexneri and Escherichia coli B/r. Mol Gen Genet. 1990;222:369–376. doi: 10.1007/BF00633842. [DOI] [PubMed] [Google Scholar]
- Zulty JJ, Barcak GJ. Structural organization, nucleotide sequence, and regulation of the Haemophilus influenzae rec-1+ gene. J Bacteriol. 1993;175:7269–7. doi: 10.1128/jb.175.22.7269-7281.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]