Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA (original) (raw)
Abstract
A cellular protein, previously described as p35/38, binds to the complementary (−)-strand of the leader RNA and intergenic (IG) sequence of mouse hepatitis virus (MHV) RNA. The extent of the binding of this protein to IG sites correlates with the efficiency of the subgenomic mRNA transcription from that IG site, suggesting that it is a requisite transcription factor. We have purified this protein and determined by partial peptide sequencing that it is heterogeneous nuclear ribonucleoprotein (hnRNP) A1, an abundant, primarily nuclear protein. hnRNP A1 shuttles between the nucleus and cytoplasm and plays a role in the regulation of alternative RNA splicing. The MHV(−)-strand leader and IG sequences conform to the consensus binding motifs of hnRNP A1. Recombinant hnRNP A1 bound to these two RNA regions in vitro in a sequence-specific manner. During MHV infection, hnRNP A1 relocalizes from the nucleus to the cytoplasm, where viral replication occurs. These data suggest that hnRNP A1 is a cellular factor that regulates the RNA-dependent RNA transcription of the virus.
Mouse hepatitis virus (MHV), a coronavirus, is an enveloped, single-stranded, (+)-sense RNA virus with an RNA genome of ≈31 kb (1, 2). MHV synthesizes multiple subgenomic mRNAs, which have a 3′-coterminal, nested-set structure (3). Each mRNA consists of a leader sequence (60–70 nt), which is derived from the 5′-end of the viral RNA genome, and coding sequences that start from the various consensus transcription-start signals, termed “intergenic” (IG) sequences. The leader RNA is often derived from a separate RNA molecule (_trans_-acting) and fused to mRNAs by a discontinuous transcription mechanism (4–6). This discontinuous RNA transcription appears to be regulated by several viral RNA elements, including _cis_- and _trans_-acting leader RNAs (4, 5), IG sequence (7), and 3′-end untranslated sequence (8). A considerable body of biochemical evidence suggests that these RNA elements may interact directly or indirectly with each other to regulate RNA transcription (9). No sequence complementarity is apparent among some of these RNA elements, so these interactions may be mediated by protein–RNA and protein–protein interactions, involving either viral or cellular proteins. Indeed, several cellular proteins that bind to these regulatory RNA regions have been detected (10, 11); the binding of these cellular proteins may be important for viral RNA transcription or replication (11, 12). Studies of other (+)-strand RNA viruses also have detected several cellular proteins that bind to viral RNAs, and these cellular proteins appear to be important for viral replication, transcription, or translation. For example, in Sindbis virus, the La protein binds to the 3′-end of (−)-strand of RNA and may regulate the efficiency of viral RNA replication (13). In HIV (HIV-1), the La protein binds to the 5′ leader RNA and alleviates translational repression (14, 15). In Rubella virus, calreticulin binds to the 3′-end of viral genome and may be required for (−)-strand RNA synthesis (16). These cellular proteins that interact with viral RNA may fulfill the roles of transcription factors for viral RNA-dependent RNA synthesis.
Previously, three cellular proteins (p35/38, p48, and p70) were detected binding to the (−) (complementary)-strand of the MHV leader RNA and IG sequences by UV crosslinking (10, 12). Because (−)-strand RNA is the template for viral RNA synthesis and both the leader and IG regions are critical for the regulation of viral mRNA synthesis, these cellular proteins may be involved in mRNA transcription. Indeed, site-specific mutagenesis of the IG sequences demonstrated that the binding of p35/38 to the (−)-strand of the IG correlates with the efficiency of subgenomic mRNA transcription from that site (12). Thus, cellular protein p35/38 may serve as a transcription factor for MHV mRNA synthesis.
In this study, we report the purification and characterization of the cellular factor p35/38. Peptide sequencing demonstrates that this protein is the heterogeneous nuclear ribonucleoprotein (hnRNP) A1, a nuclear protein involved in alternative splicing of cellular RNAs. Consistent with a role for hnRNP A1 in viral RNA biogenesis, our results show that hnRNP A1 is translocated to the cytoplasm in MHV-infected cells. The specificity of MHV RNA–hnRNP A1 binding and its correlation with mRNA transcription suggest that this nuclear protein participates in MHV RNA transcription.
MATERIALS AND METHODS
Cells and Viruses.
HeLa S3 cells (provided by Cell Culture Center, National Institutes of Health, Bethesda, MD) were grown to midlogarithmic phase for large-scale protein purification. MHV-infected and -uninfected DBT cells (17), a murine astrocytoma cell line, were used for small-scale protein extraction. MHV strain A59 (18) was propagated in DBT cells at a multiplicity of infection of 0.1, and the infected cells were harvested at different time points postinfection.
Preparation of Nuclear and Cytoplasmic Extracts.
Cells were harvested, washed, and resuspended in buffer A (10 mM _N_-2-hydroxyethylpiperazine-_N_′-2-ethanesulfulfonic acid, Hepes·KOH, pH 7.8/10 mM KCl/1.5 mM MgCl2/0.5 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/20% glycerol). After incubation on ice for 10 min, the cells were lysed with a type B glass homogenizer. The homogenate was centrifuged (1000 × g, 10 min, 4°C), and the supernatant (cytosolic fraction) and pellet (crude nuclear fraction) were collected separately. Buffer B (300 mM Hepes·KOH, pH 7.8/1.4 M KCl/30 mM MgCl2) was added to the cytosolic fraction (0.1:1 ratio); the extract was precleared by centrifugation (100,000 × g, 60 min, 4°C), and the supernatant (cytoplasmic extract S100) was stored at −70°C. The crude nuclear pellet was resuspended in buffer C (20 mM Hepes⋅KOH, pH 7.8/420 mM NaCl/1.5 mM MgCl2/0.2 mM EDTA/0.5 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/20% glycerol) for 30 min at 4°C. Nuclear debris was removed by centrifugation (4000 × g, 20 min, 4°C), and the clarified nuclear extract was dialyzed against buffer D (20 mM Hepes⋅KOH, pH 7.8/0.1 M KCl/2 mM MgCl2/0.5 mM EDTA/0.5 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/20% glycerol), and stored at −70°C. Protein concentration was determined by Bradford assay (Bio-Rad).
Purification of p35/38.
Nuclear and cytoplasmic fractions from HeLa cells (≈1010 cells) were precipitated using 20–60% ammonium sulfate, resuspended, and dialyzed against buffer D. The resulting extracts were loaded on a Q-Sepharose column (Pharmacia) that was equilibrated in buffer D. Flow-through and wash (2–3 bed volumes) fractions were collected and applied to a heparin agarose (GIBCO) column equilibrated in buffer D. Three bed volumes of buffer D-based linear gradient of 0.1 M-0.8 M KCl were used to elute proteins. UV crosslinking with 32P-labeled (−)-strand leader RNA of MHV was performed after each step to monitor the presence of p35/38. Peak fractions (0.2–0.4 M) were pooled, concentrated by ammonium sulfate precipitation, and resuspended in and dialyzed against one-tenth strength buffer D. The samples were concentrated by freeze-drying and separated by two-dimensional (2-D) electrophoresis based on the nonequilibrium pH gradient electrophoresis procedure of O’Farrell et al. (19) using Ampholine pH 3–10 (Bio-Rad) and pH 7–9 (Pharmacia) at a 1:1 ratio. The second dimension was performed on 10% SDS/PAGE separating gels. The p35/38 was identified by comparing the autoradiogram of the UV-crosslinked 32P-labeled samples with silver-stained gel.
Four protein spots (p35/38) from several 2-D gels were excised and pooled, and partial protein sequencing was performed after tryptic digestion and HPLC separation by the W. M. Keck Foundation Biotechnology Resource Laboratory of Yale University (New Haven, CT).
UV-Crosslinking of RNA–Protein Complex.
UV-crosslinking experiments were carried out as described (10).
Renaturation of Proteins from SDS/Polyacrylamide Gel.
The procedure used was as described (20).
Antibodies and Immunoblots.
hnRNP A1 mAb, 4B10 (21), and 9H10 (22) were kindly provided by Gideon Dreyfuss of the University of Pennsylvania. The polyclonal rabbit antibody against regulator of chromosome condensation 1 was obtained from Transduction Laboratories (Lexington, KY). The polyclonal rabbit antibodies against Sam68 and TFIIB were purchased from Santa Cruz Biotechnology. A chemiluminescence Western blotting kit (Boehringer Mannheim) was used for immunoblots according to the manufacturer’s instructions. For immunofluorescent histochemistry, DBT cells were grown on chamber-slides, infected with MHV-A59, and fixed with 2% formaldehyde in PBS and ice-cold acetone at the indicated times after infection. Fixed slides were incubated overnight with antibody that recognized either hnRNP A1 or Sam68. After incubation with a secondary antibody conjugated with fluorescein, the proteins were visualized by fluorescent confocal microscopy.
RESULTS
Purification of MHV (−)-Strand-Leader-RNA-Binding Protein p35/38.
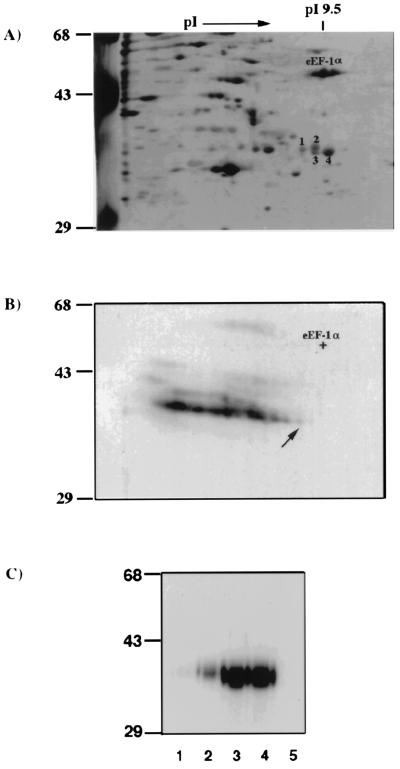
Although p35/38 originally was found in the cytoplasm of a murine cell line (DBT), a similar protein was detected in both nuclear and cytoplasmic fractions of HeLa cells (data not shown). Therefore, HeLa cell lysates were used for protein purification. After three purification steps (ammonium sulfate precipitation and Q-Sepharose and heparin agarose chromatography), partially purified proteins were UV-crosslinked to 32P-labeled (−)-strand leader RNA and analyzed by nonequilibrium 2-D gel electrophoresis. The silver-stained protein profile of the 2-D gel (Fig. 1A) was then compared with the autoradiogram of the UV-crosslinked 32P-labeled proteins (Fig. 1B). A streak of radiolabeled spots ranging from 35 to 38 kDa, each with a correspondingly decreasing pI that may represent different extents of nucleotide linking to the same protein, was detected on the autoradiogram (Fig. 1B). Nucleotide attachment will increase the molecular weight and correspondingly lower the pI values of a protein, so we assumed that the radiolabeled spot with the lowest molecular weight and the highest pI (indicated by an arrow; Fig. 1B) represents the minimally crosslinked protein. This spot corresponded to the spot no. 1 identified on the silver-stained gel (Fig. 1A) and may represent the most likely candidate for p35/38. Three other protein spots, nos. 2–4 (Fig. 1A), are in close proximity to spot no. 1 and could also be potential candidates for, or isoforms of, p35/38. Each of these four protein spots has a pI value very close to that of the translation factor eEF-1α (50-kDa, pI 9.5) (ref. 23; Fig. 1A), and all were detected in both cytoplasmic and nuclear fractions (data not shown). To establish that these four spots were indeed the MHV (−)-strand leader-RNA-binding p35/38, each protein spot and eEF-1α were eluted from the gel, denatured and renatured, and analyzed by UV-crosslinking assay. All four protein spots, but not eEF-1α, were UV-crosslinked with the MHV (−)-strand leader RNA (Fig. 1C). However, the RNA-binding affinity varied among the four protein spots; particularly, spot no. 1, which has the highest molecular weight and lowest pI, had the weakest RNA-binding activity. These results indicate that the four protein spots isolated were likely the cellular factor p35/38 and that they could be related proteins or isoforms of p35/38 because they share similar molecular weight, pI value, and RNA-binding activity.
Figure 1.
Two-dimensional PAGE analysis of the partially purified HeLa cytoplasmic extracts. (A) Silver-stained, 2-D nonequilibrium pH gradient electrophoresis gel of the partially purified p35/38 (spot nos. 1–4) that have been crosslinked to the 32P-labeled (−)-strand leader. Molecular weight markers in kDa are indicated, and eEF-1α is identified. (B) Autoradiogram of the same silver-stained 2-D gel. The arrow indicates the radiolabeled spot that matches spot no. 1 in the silver-stained gel. (C) UV-crosslinking of gel-purified spots 1–4 (lanes 1–4) and eEF-1α (lane 5) (0.5 μg each) with 32P-labeled (−)-strand leader RNA. The products were resolved by SDS/PAGE on a 10% polyacrylamide gel and autoradiographed.
Identification of p35/38 as hnRNP A1.
The identities of these four spots were determined by amino acid composition analysis and partial peptide sequencing. The amino acid composition results showed that the four proteins share over 90% amino acid similarity and that their HPLC patterns of the tryptic peptides were also very similar (data not shown), suggesting that they were indeed related proteins. Two tryptic peptides from spot no. 2, which were common to the four proteins, were selected for peptide sequencing. The sequences of these two peptides, GGNFGGRSSGPYGGGGQYFA and PRNQGGYGGSSSSSSYGSGRRF, matched the C-terminal sequence (amino acids 267–286 and 288–320) of a hnRNP A1/helix destabilizing protein (24, 25). This protein has been reported to exist in at least two isoforms (26–28), due to alternative splicing, and their molecular weight and pI range from 34–38 kDa and 9.0–9.2, respectively. The amino acid sequence of human hnRNP A1 shows 100% homology to its rat counterpart. These properties are consistent with the properties of p35/38.
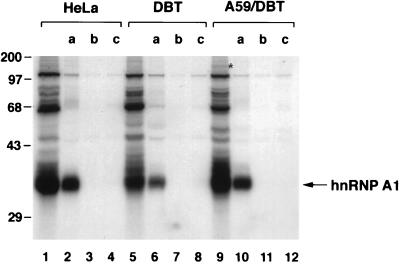
The identity of p35/38 as hnRNP A1 was further confirmed by immunoprecipitation of UV-crosslinked protein with an hnRNP A1 mAb. The cytoplasmic extracts from HeLa and DBT cells were UV-crosslinked with the MHV (−)-strand leader RNA, and the RNA–protein complex was immunoprecipitated by antibodies against different nuclear proteins (hnRNP A1, TFIIB, and Sam68). Only the hnRNP A1-specific mAb, but not the other two antibodies, could precipitate the UV-crosslinked p35/38 (Fig. 2). These data established that the MHV (−)-strand leader RNA-binding protein (p35/38) is indeed hnRNP A1. Several other minor proteins were coprecipitated; the nature of these proteins has not been investigated yet. Significantly, when the cytoplasmic extract from the A59-infected DBT cells was used, hnRNP A1 also was shown to bind to the (−)-strand leader RNA. An additional protein, p150, was found in the MHV-infected cells (Fig. 2, lane 9). It has not been determined whether this is a viral protein.
Figure 2.
Immunoprecipitation of UV-crosslinked proteins. Cytoplasmic extracts from HeLa (lanes 1–4), DBT (lanes 5–8), and A59-infected DBT cells (lanes 9–12) were UV-crosslinked with 32P-labeled (−)-strand leader RNA. Different antibodies were used for immunoprecipitation, and the precipitated proteins were analyzed by SDS/PAGE, a, mAb for hnRNP A1, 4B10; b, polyclonal antibody for TFIIB; and c, polyclonal antibody for Sam68. The asterisk indicates the potential protein specific for MHV-infected cells.
RNA-Binding Specificity of Recombinant Human and Murine hnRNP A1.
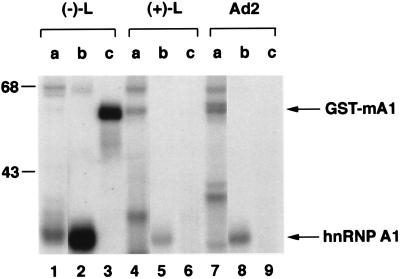
To test whether the RNA-binding of hnRNP A1 is specific to MHV (−)-strand leader RNA, we used two other RNAs, i.e., MHV (+)-strand leader RNA and adenovirus early pre-mRNA (a gift from S. Tahara), to perform a UV-crosslinking assay using recombinant proteins [human hnRNP A1 and glutathione _S_-transferase (GST)-fused murine hnRNP A1] expressed in Escherichia coli. The results showed that both of the recombinant hnRNP A1 proteins bound strongly to (−)-strand leader RNA (Fig. 3, lanes 1–3) but only minimally or not at all to the (+)-strand leader sequence (Fig. 3, lanes 4–6). This binding specificity was the same as that described for the p35/38 of HeLa or DBT cytoplasmic extracts (10). The recombinant human hnRNP A1 bound to a small degree to the adenovirus early pre-mRNA (Fig. 3, lane 8), which is consistent with the report that hnRNP A1 binds to the pre-mRNA splicing sites (22). Nevertheless, its binding to the (−)-strand leader RNA was much stronger than to the adenovirus pre-mRNA.
Figure 3.
UV-crosslinking of recombinant hnRNP A1 with various RNAs. Three different RNAs: MHV (−)-strand leader, (+)-strand leader, and adenovirus early pre-mRNA were UV-crosslinked with 60 μg of (a) HeLa nuclear extract, (b) E. coli expressed (human) hnRNP A1 lysates, and (c) GST-fused (murine) hnRNP A1 lysates.
Previously, we have demonstrated that the p35/38 also bound to the (−)-strand IG sequence (at about 15-fold lower affinity), and the RNA-binding activity was either diminished or completely abolished by minor substitutions of the IG (12). We, therefore, studied the binding activity of the recombinant hnRNP A1 to the various IG sequences.
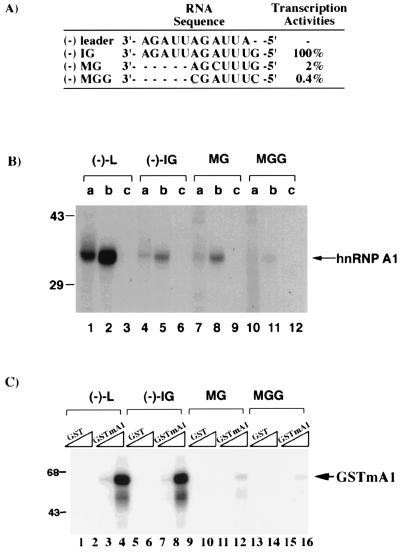
The result showed that both the recombinant human and murine hnRNP A1 bound to the wild-type IG sequence (Fig. 4 B and C). In contrast, very little recombinant hnRNP A1 bound to the MGG mutant RNA, which has two nucleotide substitutions within the IG region (Fig. 4A). The binding of the recombinant hnRNP A1 to the MG mutant RNA, which has one nucleotide substitution, was variable: The recombinant human hnRNP A1 bound the MG mutant to a similar extent to the wild-type IG (Fig. 4B) whereas the binding of GST–hnRNP A1 (murine) fusion protein was relatively weak (Fig. 4C), suggesting that the conformation of the protein may affect its binding to this mutant RNA. In the negative controls (GST protein or E. coli lysate without hnRNP A1), no RNA-binding activity was detected. These results demonstrated that the binding of hnRNP A1 to the MHV RNA is sequence-specific and that the binding specificity resides mainly in the IG consensus sequence.
Figure 4.
The specificity of binding between the recombinant hnRNP A1 and IG sequences. The sequences of the (−)-strand leader and wild-type and mutant IG are shown. (A) The relative transcription efficiencies from the various IG sites were derived from Zhang and Lai (12). (B) 32P-labeled RNA probes were UV-crosslinked with 60 μg of (a) HeLa nuclear extract, (b) E. coli expressed (human) hnRNP A1 lysates, and (c) E. coli (vector) lysates. (C) Similar to (B), except that a GST or GST-hnRNP A1 (murine) fusion protein (GSTmA1) was used. Two different concentrations (2 and 10 μg) of each protein were used for UV crosslinking.
The Similarity Between the Leader and IG Sequences and the Consensus RNA Motifs for hnRNP A1 Binding.
hnRNP A1 has been shown to contain two RNA-binding domains (RBDs) at its N terminus, followed by an RGG box and a glycine-rich C terminus (29). The consensus binding sequence for each RBD in hnRNP A1 has been determined by selection/amplification of random RNA sequences (22). Sequence comparison among some of the selected RNA aptamers and the sequences of the (−)-strand RNA of the leader and IG revealed a remarkable similarity between them (Table 1). These results further suggest that hnRNP A1 binding to the (−)-leader and (−)-IG is sequence-specific. Of interest, the sequences of RNA aptamers selected using RBD I and RBD II separately also match the (−)-leader and the (−)-IG, respectively, suggesting that an hnRNP A1 molecule has the potential to bind both RNA regions simultaneously.
Table 1.
Sequence comparison among the leader, IG sequences, and the selected RNA aptamers of hnRNP A1
| Protein domains of hnRNP A1 | RNA aptamers selected |
|---|---|
| A | |
| RBD I-RBD II-gly-rich | -UAGGGU- |
| (U) | |
| RBD I-gly-rich | -UAGUUUAUU- |
| RBD II-gly-rich | -UUUUAGGUCAG- |
| (−) leader RNA | 5′-UUAGAUUAGA-3′ |
| (−) IG | 5′-GUUUAGA-3′ |
The Relocalization of hnRNP A1 in MHV-Infected Cells.
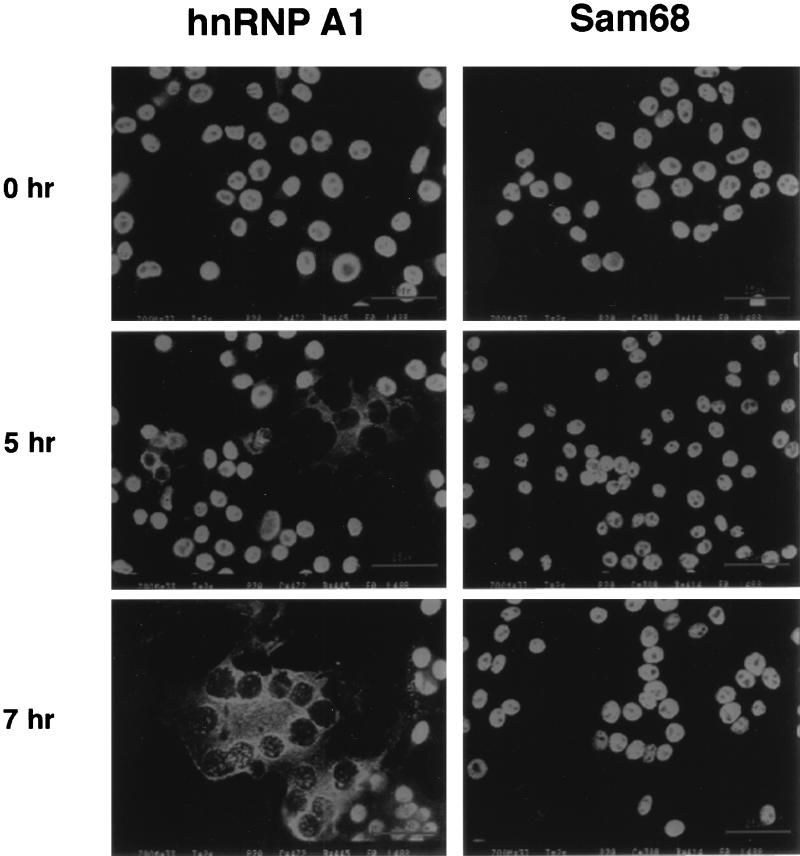
hnRNP A1 is primarily a nuclear protein although it shuttles between the nucleus and the cytoplasm (30). MHV replicates in the cytoplasm, so we were interested in knowing whether the subcellular localization of hnRNP A1 was altered by MHV infection. The immunofluorescence staining of hnRNP A1 in DBT cells at different times after infection is shown in Fig. 5. In uninfected cells, hnRNP A1 was found in the nucleus; however, by 5 h postinfection, the intensity of cytoplasmic hnRNP A1 had increased, most of which was concentrated in the perinuclear region. By 7 h postinfection, when viral RNA synthesis was at the peak level, most of hnRNP A1 was localized to the cytoplasm in the syncytia cells, with some remaining in the nucleus (Fig. 5). In contrast, another nuclear protein Src-associated in mitosis (Sam68), which has been reported to relocalize from the nucleus to cytoplasm upon poliovirus infection (31), remained in the nucleus throughout the infection (Fig. 5). Similarly, a third nuclear protein, regulator of chromosome condensation 1, also was shown to remain in the nucleus throughout the viral infection (data not shown). These results are consistent with the interpretation that hnRNP A1 relocalization occurred not as a result of general leakage of nuclear proteins in the infected cells but rather of a specific interaction with MHV RNA.
Figure 5.
The relocalization of hnRNP A1 in MHV-infected cells. Immunofluorescent staining of MHV-infected cells (×200 magnification) at various time points postinfection. hnRNP A1 or Sam68 antibodies were used.
DISCUSSION
In this report, we have demonstrated that hnRNP A1 is the protein that binds to the (−)-strand leader and IG sequence of MHV RNA. Previous studies have shown that the binding of this protein correlates with the subgenomic RNA transcription from the IG site (12), suggesting that hnRNP A1 is involved in MHV RNA synthesis. Thus, hnRNP A1 can be considered a transcription factor for MHV RNA-dependent RNA synthesis. hnRNP A1 has many characteristics compatible with its putative role in MHV RNA transcription: First, it has three RNA-binding domains, RBD-I, RBD-II, and an RGG motif, whose binding specificity matches remarkably well with the (−)-strand sequence of the leader RNA and IG. Furthermore, site-specific mutations of the IG sequence showed good correlation between hnRNP A1 binding and the efficiency of viral mRNA transcription (12). Second, it has a glycine-rich domain, which mediates protein–protein interactions. hnRNP A1 has been shown to form oligomers (32), indicating that it can interact with itself and with other proteins (33). This, as well as the presence of multiple RNA-binding elements in hnRNP A1, may allow hnRNP A1 to bring various MHV RNA components together and recruit other proteins to form an RNA–protein complex. Third, hnRNP A1 has both DNA and RNA helix destabilizing activity (25), and it can promote annealing of complementary RNA strands (34). These properties may allow the template RNA to be unwound and also promote the leader RNA binding to the (−)-strand IG sequence to carry out RNA transcription. There are at least four isoforms of hnRNP A1, all of which bind to the MHV (−)-strand leader RNA to certain extents (Fig. 1C). Whether there is any functional difference among these isoforms is not clear. It is interesting to note that spot no. 1, which is the most acidic and has the highest apparent molecular weight (Fig. 1A), crosslinked to the MHV RNA most poorly (Fig. 1C). This is probably the phosphorylated form of hnRNP A1; indeed, phosphorylation of hnRNP A1 has been shown to reduce its RNA-binding activity (35).
The fact that hnRNP A1 is primarily a nuclear protein raises an interesting question concerning the possible role of the nucleus in MHV infection. Previously, it has been shown that MHV can replicate in enucleated cells (36, 37) and also in the presence of actinomycin D (38), suggesting that nuclear functions are not required for viral replication. However, MHV replication in the enucleated cells may be inefficient at best (36). Furthermore, hnRNP A1 normally shuttles between the nucleus and cytoplasm, and actinomycin D treatment blocks the nuclear import of hnRNP A1 and thus causes increased accumulation of hnRNP A1 in the cytoplasm (30). Our studies showed that MHV infection also caused a selective accumulation of hnRNP A1 in the cytoplasm as a result of its relocalization from the nucleus to the cytoplasm. This accumulation of hnRNP A1 in the cytoplasm is not the result of general shut-off of cellular transcription by MHV infection. Previous data showed that the transcription of some cellular genes are actually enhanced during MHV infection (39). Our Northern blot analyses also indicated that the levels of three cellular mRNAs, including those encoding glyceraldehyde-3-phosphate dehydrogenase, hnRNP K, and hnRNP A1, remained constant even at 12 h postinfection (data not shown). Therefore, the hnRNP A1 relocalization in the MHV-infected cells most likely is not due to the inhibition of the transcription of a cellular factor required for hnRNP A1 nuclear import. This finding thus suggests that nuclear hnRNP A1 may be the source of a critical factor for MHV replication. In this regard, it is worth pointing out that several other viruses that replicate in the cytoplasm also bind some nuclear proteins, including poliovirus RNA, which binds to polypyrimidine tract-binding protein (40); poliovirus 3D polymerase, which binds to Sam68 (31); and Sindbis viral RNA, which binds to the La protein (13). Thus, the nucleus may play a more active role in RNA virus replication and translation than previously realized. It is possible that the relocalization of hnRNP A1 from the nucleus to the cytoplasm may perturb normal cellular functions, such as alternative RNA splicing or nuclear RNA export, thus contributing to shutdown of host functions and cytopathology of the infected cells.
The correlation between the degree of hnRNP A1 binding to MHV RNA and the efficiency of MHV mRNA transcription suggests the role of hnRNP A1 in MHV transcription. Two possible models can explain this involvement: One, the binding of hnRNP A1 to the leader and the IG sequence on the (−) template strand could bring these two different RNA domains into close proximity to form a “(−)-leader-IG-hnRNP A1 complex” through RNA–protein and protein–protein interactions. Such an interaction has been suggested from previous biochemical studies of the _cis_-acting sequence for MHV transcription (4, 6). This complex may further interact with viral RNA polymerase and other factors to form a transcription initiation complex for (+)-strand subgenomic mRNA synthesis. The strand-annealing-promoting activity of hnRNP A1 (34) may facilitate an annealing between (−)-strand IG sequence and the (+)-strand leader during initiation of (+)-strand subgenomic mRNA synthesis because these two RNA regions are complementary. Such an annealing has been proposed as the basis of MHV subgenomic mRNA transcription (3). Alternatively, the “(−)-leader-IG-hnRNP A1 complex” may interact with cellular splicing factors, causing a splicing of the full length (−)-strand RNA to generate (−)-strand subgenomic mRNAs. The resulting (−)-strand subgenomic mRNAs will become the templates for (+)-strand subgenomic mRNA synthesis. Such subgenomic (−)-strand RNAs indeed have been detected (41). However, the latter model will involve different splicing sequences from the conventional RNA splicing donor and acceptor sequences. Additional experiments will be required to distinguish between these two models. In either case, hnRNP A1 is likely to play a significant role in regulating MHV RNA transcription.
Acknowledgments
We thank Dr. Gideon Dreyfuss for valuable comments and for providing the hnRNP A1 mAbs and recombinant hnRNP A1 clone and Daphne Shimoda for assistance in preparing the manuscript. This work was supported by Grant 19244 from the National Institutes of Health. M.M.C.L. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
MHV
mouse hepatitis virus
IG
intergenic
hnRNP
heterogeneous nuclear ribonucleoprotein
2-D
two-dimensional
GST
glutathione _S_-transferase
RBD
RNA-binding domain
References
- 1.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Bagdzyahdzhyan A, Lai M M-C. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pachuk C J, Bredenbeek P J, Zoltick P W, Spaan W J M, Weiss S R. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai M M C. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 4.Liao C-L, Lai M M C. J Virol. 1994;68:4727–4737. doi: 10.1128/jvi.68.8.4727-4737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Liao C-L, Lai M M C. J Virol. 1994;68:4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong Y S, Makino S. J Virol. 1994;68:2615–2623. doi: 10.1128/jvi.68.4.2615-2623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino S, Joo M, Makino J K. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Y-J, Zhang X M, Wu R C, Lai M M C. J Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai M M C, Liao C-L, Lin Y-J, Zhang X. Infect Agents Dis. 1994;3:98–105. [PubMed] [Google Scholar]
- 10.Furuya T, Lai M M C. J Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu W, Leibowitz J L. Virology. 1995;214:128–138. doi: 10.1006/viro.1995.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X M, Lai M M C. J Virol. 1995;69:1637–1644. doi: 10.1128/jvi.69.3.1637-1644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pardigon N, Strauss J H. J Virol. 1996;70:1173–1181. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang Y N, Kenan D J, Keene J D, Gatignol A, Jeang K T. J Virol. 1994;68:7008–70020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svitikin Y V, Pause A, Sonenberg N. J Virol. 1994;69:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh N K, Atreya C D, Nakhasi H L. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano N, Fujiwara K, Hino S, Matsumoto M. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 18.Robb J, Bond C W. Virology. 1979;94:352–370. doi: 10.1016/0042-6822(79)90467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Farrell P Z, Goodman H M, O’Farrell P H. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 20.Li W W, Sistonen L, Morimoto R I, Lee A S. Mol Cell Biol. 1994;14:5533–5546. doi: 10.1128/mcb.14.8.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piñol-Roma S, Choi Y D, Matunis M J, Dreyfuss G. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 22.Burd C G, Dreyfuss G. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bravo R, Celis J E. Clin Chem. 1982;28:766–781. [PubMed] [Google Scholar]
- 24.Riva S, Morandi C, Tsoulfas P, Pandolfo M, Biamonti G, Merrill B, Williams K R, Multhaup G, Beyreuther K, Werr H, Herinrich B, Schaefer K P. EMBO J. 1986;5:2267–2273. doi: 10.1002/j.1460-2075.1986.tb04494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cobianchi F, SenGupta D N, Zmudzka B Z, Wilson S H. J Biol Chem. 1986;261:3536–3543. [PubMed] [Google Scholar]
- 26.Buvoli M, Biamonti G, Tsoulfas P, Bassi M T, Ghetti A, Riva S, Morandi C. Nucleic Acids Res. 1988;16:3751–3570. doi: 10.1093/nar/16.9.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biamonti G, Buvoli M, Bassi M T, Morandi C, Cobianchi F, Riva S. J Mol Biol. 1989;207:491–503. doi: 10.1016/0022-2836(89)90459-2. [DOI] [PubMed] [Google Scholar]
- 28.Buvoli M, Cobianchi F, Bestagno M G, Mangiarotti A, Bassi M T, Biamonti G, Riva S. EMBO J. 1990;9:1229–1235. doi: 10.1002/j.1460-2075.1990.tb08230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dreyfuss G, Mantunis M J, Pinol-Roma S, Burd C G. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 30.Piñol-Roma S, Dreyfuss G. Nature (London) 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 31.McBride A E, Schlegel A, Kirkegaard K. Proc Natl Acad Sci USA. 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnett S F, Theiry T A, LeStourgeon W M. Mol Cell Biol. 1991;11:864–871. doi: 10.1128/mcb.11.2.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 34.Amalendra K, Samuel H W. Biochemistry. 1990;29:10717–10722. [Google Scholar]
- 35.Cobianchi F, Calvio C, Stoppini M, Buvoli M, Riva S. Nucleic Acids Res. 1993;21:949–955. doi: 10.1093/nar/21.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilhelmsen K C, Leibowitz J L, Bond C W, Robb J A. Virology. 1981;110:225–230. doi: 10.1016/0042-6822(81)90027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brayton P R, Ganges R G, Stohlman S A. J Gen Virol. 1981;50:457–460. doi: 10.1099/0022-1317-56-2-457. [DOI] [PubMed] [Google Scholar]
- 38.Evans M R, Simpson R W. Virology. 1980;105:582–591. doi: 10.1016/0042-6822(80)90058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stohlman S A, Hinton D R, Cua D, Dimacali E, Sensintaffar J, Hofman F M, Tahara S M, Yao Q. J Virol. 1995;69:5898–5903. doi: 10.1128/jvi.69.9.5898-5903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellen C U, Pestova T V, Litterst M, Wimmer E. J Virol. 1994;68:941–950. doi: 10.1128/jvi.68.2.941-950.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sethna P B, Hung S L, Brian D A. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]