OSM-9, A Novel Protein with Structural Similarity to Channels, Is Required for Olfaction, Mechanosensation, and Olfactory Adaptation inCaenorhabditis elegans (original) (raw)
Abstract
Although cyclic nucleotide-gated channels mediate sensory transduction in olfaction and vision, other forms of sensory transduction are independent of these channels. Caenorhabditis elegans cyclic nucleotide-gated channel mutants respond normally to some olfactory stimuli and to osmotic stimuli, suggesting that these chemosensory responses use an alternative sensory transduction pathway. One gene that may act in this pathway is_osm-9_, which is required for each of these responses as well as a mechanosensory response to nose touch. osm-9_encodes a protein with ankyrin repeats and multiple predicted transmembrane domains that has limited similarity to the_Drosophila phototransduction channels transient receptor potential (TRP) and TRP-like (TRPL). The sequence of OSM-9 and other TRP-like genes reveals a previously unsuspected diversity of mammalian and invertebrate genes in this family. osm-9 is required for the activity of the predicted G-protein-coupled odorant receptor ODR-10, which acts in the AWA olfactory neurons; its similarity to other G-protein-regulated transduction channels suggests that OSM-9 is involved in AWA signaling. osm-9:: GFP fusion genes are expressed in a subset of chemosensory, mechanosensory, and osmosensory neurons. osm-9 also affects olfactory adaptation within neurons that require the cyclic nucleotide-gated channel for olfaction; in these neurons, the gene has a regulatory function and not a primary role in sensory transduction.
Keywords: olfaction, C. elegans, mechanosensation, sensory transduction, signaling pathways, olfactory adaptation, TRP channels
Sensory neurons transduce environmental stimuli via the activation of channels that differ by organism and by sensory system. One important class of sensory channels includes cAMP- and cGMP-gated channels. In the vertebrate visual system, photoactivation of the G-protein-coupled receptor rhodopsin results in closure of cGMP-gated channels (Stryer, 1991). Olfactory transduction in vertebrates also begins with the activation of G-protein-coupled receptors and is mediated by opening of cAMP-gated channels (Reed, 1992). Mice mutant for the olfactory cyclic nucleotide-gated channel fail to generate electrical responses to all tested odorants, although the olfactory neurons are present and have normal passive membrane properties (Brunet et al., 1996).
Cyclic nucleotide-gated channels have also been implicated in chemosensation in the nematode Caenorhabditis elegans. C. elegans senses a large number of volatile attractants with two pairs of olfactory neurons designated AWA and AWC (Bargmann et al., 1993). AWC olfactory responses may be mediated by a putative cyclic nucleotide-gated channel encoded by the tax-2 and_tax-4_ genes (Coburn and Bargmann, 1996; Komatsu et al., 1996). The TAX-2/TAX-4 channel also participates in chemotaxis to water-soluble attractants (Dusenbery et al., 1975) and in thermotaxis (Mori and Ohshima, 1995). TAX-2 and TAX-4 gene fusions are expressed in the sensory neurons that mediate these responses.
By contrast, sensory transduction in the AWA olfactory neurons seems to be independent of cyclic nucleotide-gated channels. The AWA olfactory neurons recognize attractive odorants using G-protein-coupled receptors, including the diacetyl receptor ODR-10 (Sengupta et al., 1996). The cyclic nucleotide-gated channel encoded by tax-2_and tax-4 is not expressed in the AWA olfactory neurons, and_tax-2 and tax-4 are not required for AWA olfactory responses. Thus, despite the similarity in the olfactory function of AWA and AWC neurons, different transduction molecules are required in each neuron. Identifying the alternative transduction molecules is of interest both in understanding this system and in identifying signaling molecules in other sensory neurons in which the transduction mechanisms are unknown, such as the vertebrate vomeronasal olfactory neurons.
Here we describe the molecular characterization of the _osm-9_gene, which functions in AWA-mediated olfactory transduction, in the response to osmotic and nose-touch stimuli and in adaptation to olfactory stimuli. osm-9 affects the sensory neurons that are spared in tax-2 and tax-4 mutants, suggesting that osm-9 functions in an alternative sensory transduction pathway. osm-9 encodes a predicted six-transmembrane domain protein with similarity to the transient receptor potential (TRP) channel in Drosophila. It is expressed in sensory neurons that mediate chemosensory, osmosensory, and mechanosensory functions. The AWC olfactory neurons express both OSM-9 and the predicted C. elegans cyclic nucleotide-gated channel; _osm-9_regulates olfactory adaptation but not olfactory transduction within these neurons.
MATERIALS AND METHODS
Strains and genetics. Wild-type worms were_C. elegans_ variety Bristol, strain N2. Worms were grown at 20°C using standard methods (Brenner, 1974). The following strains were used in this work: MT3567 osm-9(n1516) IV, MT3642_osm-9(n1603)_ IV, CX10 osm-9(ky10) IV, MT3665_osm-9(n1601)_ IV, CX3036 osm-9(ky161) IV, MT6317_osm-9(n2743)_ IV, CX2635 osm-9(ky10) IV; lin-15(n765ts) X, CX2851 lin-1(e1275ts) cha-1(p1152) IV, CB1896_unc-33(e204) dpy-13(e184)_ IV, and CX4 _odr-7(ky4)_X.
_Behavioral assays, statistics, and cell ablations._Population chemotaxis assays were performed as described (Bargmann et al., 1993). The chemotaxis index (CI) was calculated in the following way: CI = (number of animals at attractant) − (number of animals at diluent)/(total number). Osmotic avoidance assays were performed as described by Vowels and Thomas (1994) using 4 m fructose as the repellent. Assays were compared by t tests using the Statview II program. The nose-touch response was assayed as described by Kaplan and Horvitz (1993). At least five animals from each strain were tested for responses to three rounds of 10 nose-touch stimuli each. Wild-type animals responded to >80% of stimuli, whereas animals mutant for any of the six osm-9 alleles responded to <20% of stimuli.
Benzaldehyde avoidance was assayed as described by Troemel et al. (1995). A 20 μl microcapillary tube containing 2–3 μl of benzaldehyde was placed immediately in front of a freely moving adult animal on a thin bacterial lawn, and the time until reversal was scored. If the animal did not reverse within 20 sec, the odorant was removed. Individual animals were tested up to eight times over 2 d, with at least 15 min of rest between two assays. For statistical analysis, assays were scored as positive if the animal reversed at all within the 20 sec interval, and the fraction that reversed was compared using χ2 analysis. This analysis revealed a significant difference between wild-type and osm-9(ky10) animals (p < 0.001), but no significant difference between _osm-9(ky10)_ animals in which the ASH sensory neurons had been killed and intact _osm-9(ky10)_ animals (_p_ > 0.05). Wild-type animals in which the ASH neurons were killed responded similarly to _osm-9(ky10)_animals (Troemel et al., 1995) (data not shown). Benzaldehyde avoidance of tax-2(p691) animals was indistinguishable from that of wild-type animals (data not shown).
ASH sensory neurons were killed with a laser microbeam as described (Bargmann and Horvitz, 1991), and cell deaths were confirmed by the absence of DiO filling of the ASH neurons in adult animals after testing was complete (Herman and Hedgecock, 1990).
Genetic mapping of osm-9. Genetic mapping was performed using the diacetyl chemotaxis defect of osm-9. osm-9(ky10) was mapped to the genetic interval between_cha-1_ and unc-33 on chromosome IV by the following data: osm9(ky10)/lin-1(e1275ts) cha-1(p1152), 50 of 52 Lin non-Unc recombinants segregated_osm-9(ky10)_ animals;osm-9(ky10)/unc-33(e204) dpy-13(e184), 98 of 102 Dpy non-Unc recombinants segregated osm-9(ky10) animals.
Molecular biology methods. Subcloning and general DNA manipulations were performed as described (Sambrook et al., 1989). Nested deletions for DNA sequencing were generated using Exonuclease III (New England Biolabs, Beverly, MA). Subclones were sequenced using the fmol sequencing system (Promega, Madison, WI), as well as the Licor automated sequencing system. Sequence analysis was performed using Geneworks (IntelliGenetics, Mountain View, CA). Sequence comparisons were performed using the BLAST (basic local alignment search technique) network service to search the GenBank database (Altschul et al., 1990).
Germline transformation. Germline transformation (Mello et al., 1991) was performed by coinjecting test DNA at a concentration of 30 ng/μl [or 3 ng/μl in the case of the yeast artificial chromosome (YAC) Y44E2] and lin-15 DNA at a concentration of 50 ng/μl into the gonads of osm-9(ky10);lin-15(n765ts) or lin-15(n765ts) animals. Transgenic lines were recognized by rescue of the _lin-15_multivulval phenotype at 20°C. Multiple independent transgenic lines were established from each injection.
osm-9 genomic and cDNA clones. The YAC Y44E2 spans the gap between unc-33 and cha-1 on the physical map. Y44E2 (∼200 kb in length) was purified by CHEF pulsed field electrophoresis and electroelution and was found to rescue the diacetyl chemotaxis defect of osm-9(ky10). Y44E2 DNA was used to screen approximately 105 plaques of a mixed-stage worm genomic library (A. Kamb, personal communication). Positive phage clones (116) were identified; DNA was prepared from these clones and injected in pools of five phage. A single rescuing phage of ∼20 kb, designated λ2–12, was identified; the smallest rescuing subclone of this phage was designated p[osm-9] and was ∼14 kb in size. Three independent transgenic lines expressing_p[osm-9]_ were found to be rescued for nose-touch responses (A. Hart, personal communication), osmotic avoidance, and diacetyl chemotaxis. Subcloning of the λ2–12 phage further yielded a 7 kb partially rescuing DNA fragment designated p[osm-9Δ]. p[osm-9Δ] was subsequently sequenced on both strands using a combination of Licor automated sequencing of subclones of this fragment and fmol sequencing of nested deletions.
The osm-9 genomic DNA fragment _p[osm-9Δ]_failed to detect cDNA clones in ∼2 × 106plaques of a mixed-stage worm cDNA library. Therefore, _osm-9_cDNAs were isolated using a combination of RT-PCR and 3′-RACE (Life Technologies, Gaithersburg, MD). Total RNA was prepared from mixed-stage N2 worms by LiCl precipitation (M. Finney, personal communication). First-strand cDNA was synthesized as described (Aatsinki et al., 1994) using primers in the third ankyrin motif, the sixth putative transmembrane domain of OSM-9, and the 12th exon. These cDNAs were used as templates for PCR amplification using primers to detect SL1 _trans_-spliced transcripts (Krause and Hirsh, 1987), a primer in the third ankyrin motif, and a primer in the sixth putative transmembrane domain, respectively. Reactions were subjected to additional rounds of amplification using nested primers. The conditions used for RT-PCR were the following: 30 sec at 94°C, 1 min at 52°C, and 1 min at 72°C for 30 cycles, followed by 10 min at 72°C. Bands of the expected sizes of 900 bp, 1 kb, and 400 bp, respectively, were detected and sequenced. 3′-RACE was used to isolate the 3′ end of the OSM-9 cDNA. First strand cDNA was synthesized using an oligo(dT)-based primer; the cDNA was then used as a template for PCR amplification using a primer in the 12th exon. The reaction was subjected to additional rounds of amplification using nested primers. The conditions used for PCR were the following: 30 sec at 94°C, 1 min at 52°C, and 1 min at 72°C for 30 cycles, followed by 10 min at 72°C. A band of 600 bp was obtained and sequenced.
Sequencing of osm-9 alleles. Genomic DNA was isolated from N2, osm-9(ky10), osm-9(ky161),osm-9(n1516), osm-9(n1601),osm-9(n1603), and osm-9(n2743) worms as described (Klein and Meyer, 1993). Fragments of osm-9 genomic DNA were PCR-amplified using primers within introns and sequenced using33P end-labeled primers. At least one strand of the open reading frame of all 14 exons, their splice junctions, and ∼30 bp beyond the 3′ and 5′ boundaries of each open reading frame were sequenced. The exons containing mutations were sequenced on both strands. To define the breakpoints of the n1601 deletion, primers flanking the deletion were used to amplify and sequence the region; the n1601 deletion breakpoints reside in the first and fourth introns with a 12 nucleotide insertion between the breakpoints. The splice junctions are apparently unaffected by the deletion.
Generation of osm-9 expression constructs. All_osm-9:: GFP_ fusions were made using the green fluorescent protein (GFP) expression vector pPD95.77 (Chalfie et al., 1994) (A. Fire, S. Xu, N. Ahnn, and G. Seydoux, personal communication). The GFP gene used in these experiments contains five engineered introns and the S65C mutation (Heim et al., 1995).osm-9 sequences for osm-9:: GFP1, osm-9:: GFP2, and osm-9:: GFP4 were amplified from a subclone of the genomic region in pBluescript. For_osm-9:: GFP1_ and osm-9:: GFP2, PCR was performed using the T7 primer and an osm-9 primer designed to contain a _Bam_HI site at one end. Reaction conditions were 30 sec at 94°C, 1 min at 60°C, and 4 min at 72°C for 15 cycles, followed by 10 min at 72°C. The resulting PCR products were cloned into the _Sal_I and _Bam_HI sites of pPD95.77. Junctions were verified by sequencing.
The transcriptional osm-9:: GFP1 fusion gene was constructed by ligating ∼1.6 kb of upstream promoter sequence and part of the 5′ untranslated region into pPD95.77. The translational fusion osm-9:: GFP2 contained 80 additional nucleotides and extended through the first six amino acids of_osm-9. osm-9:: GFP4_ contained the entire_osm-9_ coding region, with GFP fused in-frame to the last amino acid of OSM-9 at an AvrII site designed into the PCR primer. The_osm-9:: GFP5_ fusion gene was constructed by cloning the Xba_I-AatII fragment of pPD95.77 into the AgeI and AatII sites of the osm-9 gene in p[osm-9]. This creates an in-frame fusion of OSM-9 to GFP at residue 832 of OSM-9 and retains ∼3 kb downstream of the osm-9 stop codon. The_osm-9:: GFP3 fusion gene was constructed by deleting an AatII to Sac_I fragment from_osm-9:: GFP5 and religating the vector.
Transgenic animals were viewed by fluorescence microscopy; cell identifications were made using Nomarski images of the same animals to determine cell position and morphology (White et al., 1986). In all cases, cells were observed in well fed animals raised under uncrowded conditions (Brenner, 1974). All transgenes were expressed in all larval stages and in adults. All neurons expressing_osm-9:: GFP5_ were identified in 12 animals with robust fluorescence; 11 of 12 animals showed expression in ASH. AWA expression but no ASH expression was observed in >20 animals expressing osm-9:: GFP3.
Generation of ZC21.2 expression constructs. Approximately 4 kb of sequence upstream of the predicted start site of the C. elegans trp homolog ZC21.2 was amplified from genomic DNA using a 5′ primer with an introduced _Sph_I site and a 3′ primer with an introduced _Xba_I site. The upstream region extended either to the 10th predicted residue of ZC21.2 or to the 111th predicted residue of ZC21.2; the latter fusion gene included the first two predicted introns of ZC21.2. A third construct included both upstream regions and 2.8 kb downstream of the stop codon of ZC21.2. The amplified fragments were cloned into the expression vector pPD95.79. Expression was examined as described above; all clones gave similar expression patterns (n > 40 animals; at least three independent lines per construct). At least 10 independent animals showing robust expression were specifically determined not to have expression in ASH and AWA.
RESULTS
osm-9 mutants are defective in AWA olfactory responses and ASH avoidance responses
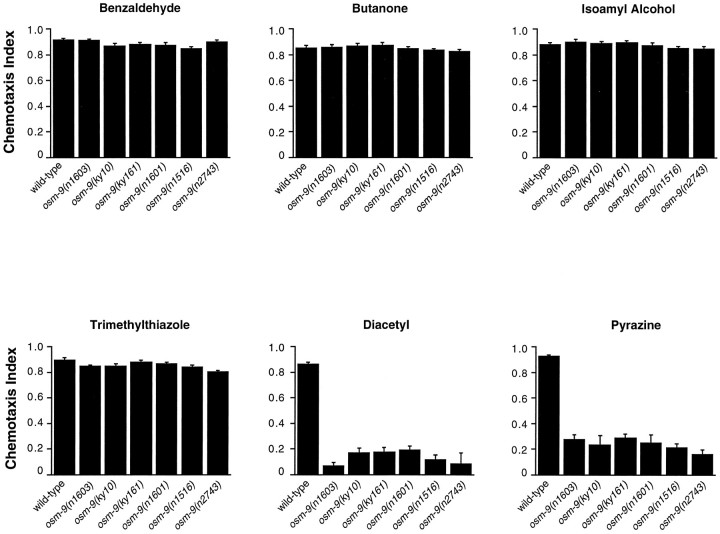
Six recessive alleles of osm-9 have been identified on the basis of defective responses to odorants, high osmotic strength, or light touch to the nose (J. Thomas, J. Kaplan, P. Sengupta, personal communication) (Colbert and Bargmann, 1995). All six_osm-9_ mutants were defective in chemotaxis to the AWA-sensed odorants diacetyl and pyrazine, but were proficient in chemotaxis to the AWC-sensed odorants benzaldehyde, butanone, and isoamyl alcohol (Fig. 1). osm-9 mutants responded normally to trimethylthiazole, which is sensed redundantly by the AWA and AWC neurons. Although osm-9 mutants have robust responses to all concentrations of AWC-sensed odorants, their olfactory adaptation after prolonged exposure to the AWC-sensed odorants isoamyl alcohol and butanone is diminished (Colbert and Bargmann, 1995).
Fig. 1.
osm-9 mutants do not respond to the AWA-sensed odorants diacetyl and pyrazine. Animals were tested for chemotaxis to a point source of each odorant. Chemotaxis Index (CI) = (number of animals at odorant) − (number of animals at diluent)/(total number of animals). A CI of 1.0 indicates complete attraction; a CI of 0 indicates a random distribution of worms on the assay plate. Each data point represents the average of at least six independent assays using 100–200 animals per assay. Error bars represent SEM. Odorants were diluted in ethanol; 1 μl of diluted odorant was used in each assay. Odorants dilutions were 1:200 benzaldehyde, 1:1000 butanone, 1:100 isoamyl alcohol, 1:1000 2,4,5-trimethylthiazole, 1:1000 diacetyl, and 10 mg/ml pyrazine. AWC responses of osm-9(ky10) were normal across a full range of odorant concentrations (Colbert and Bargmann, 1995; and data not shown).
osm-9 mutants displayed normal chemotaxis to the water-soluble attractants sodium chloride and lysine (data not shown). Chemotaxis to sodium chloride is mediated mainly by the ASE sensory neurons, whereas chemotaxis to lysine also involves the ASK sensory neurons (Bargmann and Horvitz, 1991).
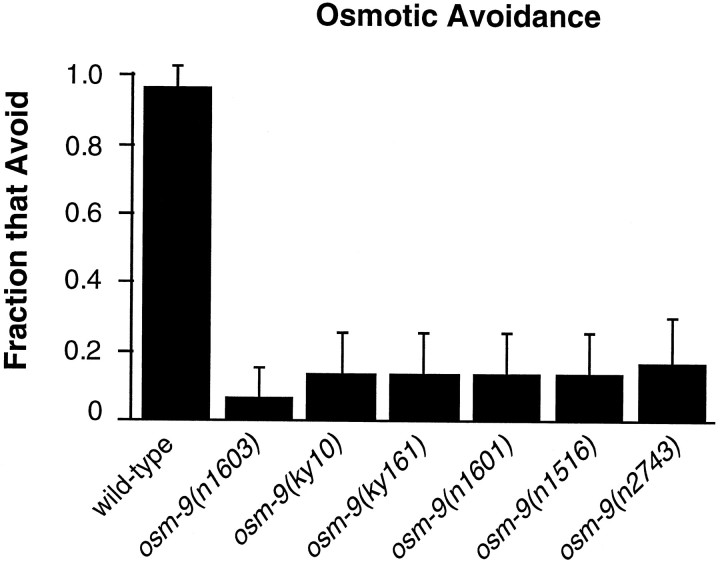
The ASH sensory neurons mediate avoidance responses to high osmotic strength, light touch to the nose, and volatile repellents (Bargmann et al., 1990; Kaplan and Horvitz, 1993; Troemel et al., 1995). Three_osm-9_ alleles were identified in screens for osmotic avoidance-defective mutants (J. Thomas, personal communication), and all six osm-9 mutants displayed defects in the osmotic avoidance response (Fig. 2). One_osm-9_ allele was identified in a screen for nose-touch defective mutants (J. Kaplan, personal communication), and all six_osm-9_ mutants fail to respond to nose-touch stimuli (see Materials and Methods). Avoidance of body touch is mediated by six nonciliated sensory neurons (Chalfie and Sulston, 1981); this response is normal in osm-9 mutants.
Fig. 2.
osm-9 mutants do not avoid high osmotic strength. Animals were placed on an agar plate within a 1-cm-high osmotic strength ring consisting of 10–15 μl of 4m fructose. Assays were scored after 10 min for retention or escape from the ring. Fraction that Avoid = (number of animals retained by the fructose ring)/(total number of animals assayed). A fraction of 1.0 represents complete osmotic avoidance; a fraction of 0 indicates that all animals escaped the ring. Each data point represents responses of at least 30 animals. Error bars denote the 95% confidence intervals.
The avoidance of high concentrations of benzaldehyde depends on the ASH sensory neurons (Troemel et al., 1995), and this response was defective in osm-9(ky10) animals (Table1). Ninety percent of wild-type animals reversed their movement within 20 sec of exposure to a source of undiluted benzaldehyde, whereas only 50% of osm-9 mutants avoided the odorant. Killing the ASH neurons in _osm-9_mutants did not affect their residual benzaldehyde response, indicating that the ASH component of avoidance is absent in the mutants.
Table 1.
Benzaldehyde avoidance in wild-type and _osm-9_animals
| Number of animals | Avoidance responses | p value |
|---|---|---|
| Wild-type | 5 | 0.90 (30/33) |
| p < 0.001 | ||
| osm-9(ky10) | 6 | 0.50 (21/42) |
| osm-9(ky10) | 16 | 0.59 (76/128) |
| p > 0.05 | ||
| osm-9(ky10), ASH killed | 8 | 0.45 (29/64) |
To determine whether osm-9 affects the development or survival of sensory neurons, AWA and ASH neurons were visualized in_osm-9(ky10)_ mutants by using an_odr-7:: GFP_ fusion gene that is expressed only in AWA (Sengupta et al., 1994) and by using DiO filling, which stains ASH and five other anterior sensory neurons (Herman and Hedgecock, 1990). The cell bodies, axons, and dendrites of these neurons were superficially normal in osm-9 mutants (data not shown). We conclude that osm-9 is required for all known functions of AWA and ASH neurons but not for the survival or differentiation of those neurons. However, osm-9 is not essential for the sensory functions of the AWC, ASE, or ASK neurons.
osm-9 encodes a potential channel related to TRP channels
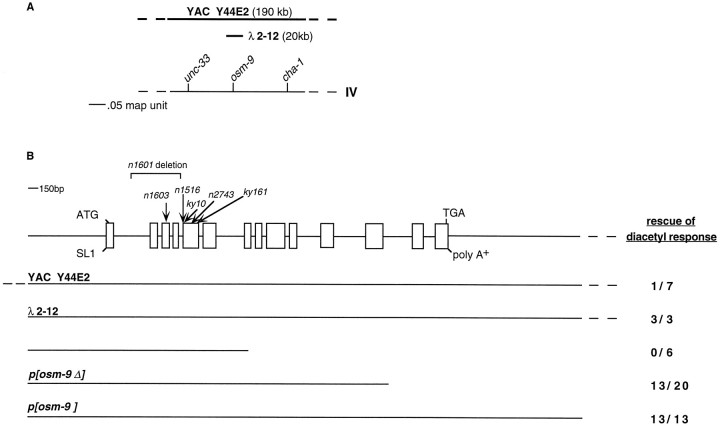
osm-9 was mapped to chromosome IV in the genetic interval between cha-1 and unc-33 (see Materials and Methods). This interval is spanned by the YAC Y44E2, which conferred germline rescue of the osm-9 diacetyl chemotaxis defect (Fig. 3A,B). A genomic phage clone from this region also rescued the olfactory defect of_osm-9_ mutants (λ2–12), as did a 14 kb fragment of this phage (p[osm-9]) (Fig. 3B). A smaller 7 kb fragment designated p[osm-9Δ] partially rescued the diacetyl chemotaxis defect of osm-9. Sequencing and RT-PCR cDNA analysis revealed a single potential osm-9 transcript in this 7 kb region.
Fig. 3.
Cloning of osm-9. A, Genetic map position of osm-9 on chromosome IV. The YAC Y44E2 spans the region between unc-33 and_cha-1_ on the physical map. The phage clone λ2–12 contains the osm-9 coding sequence. B, Localization of osm-9 coding sequence. The genomic organization of osm-9 is depicted, along with the location of the n1601 deletion and the_n1603_, n1516, ky10,n2743, and ky161 point mutations.Boxes indicate exons; SL1 refers to the site of attachment of the trans_-splice leader SL1 (Krause and Hirsh, 1987). The YAC Y44E2, the phage λ2–12, and three subclones of the phage were assayed for their ability to rescue the_osm-9 diacetyl chemotaxis defect. Rescue of diacetyl response = (number of independent transformed lines with a diacetyl chemotaxis index >0.4)/(total number of lines assayed). The subclone p[osm-9] also rescued the_osm-9_ nose-touch defects (A. Hart, personal communication; data not shown) and osmotic avoidance defects (Fig.5E). Rescuing phage and phage subclones contain ∼1.6 kb of upstream sequence; p[osm-9] contains 3 kb of downstream sequence.
osm-9 encodes a predicted protein of 937 amino acids (Fig.4A); the genomic organization of the osm-9 coding region is shown in Figure3B. The OSM-9 protein has six hydrophobic regions that may correspond to membrane-spanning domains but no signal sequence, suggesting that the long amino and C termini are cytoplasmic (Fig.4B). A BLAST search of the GenBank database revealed similarity between osm-9 and the Drosophila TRP channel family in the vicinity of the sixth putative transmembrane domain (Fig. 4C). This region is strongly conserved between TRP family members. The overall structure of OSM-9 is also reminiscent of TRP. Like the Drosophila TRP channel (Montell and Rubin, 1989; Phillips et al., 1992; Wes et al., 1995), OSM-9 contains ankyrin motifs in its amino terminus (Fig. 4D), six predicted membrane-spanning regions, and a hydrophilic C terminal domain. The ankyrin motifs of OSM-9 are no more similar to those of TRP than to the ankyrin consensus motif.
Fig. 4.
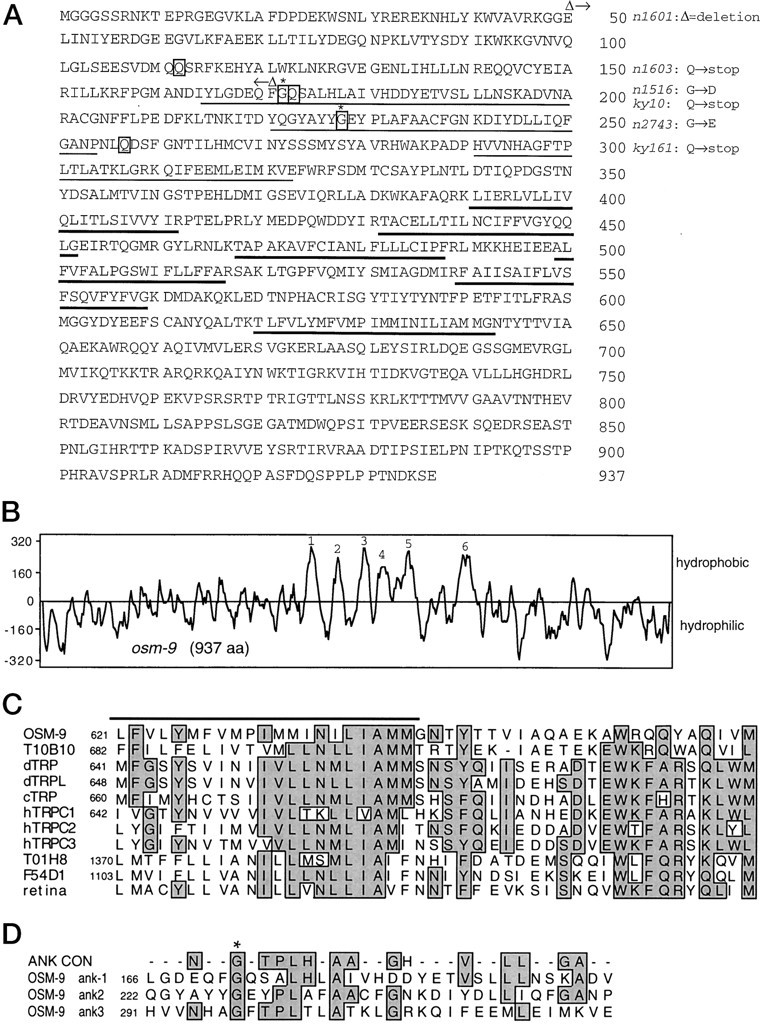
Sequence analysis of OSM-9. A, Predicted amino acid sequence of the osm-9 cDNA. Amino acids are numbered beginning at the first methionine. Underlined regions correspond to the three ankyrin motifs found in_osm-9_. Bold underlined regions correspond to the six putative transmembrane domains identified by hydrophobicity analysis. Boxed amino acid residues denote sites of amino acid substitutions in osm-9 mutant alleles. The corresponding substitutions are identified in the_margin_. The amino acid residues deleted in the_n1601_ allele are bracketed by Δs.B, Hydrophobicity plot of OSM-9 derived by Kyte-Doolittle hydropathic analysis (Kyte and Doolittle, 1982). Predicted transmembrane domains are numbered 1 through_6_. C, Comparison of the sixth predicted transmembrane domains of OSM-9,Drosophila TRP (dTRP),Drosophila TRPL (dTRPL), the C. elegans TRP homolog ZC21.2 (cTRP), and three human TRP genes hTRPC1, -2, and -3 (Wes et al., 1995), the predicted C. elegans genes T10B10.7, T01H8.5, and F54D1.5, and human EST zf57d10.s1 (Soares human retina cDNA). Numbers indicate the amino acid residues shown. Residues shared between at least 5 of the 11 genes are_shaded_. The hydrophobic putative transmembrane domain is_underlined_. All of the predicted C. elegans clones have an overall structure reminiscent of TRP, with six hydrophobic domains; the human cDNAs share this structure as far as they extend. D, Sequence alignment of the OSM-9 ankyrin motifs. A comparison of the three OSM-9 ankyrin motifs with the ankyrin consensus motif (designated ANK CON) is shown (Hatada et al., 1992). Numbers indicate the amino acid residue number at the start of each ankyrin motif. Residues identical with those of the consensus are shaded. An_asterisk_ is placed over the consensus glycine residue that is altered in the first and second ankyrin motifs, respectively, in the n1516 and n2743 alleles.
Additional sequence searches with osm-9 reveal that the TRP channel family is much larger and more divergent than was previously known (Fig. 4C). In addition to other clones from C. elegans, we identified a human retinal expressed sequenced tag (EST) that seems to encode a divergent TRP-like gene. All of these genes have numerous membrane-spanning domains, and all share structural and sequence similarity in the region that is conserved between TRP and OSM-9.
To confirm the identity of osm-9, the coding regions and intron/exon boundaries of the six mutant osm-9 alleles were sequenced (Figs. 3B, 4A). Three potential null alleles result in early termination of the OSM-9 protein. The_n1603_ mutation converts glutamine 112 in the third exon to a stop codon, the ky10 mutation converts glutamine 173 in the fifth exon to a stop codon, and the ky161 mutation converts glutamine 257 in the fifth exon to a stop codon. The n1601_allele represents a deletion of the second, third, and fourth exons of the osm-9 gene. The n1516 and the_n2743 alleles have missense mutations that convert a glycine residue to an aspartate or a glutamate, respectively; these glycine residues occupy analogous conserved positions in the first and second ankyrin motifs of osm-9 (Hatada et al., 1992) (Fig.4D, marked with an asterisk). The missense mutations confer a phenotype similar to that seen in the putative null alleles.
osm-9:: GFP fusion genes are expressed in a subset of sensory neurons
To determine where osm-9 acts to affect sensory responses, we examined its expression pattern by constructing fusions between the GFP reporter gene and osm-9 (Chalfie et al., 1994). These experiments defined three regulatory regions in the_osm-9_ gene: an upstream region that directed expression in ADL and OLQ neurons, a region near the start site that was necessary for expression in AWA neurons, and a region 3′ of the stop codon that directed expression in several additional neurons (Fig.5A; and data not shown). Thus, a transcriptional fusion of GFP with 1.5 kb of the_osm-9_ upstream region was expressed in the two ADL chemosensory neurons and the four OLQ mechanosensory neurons (osm-9:: GFP1) (Fig. 5A; and data not shown), whereas a slightly longer fusion gene including the first six amino acids of OSM-9 was expressed in ADL, OLQ, and the AWA olfactory neurons (osm-9:: GFP2) (Fig. 5A,B). Including the entire osm-9 coding region in the_osm-9:: GFP_ fusion gene gave the same pattern of AWA, ADL, and OLQ expression observed with_osm-9:: GFP2_ (osm-9:: GFP3, osm-9:: GFP4) (Fig. 5A,C; and data not shown). However, an osm-9 fusion gene with ∼3 kb of sequence downstream of the osm-9 stop codon was expressed in numerous additional sensory neurons, including the OLQ and IL2 neurons in the anterior ganglion, the AWA, AWC, ASE, ADF, ASG, ASH, ASI, ASJ, ASK, and ADL neurons in the amphid sensory structure, the FLP and PVD neurons in the body, and the PHA and PHB neurons in the phasmid sensory structure (osm-9:: GFP5) (Fig. 5A,D). OLQ, ASH, FLP, and PVD have mechanosensory functions, whereas the other neurons are either presumed or known to be chemosensory.osm-9:: GFP5 was also expressed in the non-neuronal rectal gland cells and in a few cells in the ventral uterine region.
Fig. 5.
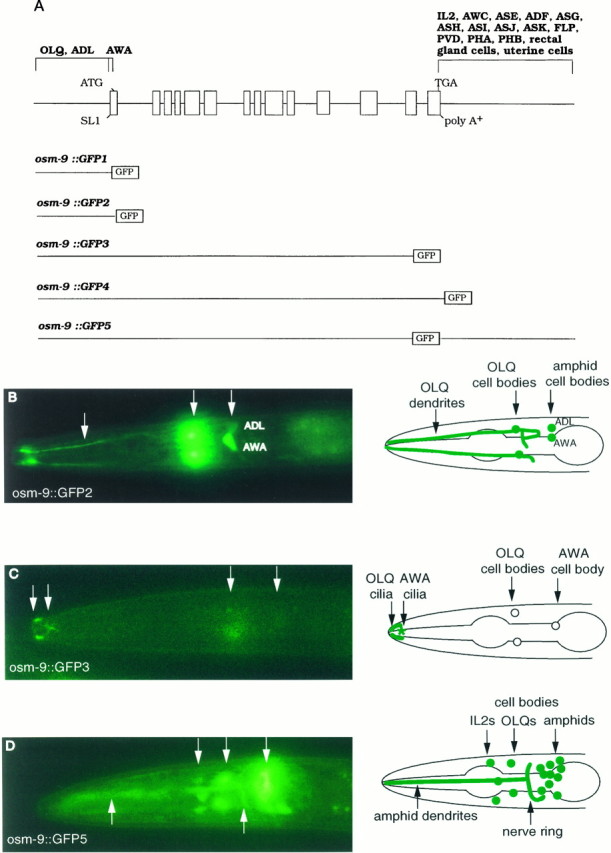
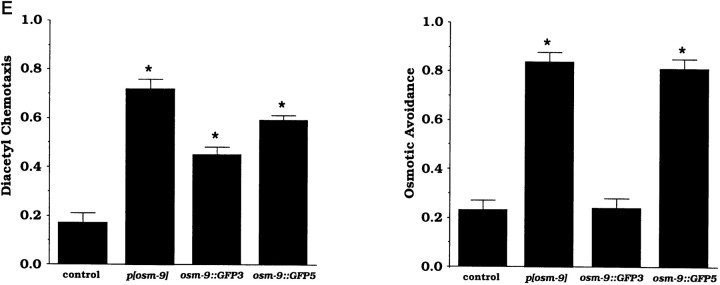
osm-9:: GFP fusion genes are expressed in a subset of sensory neurons. A,osm-9:: GFP fusion genes. Three regions that regulate GFP expression in particular cell types are indicated. All_osm-9:: GFP_ fusion genes were examined in at least three independent transgenic lines. B, A lateral view of the head of a transgenic animal expressing the_osm-9:: GFP2_ fusion gene. GFP is expressed in the four OLQ mechanosensory neurons and in the AWA and ADL amphid chemosensory neurons. All neurons are bilaterally symmetric, and only those on the left side are visible in this focal plane. The fusion protein is present at high levels at the base of the OLQ cilia, but it is only weakly expressed in the cilia themselves.C, A lateral view of the head of a transgenic animal expressing the osm-9:: GFP3 fusion gene, which encodes a GFP-tagged OSM-9 protein. GFP is localized to the OLQ and AWA cilia at the tip of the nose (arrows), but little GFP is present in the cell bodies, axons, or dendrites. ADL expression is weaker or absent in these lines. The fusion gene deletes the last 105 amino acids of OSM-9; when GFP is fused to the last amino acid of OSM-9 (osm-9:: GFP4), a similar but fainter expression pattern is observed. D, A lateral view of the head of a transgenic animal expressing the_osm-9:: GFP5_ fusion gene. GFP expression is present in the OLQ and IL2 sensory neurons and in the AWA, AWC, ASE, ADF, ASG, ASH, ASI, ASJ, ASK, and ADL amphid chemosensory neurons. In more posterior parts of the animal, the FLP, PVD, PHA, and PHB sensory neurons, the ventral uterine cells, and the rectal gland cells also express osm-9:: GFP5. E, Rescue of_osm-9_ chemotaxis and osmotic avoidance by_osm-9:: GFP_ fusion genes. osm-9(ky10); lin-15(n765) animals were transformed with the_lin-15_ plasmid alone (control) or lin-15_plasmid with p[osm-9], osm-9:: GFP3, or_osm-9:: GFP5. Each data point represents 8–24 independent assays. Three independently derived lines of transgenic animals were characterized for osm-9:: GFP3 and six lines for osm-9:: GFP5; all lines gave comparable results (n = 3–4 assays per transgenic line). Error bars represent SEM. Asterisks denote values that are different from the lin-15 control plasmid injection at p < 0.001.
The importance of the 3′ regulatory region was tested further by examining its role in two functions of osm-9: olfaction and osmotic avoidance. The osm-9:: GFP3 and_osm-9:: GFP5_ clones differ from one another only by the presence of the 3′ regulatory region; these clones were tested for rescue of two osm-9 mutant phenotypes (Fig. 5E).osm-9:: GFP5 rescued both diacetyl chemotaxis and osmotic avoidance, consistent with its expression in both AWA and ASH, whereas osm-9:: GFP3 rescued only diacetyl chemotaxis, consistent with its expression in AWA but not ASH. These results indicate that different functions of osm-9 can be rescued by different transgenes and suggest that osm-9 acts cell-autonomously within chemosensory neurons.
The subcellular localization of GFP-tagged OSM-9 protein in the sensory neurons was also examined. The AWA and OLQ neurons extend sensory processes that terminate in cilia near the tip of the nose.osm-9:: GFP3 and osm-9:: GFP4_were strongly localized to the sensory cilia of these two cell types (Fig. 5C; and data not shown). This localization is consistent with a direct role of osm-9 in sensory transduction. However, many of the neurons that expressed_osm-9:: GFP5 did not show the strong enrichment of OSM-9:: GFP protein in the cilia that was observed in OLQ and AWA (compare Fig. 5, C and D).osm-9:: GFP3 and _osm-9:: GFP5_should encode identical proteins, but these proteins may not be localized to the same degree in all sensory neurons.
A C. elegans trp homolog is not coexpressed with osm-9
Many ion channels are heteromultimers composed of several related subunits, leading us to examine the C. elegans gene ZC21.2, which is highly similar to the Drosophila TRP and TRP-like (TRPL) genes (Sulston et al., 1992; Wes et al., 1995) (Fig. 4). To determine whether ZC21.2 might also function in olfactory transduction, expression of several ZC21.2:: GFP fusion genes was analyzed (see Materials and Methods). The upstream and downstream regions of this gene drove GFP expression in a set of cells that did not overlap with those expressing osm-9:: GFP fusions, including motor neurons, interneurons, vulval and intestinal muscles, and a single putative sensory neuron, BAG (Fig.6). Although these fusion gene patterns may be incomplete, the lack of overlap between their expression patterns indicates that osm-9 and ZC21.2 are unlikely to encode two subunits of the same channel.
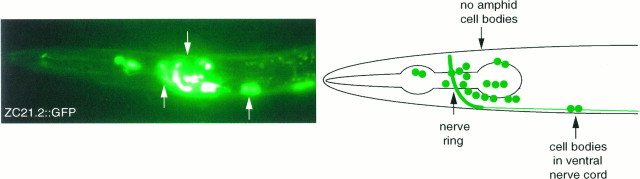
Fig. 6.
A C. elegans trp gene is expressed in motor neurons and muscles. A, Expression of ZC21.2:: GFP. Strong expression is present in all RMD, SMD, SMB, RIA, RIB, and RIM motor neurons in the head, many classes of motor neurons in the ventral nerve cord, the BAG sensory neurons, the AVA and SIA interneurons, two pharyngeal neurons, and the vulval and intestinal muscles. No expression was observed in cells that expressed_osm-9:: GFP_ fusion genes.
DISCUSSION
The TRP-related protein OSM-9 is required for an alternative olfactory transduction pathway
OSM-9 is required for all known functions of the AWA olfactory neurons and the ASH polymodal sensory neurons. Interestingly, ciliated sensory neurons require either osm-9 or the cyclic nucleotide-gated channel genes tax-2 and tax-4 to perform their function (Table 2). Each gene acts in multiple sensory modalities: osm-9 in olfaction, osmosensation, and touch, and tax-2 and_tax-4_ in olfaction, thermosensation, and taste.
Table 2.
osm-9 and tax-2 mutants have complementary behavioral defects
| Sensory modality | Sensory neuron | Defective in_osm-9_ | Defective in tax-2 |
|---|---|---|---|
| Olfaction: diacetyl, pyrazine2-a | AWA | X | |
| Osmotic avoidance2-b | ASH | X | |
| Mechanosensation: nose touch2-c | ASH | X | |
| Volatile avoidance2-d | ASH | X | |
| Olfaction: benzaldehyde, butanone, isoamyl alcohol2-a | AWC | Adaptation to butanone, isoamyl alcohol2-g | X |
| Water-soluble attractants (taste)2-e | ASE, ASK | X | |
| Thermosensation2-f | AFD | X |
The similarity of OSM-9 to TRP channels suggests that it might function as an alternative sensory channel in AWA olfactory neurons. C. elegans olfactory neurons recognize odorants using seven transmembrane domain receptors (Troemel et al., 1995; Sengupta et al., 1996). For example, the AWA neurons sense diacetyl through the predicted G-protein-coupled receptor en- coded by the_odr-10_ gene (Sengupta et al., 1996). ODR-10 fusion proteins are localized to the AWA olfactory cilia, where the OSM-9 protein also resides. These data suggest that OSM-9 acts with ODR-10 and other olfactory receptors in a G-protein-mediated olfactory pathway; however, these genetic experiments do not demonstrate a direct action of OSM-9 in sensory transduction.
In C. elegans, the two major types of olfactory receptor neurons respond to odorants in different ways. The AWC neurons require the TAX-2/TAX-4 cyclic nucleotide-gated channel (Coburn and Bargmann, 1996; Komatsu et al., 1996), whereas the AWA neurons do not require TAX-2 and TAX-4 but do require OSM-9. It is striking that OSM-9 is related to the only other known G-protein-regulated sensory transduction channel, the TRP/TRPL channel that mediates the light-activated conductance in the Drosophila visual system (Peretz et al., 1994a,b; Vaca et al., 1994; Niemeyer et al., 1996). Like OSM-9, the TRP and TRPL proteins have six transmembrane domains and several ankyrin repeats. During phototransduction, activation of the G-protein-coupled receptor rhodopsin leads to stimulation of phospholipase C and an increase in inositol triphosphate (IP3) (Ranganathan et al., 1995). The subsequent mechanism of TRP and TRPL activation is controversial. Some evidence suggests that TRP participates in a capacitative (or store-operated) process, in which depletion of internal IP3-gated calcium stores activates a replenishing plasma membrane conductance (Berridge, 1995; Zhu et al., 1996). When TRP or its human homologs are expressed in heterologous cells, they can induce channels that are activated by thapsigargin, which depletes internal calcium stores (Vaca et al., 1994; Peterson et al., 1995; Zitt et al., 1996). However, thapsigargin treatment does not mimic or abolish photoexcitation in_Drosophila_ photoreceptors (Ranganathan et al., 1994; Hardie, 1996). The precise relationship between TRP, IP3, and the store-operated channels remains elusive.
OSM-9 is unlikely to be the C. elegans TRP homolog, because the C. elegans ZC21.2 gene is much more similar to_trp_ than either is to osm-9 (Wes et al., 1995). Moreover, analysis of C. elegans sequences and mammalian ESTs reveals a greater variety of TRP family members than was previously described. We speculate that _trp_-related genes might share common regulatory mechanisms. One possibility is that OSM-9 is involved in an IP3 signaling pathway, a model that is especially intriguing because IP3 pathways are implicated in some forms of chemosensation. In Drosophila, phospholipase C is required for odor responses (Riesgo-Escovar et al., 1995). Individual lobster olfactory receptor neurons seem to express two different transduction channels: odorant-induced opening of cyclic nucleotide-gated channels results in a hyperpolarizing conductance, whereas odorant-induced opening of plasma membrane IP3-gated channels results in a depolarizing conductance (Fadool and Ache, 1992; Ache and Zhainazarov, 1995). Vertebrate olfactory receptor neurons have odorant-stimulated IP3signaling pathways (Boekhoff et al., 1990; Miyamoto et al., 1992), but their function is unclear (Jaworsky et al., 1995; Brunet et al., 1996). In addition, bitter-tasting compounds stimulate the production of IP3 in vertebrate taste cell membranes (Spielman et al., 1996). Although these data raise interesting possibilities for OSM-9 function, expression of OSM-9 in Xenopus oocytes does not result in the appearance of IP3-gated, thapsigargin-gated, or voltage-gated channels (E. Reuveny, T.L.S., Liqin Tong, C.I.B., and L. Jan, unpublished data).
OSM-9 may participate in olfactory adaptation, but not transduction, in AWC olfactory neurons
osm-9 seems to play a regulatory role in the AWC olfactory neurons, where the cyclic nucleotide-gated channel encoded by_tax-2_ and tax-4 has a more central role in olfaction. When C. elegans is exposed to high concentrations of an odorant for minutes or hours, its response to that odorant is diminished by olfactory adaptation (Colbert and Bargmann, 1995).osm-9 mutants are defective in adaptation to two AWC-sensed odorants, butanone and isoamyl alcohol, but normal in adaptation to the AWC-sensed odorant benzaldehyde (Colbert and Bargmann, 1995). For these responses, osm-9 affects only adaptation and not olfactory acuity: the AWC olfactory responses of naive osm-9 animals are normal across the entire range of odorant concentrations. This_osm-9_ function is reminiscent of the light adaptation function of the Drosophila TRP channel (Minke, 1982). The TRP channel contributes to adaptation of the photoresponse by mediating calcium influx into photoreceptors (Minke, 1982; Peretz et al., 1994a,b; Hardie, 1996). _osm-9-_dependent adaptation differs from TRP adaptation in two respects. First, the light adaptation affected by trp occurs within seconds of light exposure, whereas OSM-9 affects adaptation processes occurring over minutes or hours of odorant exposure. Second, trp contributes both to phototransduction and to adaptation, whereas osm-9 affects only adaptation in AWC neurons.
osm-9:: GFP fusion genes are coexpressed with_tax-2_ and tax-4 in the ASE, ASG, ASI, ASJ, and ASK chemosensory neurons. ASE- and ASK-mediated chemotaxis depend on_tax-2/tax-4_ but not on osm-9; these cells might also use osm-9 for a regulatory function such as adaptation.
Ciliated mechanosensory neurons require_osm-9_ function
osm-9 also functions in one of two alternative pathways for mechanosensation. C. elegans responds to at least two distinct classes of mechanosensory stimuli: body touch and nose touch. The response to body touch is mediated by six nonciliated sensory neurons with processes that run along the body of the worm, under the epidermis (Chalfie and Sulston, 1981). The mechanosensory channel for body touch may be encoded by the mec-4 and _mec-10_genes; MEC-4 and MEC-10 belong to a family of ion channels that includes the mammalian amiloride-sensitive epithelial sodium channel (Driscoll and Chalfie, 1991; Hong and Driscoll, 1994; Huang and Chalfie, 1994). The nose-touch response is primarily mediated by the ciliated amphid neuron ASH, with minor components of the response contributed by the FLP and OLQ ciliated neurons (Kaplan and Horvitz, 1993); all of these neurons express _osm-9:: GFP_fusion genes. mec-4 and mec-10 are not required to sense nose touch, and osm-9 is not required for responses to body touch. On the basis of its phenotype and expression pattern, OSM-9 represents a candidate mechanosensory channel in ciliated touch neurons. It might be directly gated by mechanical stimuli, or it could be regulated by G-protein-coupled receptors; mechanosensation by muscle cells in rat cerebral arteries may proceed through a G-protein-mediated pathway (Osol et al., 1993). Alternatively, osm-9 could indirectly regulate touch responses, for example, by regulating the ionic concentrations in sensory cilia. The localization of the OSM-9:: GFP protein to the OLQ cilia is intriguing, because it is the first potential channel protein seen to be enriched in the sensory apparatus of mechanosensory neurons.
The ASH neurons also sense both volatile repellents and osmotic stimuli, and osm-9 mutants are defective in both of these responses. osm-9 is not related to other putative osmosensory proteins, such as the mscL gene in E. coli, which encodes a mechanosensitive channel (Sukharev et al., 1994), or the yeast SLN1 gene, which encodes a membrane protein with regions of homology to the histidine kinase sensor and the response regulator of bacterial two-component systems (Maeda et al., 1994). In the ASH neurons, osmosensation and mechanosensation can be separated genetically, so they are not identical sensory modalities (Hart et al., 1995; Maricq et al., 1995).
Our observations and those in Drosophila indicate that TRP-related genes have key roles in four invertebrate sensory modalities: vision, olfaction, osmosensation, and mechanosensation. In_C. elegans_, the same gene, osm-9, can have either an essential function or a modulatory function in different sensory neurons. It will be interesting to determine whether related proteins function in some of the sensory modalities that remain poorly understood in vertebrates, such as vomeronasal olfaction, mechanotransduction, and taste.
Footnotes
This work was supported by grants from the Human Frontiers Science Program and the Howard Hughes Medical Institute. C.I.B. is an Assistant Investigator and T.L.S. is a Fellow of the Howard Hughes Medical Institute. We thank Liqin Tong and Shannon Grantner for excellent technical support, Anne Hart for assaying transgenic animals for their nose-touch response, Piali Sengupta for isolating ky161, Josh Kaplan for providing n2743, Jim Thomas for providing n1516, n1603, and_n1601_, Alan Coulson for mapping phage clones, the Kenyon lab for the use of their CHEF pulsed field electrophoresis apparatus, Lisa Wrischnik for advice on YAC isolation, Chen-Ming Fan for cloning advice, Sasha Kamb for the genomic phage library, Andrew Fire for expression vectors, and Noelle Dwyer for injection assistance. Some strains were provided by the Caenorhabditis Genetics Center.
Correspondence should be addressed to Cori Bargmann, Box 0452, Department of Anatomy, The University of California, San Francisco, CA 94143.
REFERENCES
- 1.Aatsinki JT, Lakkakorpi JT, Pietila EM, Rajaniemi HJ. A coupled one-step reverse transcription PCR procedure for generation of full-length open reading frames. Biotechniques. 1994;16:282–288. [PubMed] [Google Scholar]
- 2.Ache BW, Zhainazarov A. Dual second-messenger pathways in olfactory transduction. Curr Opin Neurobiol. 1995;5:461–466. doi: 10.1016/0959-4388(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 3.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 5.Bargmann CI, Thomas JH, Horvitz HR. Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harbor Symp Quant Biol. 1990;55:529–538. doi: 10.1101/sqb.1990.055.01.051. [DOI] [PubMed] [Google Scholar]
- 6.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 7.Berridge MJ. Capacitative calcium entry. Biochem J. 1995;312:1–11. doi: 10.1042/bj3120001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boekhoff I, Tareilus E, Strotmann J, Breer H. Rapid activation of alternative second messenger pathways in olfactory cilia from rats by different odorants. EMBO J. 1990;9:2453–2458. doi: 10.1002/j.1460-2075.1990.tb07422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 11.Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 12.Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 13.Coburn CM, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 14.Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14:803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- 15.Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- 16.Dusenbery DB, Sheridan RE, Russell RL. Chemotaxis-defective mutants of the nematode Caenorhabditis elegans. Genetics. 1975;80:297–309. doi: 10.1093/genetics/80.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fadool D, Ache B. Plasma membrane inositol 1,4,5-trisphosphate-activated channels mediate signal transduction in lobster olfactory receptor neurons. Neuron. 1992;9:907–918. doi: 10.1016/0896-6273(92)90243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardie RC. INDO-1 measurements of absolute resting and light-induced Ca2+ concentration in Drosophila photoreceptors. J Neurosci. 1996;16:2924–2933. doi: 10.1523/JNEUROSCI.16-09-02924.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- 20.Hatada EN, Nieters A, Wulczyn FG, Naumann M, Meyer R, Nucifora G, McKeithan TW, Scheidereit C. The ankyrin repeat domains of the NF-kB precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-kB DNA binding. Proc Natl Acad Sci USA. 1992;89:2489–2493. doi: 10.1073/pnas.89.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 22.Herman R, Hedgecock E. Limitation of the size of the vulval primordium of Caenorhabditis elegans by lin-15 expression in surrounding hypodermis. Nature. 1990;348:169–171. doi: 10.1038/348169a0. [DOI] [PubMed] [Google Scholar]
- 23.Hong K, Driscoll M. A transmembrane domain of the putative channel subunit MEC-4 influences mechanotransduction and neurodegeneration in C. elegans. Nature. 1994;367:470–473. doi: 10.1038/367470a0. [DOI] [PubMed] [Google Scholar]
- 24.Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- 25.Jaworsky DE, Matsuzaki O, Borisy FF, Ronnett GV. Calcium modulates the rapid kinetics of the odorant-induced cyclic AMP signal in rat olfactory cilia. J Neurosci. 1995;15:310–318. doi: 10.1523/JNEUROSCI.15-01-00310.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan JM, Horvitz HR. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1993;90:2227–2231. doi: 10.1073/pnas.90.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein RD, Meyer BJ. Independent domains of the Sdc-3 protein control sex-determination and dosage compensation in C. elegans. Cell. 1993;72:349–364. doi: 10.1016/0092-8674(93)90113-5. [DOI] [PubMed] [Google Scholar]
- 28.Komatsu H, Mori I, Rhee J-S, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 29.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 31.Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 32.Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 33.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minke B. Light-induced reduction in excitation efficiency in the trp mutant of Drosophila. J Gen Physiol. 1982;79:361–385. doi: 10.1085/jgp.79.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto T, Restrepo D, Cragoe EJ, Teeter JH. IP3 and cAMP-induced responses in isolated olfactory receptor neurons from the channel catfish. J Membr Biol. 1992;127:173–183. doi: 10.1007/BF00231505. [DOI] [PubMed] [Google Scholar]
- 36.Montell C, Rubin GM. Molecular characterization of the Drosophila trp locus: a putative integral membrane protein required for phototransduction. Neuron. 1989;2:1313–1323. doi: 10.1016/0896-6273(89)90069-x. [DOI] [PubMed] [Google Scholar]
- 37.Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- 38.Niemeyer BA, Suzuki E, Scott K, Jalink K, Zuker CS. The Drosophila light-activated conductance is composed of the two channels TRP and TRPL. Cell. 1996;85:651–659. doi: 10.1016/s0092-8674(00)81232-5. [DOI] [PubMed] [Google Scholar]
- 39.Osol G, Laher I, Kelley M. Myogenic tone is coupled to phospholipase C and G protein activation in small cerebral arteries. Am J Physiol. 1993;265:H415–H420. doi: 10.1152/ajpheart.1993.265.1.H415. [DOI] [PubMed] [Google Scholar]
- 40.Peretz A, Sandler C, Kirschfeld K, Hardie RC, Minke B. Genetic dissection of light-induced Ca2+ influx into Drosophila photoreceptors. J Gen Physiol. 1994a;104:1057–1077. doi: 10.1085/jgp.104.6.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peretz A, Suss-Toby E, Rom-Glas A, Arnon A, Payne R, Minke B. The light response of Drosophila photoreceptors is accompanied by an increase in cellular calcium: effects of specific mutations. Neuron. 1994b;12:1257–1267. doi: 10.1016/0896-6273(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 42.Peterson CCH, Berridge MJ, Borgese MF, Bennett DL. Putative capacitative calcium entry channels: expression of Drosophila trp and evidence for the existence of vertebrate homologues. Biochem J. 1995;311:41–44. doi: 10.1042/bj3110041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips AM, Bull A, Kelly LE. Identification of a Drosophila gene encoding a calmodulin-binding protein with homology to the trp phototransduction gene. Neuron. 1992;8:631–642. doi: 10.1016/0896-6273(92)90085-r. [DOI] [PubMed] [Google Scholar]
- 44.Ranganathan R, Bacskai BJ, Tsien RY, Zuker CS. Cytosolic calcium transients: spatial localization and role in Drosophila photoreceptor cell function. Neuron. 1994;13:837–848. doi: 10.1016/0896-6273(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 45.Ranganathan R, Malicki DM, Zuker CS. Signal transduction in Drosophila photoreceptors. Annu Rev Neurosci. 1995;18:283–317. doi: 10.1146/annurev.ne.18.030195.001435. [DOI] [PubMed] [Google Scholar]
- 46.Reed RR. Signaling pathways in odorant detection. Neuron. 1992;8:205–209. doi: 10.1016/0896-6273(92)90287-n. [DOI] [PubMed] [Google Scholar]
- 47.Riesgo-Escovar J, Raha D, Carlson JR. Requirement for a phospholipase C in odor response: overlap between olfaction and vision in Drosophila. Proc Natl Acad Sci USA. 1995;92:2864–2868. doi: 10.1073/pnas.92.7.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 49.Sengupta P, Colbert HA, Bargmann CI. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell. 1994;79:971–980. doi: 10.1016/0092-8674(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 50.Sengupta P, Chou JH, Bargmann CI. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- 51.Spielman AI, Nagai H, Sunavala G, Dasso M, Breer H, Boekhoff I, Huque T, Whitney G, Brand JG. Rapid kinetics of second messenger production in bitter taste. Am J Physiol. 1996;270:C926–C931. doi: 10.1152/ajpcell.1996.270.3.C926. [DOI] [PubMed] [Google Scholar]
- 52.Stryer L. Visual excitation and recovery. J Biol Chem. 1991;266:10711–10714. [PubMed] [Google Scholar]
- 53.Sukharev SI, Blount P, Martinac B, Blattner FR, Kung C. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature. 1994;368:265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- 54.Sulston J, Du Z, Thomas K, Wilson R, Hillier L, Staden R, Halloran N, Green P, Thierry-Mieg J, Qiu L, Dear S, Coulson A, Craxton M, Durbin R, Berks M, Metzstein M, Hawkins T, Ainscough R, Waterston R. The C. elegans genome sequencing project: a beginning. Nature. 1992;356:37–41. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]
- 55.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 56.Vaca L, Sinkins WG, Hu Y, Kunze DL, Schilling WP. Activation of recombinant TRP by thapsigargin in Sf9 insect cells. Am J Physiol. 1994;267:C1501–C1505. doi: 10.1152/ajpcell.1994.267.5.C1501. [DOI] [PubMed] [Google Scholar]
- 57.Vowels JJ, Thomas JH. Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics. 1994;138:303–316. doi: 10.1093/genetics/138.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wes PD, Chevesich J, Jeromin A, Rosenberg C, Stetten G, Montell C. TRPC1, a human homolog of a Drosophila store-operated channel. Proc Natl Acad Sci USA. 1995;92:9652–9656. doi: 10.1073/pnas.92.21.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of Caenorhabditis elegans. Philos Trans R Soc Lond [Biol] 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 60.Zhu X, Jiang M, Peyton M, Boulay G, Hurst R, Stefani E, Birnbaumer L. trp, a novel mammalian gene family essential for agonist-activated capacitative Ca2+ entry. Cell. 1996;85:661–671. doi: 10.1016/s0092-8674(00)81233-7. [DOI] [PubMed] [Google Scholar]
- 61.Zitt C, Zobel A, Obukhov AG, Harteneck C, Kalkbrenner F, Luckhoff A, Schultz G. Cloning and functional expression of a human Ca2+-permeable cation channel activated by calcium store depletion. Neuron. 1996;16:1189–1196. doi: 10.1016/s0896-6273(00)80145-2. [DOI] [PubMed] [Google Scholar]