Molecular Characterization of a Novel, Widespread Nuclear Protein That Colocalizes with Spliceosome Components (original) (raw)
Abstract
We report the identification and molecular characterization of a novel type of constitutive nuclear protein that is present in diverse vertebrate species, from Xenopus laevis to human. The cDNA-deduced amino acid sequence of the Xenopus protein defines a polypeptide of a calculated mass of 146.2 kDa and a isoelectric point of 6.8, with a conspicuous domain enriched in the dipeptide TP (threonine-proline) near its amino terminus. Immunolocalization studies in cultured cells and tissues sections of different origin revealed an exclusive nuclear localization of the protein. The protein is diffusely distributed in the nucleoplasm but concentrated in nuclear speckles, which represent a subnuclear compartment enriched in small nuclear ribonucleoprotein particles and other splicing factors, as confirmed by colocalization with certain splicing factors and Sm proteins. During mitosis, when transcription and splicing are downregulated, the protein is released from the nuclear speckles and transiently dispersed throughout the cytoplasm. Biochemical experiments have shown that the protein is recovered in a ∼12S complex, and gel filtration studies confirm that the protein is part of a large particle. Immunoprecipitation and Western blot analysis of chromatographic fractions enriched in human U2 small nuclear ribonucleoprotein particles of distinct sizes (12S, 15S, and 17S), reflecting their variable association with splicing factors SF3a and SF3b, strongly suggests that the 146-kDa protein reported here is a constituent of the SF3b complex.
INTRODUCTION
Biochemical fractionations and the use of antibodies to analyze the distribution of proteins in situ as well as recombinant DNA technologies have led to the identification of macromolecular domains within the mammalian cell nucleus. Beyond such obvious features as the nucleolus, heterochromatin, and the nuclear membrane, several particulate nuclear elements (termed “nuclear granules” or “nuclear dots”) have been described that can be correlated with fundamental nuclear processes, e.g., transcription, RNA splicing, and processing of mature mRNA (reviewed by Spector, 1993).
Splicing occurs in a multicomponent complex termed the spliceosome. Many of the detailed biochemical steps involved in the pre-mRNA splicing reaction have been extensively studied in vitro and are well understood (reviewed by Green, 1991; Moore et al., 1993). The major constituents of the spliceosome are the U1, U2, U4/U6, and U5 small nuclear ribonucleoprotein particles (snRNPs; reviewed by Baserga and Steitz, 1993). Moreover, spliceosomes are associated with numerous non-snRNP splicing factors, several of which have been purified and cloned (reviewed by Krämer, 1996; Will and Lührmann, 1997). Immunolocalization studies have revealed that proteins involved in pre-mRNA maturation tend to be heterogeneously distributed in the nucleus, suggesting that the processing reactions might be compartmentalized in vivo (Carter et al., 1993; however, see also Huang and Spector, 1996). In addition to the widespread nucleoplasmic distribution of several constituents of the spliceosome, reflecting their association with perichromosomal fibrils that are believed to represent nascent transcripts with associated spliceosomes, some proteins of the splicing machinery (e.g., the Sm proteins) appear to be concentrated in nuclear speckles and foci. At the electron microscopic level these intranuclear structures correspond to clusters of interchromatin granules and coiled bodies, respectively (Lamond and Carmo-Fonseca, 1993; Spector, 1993; Bohmann et al., 1995). The latter ones can be regarded as the homologous structures to the sphere organelles found in nuclei of urodele oocytes (reviewed by Gall et al., 1995). Splicing factors belonging to the family of serine/arginine-rich (SR) proteins are detected predominantly in speckles, i.e. interchromatin granules, but are absent from coiled bodies (Fu and Maniatis, 1990; Spector et al., 1991). The essential splicing factor U2AF65 (U2 snRNP auxiliary factor) appears to have a similar nuclear distribution (Gama-Carvalho et al., 1997) although it had initially been shown to have a diffuse nucleoplasmic localization with additional concentration in coiled bodies (Zamore and Green, 1992; Carmo-Fonseca et al., 1991).
Other nuclear bodies have been identified, but their structural organization, composition, and function have often remained less well defined. Nuclear dots 0.2–0.3 μm in diameter, termed ND10, have been described and shown to be composed of several proteins (Ascoli and Maul, 1991; Dyck et al., 1994; Korioth et al., 1995). Another subnuclear compartment with dramatic morphological changes during the cell cycle was identified (Saunders et al., 1991) and named PIKA (polymorphic interphase karyosomal association).
In the course of our studies aimed at the identification of nuclear structural (karyophilic) proteins of Xenopus oocytes and/or somatic cells (e.g., Franke et al., 1981; Schmidt-Zachmann et al., 1984, 1987; Krohne et al., 1987; Ankenbauer et al., 1989; Cordes et al., 1991, 1993, 1997), we have cloned the cDNA of a novel 146-kDa protein from Xenopus laevis. Localization studies suggest that this protein is a widespread nuclear constituent and that its intranuclear distribution is independent of replication and transcription. This protein is present in large RNP structures. Immunolocalization studies on cultured cells and tissues derived from different species indicate that the protein has been highly conserved during evolution. Moreover, it colocalizes with well characterized marker proteins of the splicing machinery and is specifically enriched in chromatographic fractions containing the mammalian splicing factor SF3b, suggesting that it represents a component of the SF3b complex.
MATERIALS AND METHODS
Animals, Tissues, and Cell Culture
X. laevis were purchased from the South African Snake Farm (Krysna, Republic of South Africa). Tissue samples from X. laevis (skin, intestine, liver, ovary, heart), rat (liver), cow (liver), and human (esophagus, heart, liver) were snap-frozen in isopentane cooled by liquid nitrogen to about −140°C and stored at −80°C. For X. laevis blood smear preparations blood was obtained from a toe vein of living animals or from larger vessels of decapitated toads and smear-spread on glass slides.
Cell culture lines used included X. laevis kidney epithelium XLKE, line A6, chicken embryonic fibroblasts line CEF, rat kangaroo PtK2, embryonic mouse line 3T3-L1, rat vascular smooth muscle-derived line RV, bovine kidney epithelial line MDBK, bovine mammary gland-derived line BMGE+H, human primary liver carcinoma line PLC, and human cervical adenocarcinoma line HeLa (for sources of all cell lines see American Tissue Culture Collection, Rockville, MD, and previous reports from this laboratory: Franke et al., 1979, 1980; Schmid et al., 1983; Cordes et al., 1996). Cells were maintained under standard conditions.
Isolation of Monoclonal Antibody (mAb) B2
Fractions enriched in cyto- and karyoskeletal proteins derived from Xenopus A6 cells were prepared as described earlier (Herrmann and Wiche, 1983; see also Fouquet, 1991).
Monoclonal antibodies were raised against these preparations essentially according to the method of Köhler and Milstein (1975). A 7 wk-old female BALB/c mouse was immunized with 150 μg of antigen. After three booster injections at days 28, 56, and 84, respectively, the spleen cells were harvested at day 92 and fused with cells of the mouse myeloma line P3X63-Ag8.653 at a ratio of 3:1 in the presence of 40% PEG 4000. Antibody-producing hybridoma cell lines were selected essentially as described by Schmidt-Zachmann et al. (1984). Immunoglobulin subclasses were determined by enzyme-linked immunosorbent assay with subclass-specific secondary antibodies (Sigma, Munich, Germany). The hybridoma cell line B2 was also propagated as peritoneal ascites in BALB/c mice.
One of the antibodies, mAb B2 (IgG1), showed a strong nucleolar staining on Xenopus A6 cells when analyzed by immunofluorescence microscopy. This antibody was used for the initial screening of a λ unizap cDNA expression library from X. laevis kidney (see below).
Generation of Peptide-specific Antibodies against the Xenopus 146-kDa Protein
Guinea pig antibodies specific for the Xenopus 146-kDa protein were obtained by immunization with synthetic peptides (Schnölzer et al., 1992) representing various parts of the amino acid (aa) sequence deduced from cDNA sequencing (indicated in Figure 1B). In the experiments reported here, antibodies B2.4–1 against the sequence KIANREDEYKQQRRKMI (aa 109–125) were routinely used after affinity purification on iodoacetyl-immobilized peptide (Mertens et al., 1996). Antibodies B2.4–3 (aa 471–488; KPDDIQYFDKLLVDVDES) and B2.4–4 (aa 1022–1038; RLTPILKNRHEKVQENC) showed the same specificities.
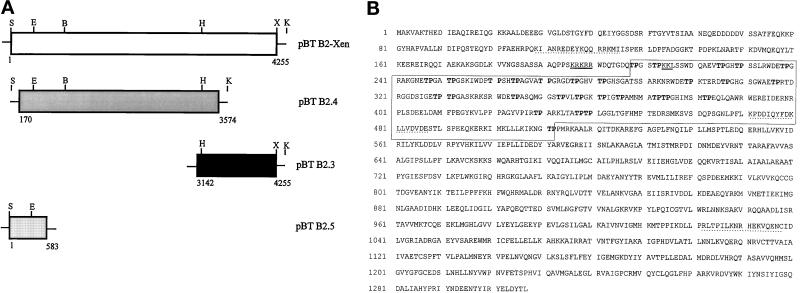
Figure 1.
Isolation of a cDNA clone coding for a novel nuclear protein from X. laevis. (A) Schematic representation of the assembled pBT B2-Xen full-length cDNA and overlapping subclones (pBT B2.4; pBT B2.3; pBT B2.5) thereof. Partial restriction maps indicate the enzymes used for generating the full-length cDNA: S, _Spe_I; E, _Eco_RV; B, _Bam_HI; H, _Hin_cII; X, _Xho_I; K, _Kpn_I (see also MATERIALS and METHODS). Numbers give the nucleotide positions with reference to the assembled full-length cDNA. (B) Amino acid (aa) sequence deduced from cDNA clone pBT B2-Xen. The open reading frame comprises 1307 aa coding for a 146-kDa protein. An internal domain characterized by the presence of several TP-motifs (in bold letters) is denoted by a box (aa 208–512). A putative bipartite nuclear localization signal (NLS) is underlined, and sequences used for generating antibodies are marked by dotted lines.
Other Antibodies
Rabbit antibodies to coilin (Bohmann et al., 1995) were generously provided by Dr. A. Lamond (University of Scotland, Dundee, Scotland), mAb Y12 to Sm proteins (Lerner et al., 1981) was kindly provided by Dr. J. Gall (Carnegie Institution of Washington, Baltimore, MD) and mAb 66 against the 66-kDa subunit of mammalian splicing factor SF3a has been described (Brosi et al., 1993b). The mAb 9E10 (ATCC CRL 1729) specifically recognizes an epitope in the decapeptide EQKLISEEDL of the human c-myc protein (Evan et al., 1985). Mab No-185 directed against the major nucleolar protein NO38 and anti-NO29 antibodies have been described (Schmidt-Zachmann et al., 1987; Zirwes et al., 1997b).
Secondary Antibodies
Secondary antibodies used for immunofluorescence microscopy were Texas Red-, Cy2- and Cy3-conjugated goat antibodies to immunoglobulins of mouse, guinea pig, or rabbit, respectively. For immunoblotting, horseradish peroxidase-conjugated antibodies to mouse or guinea pig were used (Dianova, Hamburg, Germany).
Isolation of cDNA Clones and Polymerase Chain Reaction (PCR) Products
A λ unizap cDNA expression library from X. laevis kidney cells (Stratagene, Heidelberg, Germany) was screened with mAb B2. One of the cDNA clones obtained was selected, plaque-purified, and released from phages by in vivo excision according to the manufacturer’s protocol. The resulting cDNA of ∼3.4 kilobases (kb) (clone pBT B2.4) was characterized by restriction mapping and further analyzed by constructing deletion clones with the double-stranded nested deletion kit (Pharmacia, Freiburg, Germany) and sequencing from both directions (Sanger et al., 1977).
Since clone pBT B2.4 did not contain the complete cDNA, the library was rescreened with i) the 415-base pair (bp) _Hin_cII/_Xho_I fragment representing the 3′-end and ii) the 691-bp _Eco_RI/_Bam_HI fragment representing the 5′-end of clone pBT B2.4. The screen with the 3′-end probe led to the isolation of two cDNA clones of ∼1.2 kb and ∼0.75 kb. Sequencing revealed that the ∼0.75-kb cDNA was identical to the 3′ end of the ∼1.2-kb cDNA. The latter cDNA, termed pBT B2.3, showed an overlap of 490 bp with pBT B2.4 and contained 694 bp in addition. Several additional clones were isolated with the 5′ probe and partially sequenced. None of them extended the sequence of pBT B2.4. Therefore, the missing 5′-end was cloned by the rapid amplification of cDNA ends (RACE) procedure essentially according to Frohman et al. (1988). Starting from poly (A)+ RNA isolated from Xenopus A6 cells, cDNA synthesis and subsequent PCR reactions were performed with the “5′ RACE” kit from Life Technologies (Eggenstein, Germany) according to the manufacturer’s protocol. Two antisense gene-specific primers complementary to positions 446–466 (GSP1) and 396–416 (GSP2) of clone pBT B2.4 as well as the sense anchor primer provided with the kit were used. The resulting PCR product of 583 bp was subcloned into the pBluescript as a _Spe_I/_Eco_RI fragment (clone pBT B2.5).
The overlapping clones pBT B2.4, pBT B2.3, and pBT B2.5 were used to construct a full-length cDNA clone coding for the Xenopus protein (Figure 1A). First, the 3′ portion of pBT B2.3 was excised with _Hin_cII and _Kpn_I and inserted into clone pBT B2.4 digested with the same enzymes. The resulting clone pBT B2.4.3 was digested with _Spe_I and _Bam_HI, and two fragments of ∼6.4 kb (fragment 1) and ∼0.64 kb (fragment 2) were isolated. Fragment 2 was further digested with _Eco_RV, and another fragment encoding the 5′-end of the 146-kDa protein was obtained from clone pBT B2.5 digested with _Spe_I and _Eco_RV (fragment 3). All three fragments were ligated together to generate the final construct pBT B2-Xen (see also Figure 1A). The entire ∼4.2-kb cDNA sequence was sequenced from both strands with internal oligonucleotide primers. Sequence alignments, analyses, and data base searches were performed with the software program package HUSAR (Heidelberg Unix Sequence Analysis Resources, Heidelberg, Germany).
For the isolation of a human cDNA clone encoding the 146-kDa protein, a λ zapII human fetal brain library (Stratagene, Heidelberg, Germany) was screened with a 32P-labeled random-primed 360-bp fragment derived from pBT B2-Xen by _Spe_I/_Eco_RV digestion, representing the extreme amino-terminal portion of the Xenopus protein. Two positive human cDNA clones were identified and analyzed further. Restriction maps and sequencing data demonstrated the identity of both human cDNAs. cDNA clone pBT B2-hum (∼1.5 kb) encodes the amino-terminal portion of the 146 kDa protein (aa 1–499).
RNA Isolation, Northern Blot Hybridization, and Coupled In Vitro Transcription/Translation
Total RNA from Xenopus ovaries or A6 cells was prepared as described by Chomczynski and Sacchi (1987) or Chirgwin et al. (1979). Poly(A)+-RNA was prepared using the mRNA Purification Kit (Pharmacia). RNAs (5 μg) were separated on 1% agarose gels containing 0.6% formaldehyde, transferred to Biodyne A filters (Pall, Dreieich, Germany), hybridized with antisense riboprobes derived from clone pBT B2-Xen, washed, and processed by autoradiography essentially as described by Heid et al. (1994).
Northern blot analysis of small nuclear RNAs (snRNAs) was performed according to Utans et al. (1992). The blot was hybridized with a [32P]UTP-labeled antisense U2 snRNA transcript (2 × 106 Cerenkov cpm) for 16 h at 42°C followed by two 20-min washes with 2× SSC/1% SDS and one wash with 0.2× SSC/1% SDS.
For the production of [35S]methionine-labeled 146-kDa protein in vitro using the TNT Coupled Reticulocyte Lysate (Promega via Boehringer Ingelheim Bioproducts, Heidelberg, Germany) the entire B2-Xen cDNA was subcloned as a _Hin_dIII/_Kpn_I-fragment into the mammalian expression vector pRc/CMV (Invitrogen via ITC Biotechnology, Heidelberg, Germany).
DNA Transfection
For transient expression in human PLC cells, the B2-Xen cDNA was subcloned into the linearized BT-myc-vector (Schmidt-Zachmann and Nigg, 1993), and the resulting construct was further subcloned into the eukaryotic expression vector pRc/CMV (Invitrogen). Transfections were carried out as described (Zirwes et al., 1997a), and transfected cells were analyzed 24 h after removal of the DNA-calcium phosphate precipitate by immunofluoresence with the myc-specific mAb 9E10.
Isolation and Fractionation of Xenopus Oocyte Nuclei and Egg Extract
Large-scale isolation of nuclei of mature (stages IV-VI; Dumont, 1972) X. laevis oocytes was carried out as described by Scalenghe et al. (1978) with the modifications of Kleinschmidt and Franke (1982). Subsequent fractionation of nuclear contents into low-speed pellet (LSP), high-speed pellet (HSP), and high-speed supernatant (HSS) was as described by Hügle et al. (1985). LSP fractions were cleared from yolk proteins by Freon extraction (Evans and Kay, 1991). The preparation of egg extracts was as described by Cordes et al. (1993).
Preparation of Cell Lysates and Nuclear Extract from Cultured Cells
Total cellular lysates of cultured cells were obtained as follows: the medium of a confluent 5-cm culture dish was removed and the cells were washed twice with phosphate-buffered saline (PBS). Subsequently, the cells were scraped off in 0.5 ml SDS sample buffer, transferred to a 1.5-ml tube, and incubated on ice for 15 min with 50 U Benzonase (Merck, Darmstadt, Germany) for digestion of nucleic acids. Finally the samples were boiled and analyzed by SDS-PAGE and immunoblotting.
Small-scale preparation of nuclear extracts from HeLa cells was done according to the method described by Lee and Green (1990).
Cellular extract of Xenopus A6 cells used for immunoprecipitation, gel filtration, and sucrose gradient centrifugation was prepared by the following method. Confluently grown cells were washed three times with PBS and lysed directly onto the culture dishes with 1 ml A6-lysis buffer (0.5% Triton X-100, 0.02% NaN3, 100 mM Na-2-(_N_-morpholino)ethanesulfonic acid pH 6.25, 250 mM NaCl, 0.5 mM MgCl2, 1 mM dithiothreitol, and, for inhibition of proteases, 100 μM Pefablock, 20 μM pepstatin, and 20 μM leupeptin) per 10-cm dish for 2 min at room temperature. The lysates were transferred to a 1.5-ml tube and incubated for 15 min on ice. After centrifugation of sedimentable material (15 min, 14,000 × g, 4°C), the supernatant was either snap-frozen in liquid nitrogen and stored at −80°C or used directly for biochemical studies.
Sucrose Gradient Density Centrifugation and Gel Filtration
Xenopus egg extract (150 μl) and 250 μl cell lysate obtained from Xenopus A6 cells were fractionated by centrifugation in a 5–30% linear sucrose gradient (in 5:1-buffer: 10 mM Tris-HCl, pH 7.4, 83 mM KCl, 17 mM NaCl, 2 mM MgCl2, 2.5 mM dithiothreitol, 50 μM Pefa-Block or in A6 lysis buffer without detergent). Fourteen fractions of 0.8 ml were collected from the top (light) to the bottom (heavy) of the gradient and tested by immunoblotting. Parallel gradients were used to calibrate the sedimentation of size reference proteins (bovine serum albumin, catalase, thyroglobulin; all from Pharmacia).
For gel filtration, 200 μl Xenopus A6 cell lysate were loaded onto a Superose 6 HT 10/30 column (Pharmacia) at room temperature. For calibration, dextran blue and reference proteins (thyroglobulin, ferritin, and catalase, all from Pharmacia) were fractionated in parallel. Proteins were eluted in A6 lysis buffer without detergent and, after an excluded volume of 7.4 ml, 40 fractions of 0.4 ml were collected. The first 30 fractions were analyzed by SDS-PAGE and immunoblotting.
Fractions Containing U2 snRNPs and Associated Splicing Factors
Mono Q fractions enriched in 12S U2 snRNP, 15S U2 snRNP (U2 snRNP associated with splicing factor SF3b), and 17S U2 snRNP (U2 snRNP associated with splicing factors SF3a and SF3b), as well as Mono Q and Mono S fractions enriched in SF3a and SF3b, were obtained from nuclear extracts of HeLa cells as described (Brosi et al., 1993a,b).
Immunoprecipitation Experiments
For immunoprecipitation of Xenopus A6 cell lysates and total egg extracts, affinity-purified antibody B2.4–1 (40 μl, diluted with 450 μl PBS) was coupled to 50 μl preswollen protein G-Sepharose (Pharmacia) for 1 h at 4°C. Protein solutions were preincubated with protein G-Sepharose (1 h, 4°C) to avoid nonspecific binding during the immunoprecipitation process. Subsequently, the precleared protein sample was incubated with the antibody–protein G complex by end-over-end rotation (2 h, 4°C). After low-speed centrifugation (800 × g, 5 min) the resulting supernatant was precipitated with acetone and prepared for SDS-PAGE. The Sepharose beads with the bound immune complexes were washed five times with PBS, and then once each with PBS containing 0.1% Triton X-100 and with PBS, and finally boiled in SDS sample buffer. The solubilized proteins from the precipitate were analyzed by SDS-PAGE together with the reserved supernatant. In some experiments, protein samples were incubated with protein G-Sepharose in the absence of prebound antibodies to identify nonspecific binding proteins. As further controls, antibodies were bound to protein G-Sepharose and subsequently incubated with PBS, or an irrelevant antiserum directed against the nucleolar protein NO29 (Zirwes et al., 1997b) was coupled to protein G-Sepharose. The control precipitations were processed as described above.
For immunoprecipitation of U2 snRNP-containing fractions, either affinity-purified B2.4–1 antibody (as above) or 0.5 ml mAb 66 hybridoma supernatant was bound to protein G-Sepharose. Mono Q fractions containing equivalent amounts of U2 snRNA were incubated with the antibody–protein G-Sepharose. Incubation and wash steps were performed in the presence of NET-2 (50 mM Tris-HCl, pH 7.9, 150 mM NaCl, 0.05% Nonidet-P-40, and 0.5 mM dithiothreitol). Samples were treated for SDS-PAGE as above. For Northern blot analysis of snRNAs, samples were treated with proteinase K, extracted with phenol and chloroform, and precipitated with ethanol. RNAs were separated on a 10% polyacrylamide/8.3 M urea gel.
Gel Electrophoresis and Immunoblotting
Protein fractions were separated by SDS-PAGE according to Thomas and Kornberg (1975). After electroblotting of the proteins to nitrocellulose, the filters were blocked for 45 min in Tris-buffered saline (TBS) containing 0.05% Tween (TBST) and 5% nonfat dry milk. Affinity-purified B2.4–1 antibody (diluted 1:1000) or mAb 66 (hybridoma supernatant diluted 1:10) were incubated with the membranes for 1–2 h in TBST/5% nonfat dry milk. Bound antibodies were detected by chemiluminescence using the ECL-system (Amersham, Braunschweig, Germany) after incubation with horseradish peroxidase-coupled secondary antibodies diluted 1:10,000 in TBST/5% nonfat dry milk for 1 h.
Immunofluorescence Microscopy
For immunofluorescence microscopy studies of cultured cells, cells grown on coverslips were either fixed in methanol (7 min, −20°C) followed by acetone (30 sec, −20°C) or in PBS containing 2% formaldehyde (10–20 min, room temperature) followed by incubation in PBS containing 50 mM NH4Cl (5 min). For permeabilization, formaldehyde-fixed cells were incubated for 10 min with PBS containing 0.5% Triton X-100.
For actinomycin D (AMD) experiments, the cultured cells were incubated for 4 h in fresh medium containing 5 μg/ml of the drug. For heat shock experiments Petri dishes containing coverslips and prewarmed medium (45°C) were transferred into a 45°C water bath for 15 min before fixation. Nuclease digestion was performed according to Spector et al. (1991).
After fixation, cells were washed twice in PBS and incubated with purified guinea pig antibodies (1:100 in PBS), rabbit antibodies (1:100 in PBS), or mAbs (undiluted supernatant or purified immunoglobulin 1:50 in PBS) for 20 min at room temperature. After several PBS washes, cells were incubated for 20 min with the appropriate secondary antibodies (1:100 to 1:300 in PBS), washed in PBS, dehydrated in ethanol, air dried, and mounted in Elvanol (Hoechst, Frankfurt, Germany).
Cryosections (5 μm) of frozen tissues were fixed either in acetone (−20°C) for 10 min or in PBS containing 2% formaldehyde for 10–20 min at room temperature. Formaldehyde-fixed samples were washed in PBS containing 50 mM NH4Cl for 5–10 min and 2 × 5 min in PBS before incubation with antibodies. Incubation times of primary and secondary antibodies were as described above. Micrographs were taken with an Axiophot microscope (Zeiss, Jena, Germany).
Confocal laser-scanning immunofluorescence microscopy was done on a Zeiss LSM 410 UV instrument (Zeiss). For simultaneous double-label fluorescence, an argon ion laser operating at 488 nm and a helium-neon laser operating at 543 nm were used together with a band-pass filter combination of 510–525 nm and 590–610 nm for visualization of Cy-2 and Cy-3 fluorescence, respectively.
RESULTS
Isolation and Analysis of a cDNA Clone Encoding a Novel Nuclear 146-kDa Protein from X. laevis
To identify structural, karyophilic proteins, we raised monoclonal antibodies against total karyoskeletal residue material of Xenopus kidney epithelial cells (XLKE-A6) and Xenopus oocyte nuclei. Among others, we obtained one monoclonal antibody (mAb B2) that showed a specific labeling of nucleoli by immunofluorescence microscopy and immunoprecipitated a ∼70-kDa protein from XLKE-A6 cellular lysates (our unpublished observations). This antibody was used to screen a λ-Uni-Zap X. laevis kidney expression library. We initially cloned a positive phage recombinant (clone pBT B2.4) of 3405 bp with a continuous open reading frame. Sequencing analysis revealed that the 5′- and 3′-ends of the cDNA were missing. The same cDNA library was rescreened with a random prime-labeled 415-bp fragment representing the 3′-terminus of clone pBT B2.4, and two positive recombinants of ∼1.2 kb and ∼0.75 kb were obtained. The sequence of the 0.75 kb fragment was identical to the 3′-end of the 1.2 kb cDNA clone. The 1.2- kb clone (pBT B2.3) overlapped with the 3′-end of clone pBT B2.4 and apparently contained the lacking 3′-end of the cDNA.
Several attempts to isolate a cDNA clone containing the 5′-end failed; therefore, this part of the coding region was cloned with the anchor-PCR method (see MATERIALS AND METHODS; Frohman et al., 1988). The combined sequence of the 5′ RACE PCR product and the cDNA library inserts was considered to be the full-length cDNA clone. Using appropriate restriction enzymes the corresponding full-length cDNA clone pBT B2-Xen was generated (Figure 1A), and its nucleotide sequence of both strands was determined (see EMBL data base, accession no. Y08997). The deduced aa sequence is shown in Figure 1B.
Clone pBT B2-Xen (4255 bp) contained an initiation codon at position 88, an open reading frame of 4011 bp, and a 3′ untranslated region of 244 bp with a consensus AATAAA polyadenylation signal 12 bp upstream of the poly(A) tail of 16 bp. The open reading frame encoded a polypeptide of 1307 aa with a calculated molecular mass of 146.2 kDa and a isoelectric point of 6.8. The presumptive start codon lies within the 5′ region generated by the RACE system. Although the open reading frame continues to the 5′ end of the cDNA, this ATG is likely to be the initiation codon because the surrounding sequence (AAAATGGC) perfectly matches the optimal sequence for eukaryotic initiation of translation (Kozak, 1989).
The most conspicuous feature of the encoded protein is the clustering of the dipeptide TP (threonine, proline) in a domain located between aa 209 to 512 (boxed in Figure 1B) where the TP-dipeptides, which probably represent CDK phosphorylation sites (Nigg, 1995), are repeated 29 times. We also noted a putative bipartite nuclear localization signal (NLS) between aa positions 196–215 (KRKRR(x)13KK, underlined in Figure 1B; Dingwall and Laskey, 1991). Other notable features, e.g., sequence elements involved in nucleic acid binding or protein-protein interactions, were not detected. Potential phosphorylation sites are found for cAMP-dependent kinase, protein kinase C, and casein kinase II, although the biological significance of these sites is unknown.
Searches of current data bases did not reveal significant homologies of the identified Xenopus protein with other vertebrate proteins, but several expressed sequence tags identical to pBT B2-Xen were found. In addition, these data base searches disclosed a striking homology of the 146-kDa protein with hypothetical proteins of 137, 110, and 130 kDa from Schizosaccharomyces pombe, Saccharomyces cerevisiae, and Caenorhabditis elegans, respectively (accession nos. Q10178, P49955, and Z50875). These proteins of unknown function consist of 1205, 971, and 1166 aa residues, i.e., they are significantly smaller than the Xenopus protein and lack the amino-terminal portion. The overall aa sequence identity between the Xenopus 146-kDa protein and the homologues in S. cerevisiae, S. pombe, and C. elegans are 53.8, 67.8, and 76.1%, respectively (our unpublished observations). An attempt to clone the putative human homologue resulted in the isolation of a partial cDNA clone (pBT B2-hum) encoding the first 499 aa of the 146-kDa protein. The identity between the corresponding Xenopus and human aa sequences is 91.75%. This extremely high conservation suggests that these proteins represent homologous molecules that most likely participate in a fundamental cellular process.
Poly(A)+ RNA from Xenopus ovary tissue and XLKE-A6 cells was probed in Northern blot experiments with a ∼1.6-kb antisense cRNA derived from pBT B2-Xen. A strong signal corresponding to a mRNA of ∼4.4 kb was detected (Figure 2A), indicating that the pBT B2-Xen cDNA clone was of full or nearly full length. In vitro transcription and translation of pBT B2-Xen in a reticulocyte lysate yielded a polypeptide of ∼140 kDa, consistent with the calculated molecular mass of the encoded protein. The in vitro translation product is specifically precipitated with antibody B2.4–1 raised against aa 109–205 of the 146 kDa protein, demonstrating that the antibody recognizes the protein encoded by the pBT B2-Xen cDNA (Figure 2, B and B′).
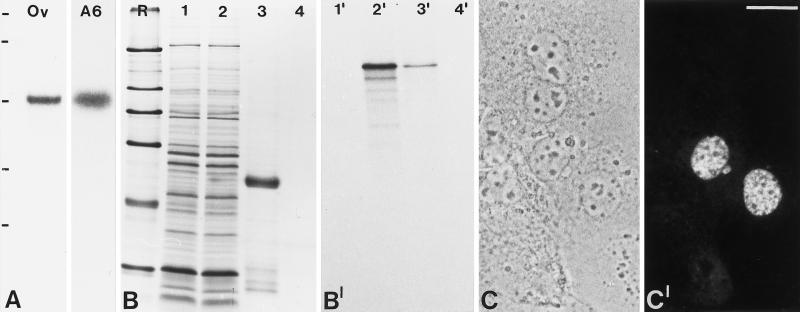
Figure 2.
Molecular characterization of the cDNA clone encoding the 146-kDa protein. (A) Identification of specific mRNAs by Northern blot analysis. Poly(A)+-RNA from Xenopus oocytes (Ov) and Xenopus A6 cells (A6) was separated in an agarose gel, transferred to a membrane, and probed with antisense cRNA derived from clone pBT B2-Xen. Note the specific reaction with a ∼4.4 kb RNA. RNA-size markers of 7.5, 4.4, 2.4, and 1.4 kb are indicated on the left (from top to bottom). (B) Coomassie blue staining of SDS-PAGE–separated rabbit reticulocyte lysates after in vitro transcription/translation in the absence (lane 1) or presence of the pBT B2-Xen template (lane 2). Lanes 3 and 4 show immunoprecipitates of translated protein in the presence or absence of antibody B2.4–1, respectively. R, reference proteins: 205, 116, 97.4, 66, 45, and 29 kDa (from top to bottom). (B′) Corresponding autoradiograph of translation products and immunoprecipitates. (C) Phase-contrast microscopy of cultured human hepatocellular carcinoma (PLC) cells used to determine the subcellular localization of the 146-kDa Xenopus protein carrying an amino-terminal myc tag in transfection experiments. (C′) Corresponding immunofluorescence using mAb 9E10 recognizing the myc tag. Bar, 20 μm
Since the molecular mass of the protein encoded by the isolated cDNA clone pBT B2-Xen differed significantly from the size of the nucleolar antigen detected by mAb B2 used for the initial screening procedure (see above), we analyzed the intracellular location of the 146-kDa protein. Human carcinoma cells (PLC) were transiently transfected with a cDNA encoding the Xenopus 146- kDa protein with a myc tag at its amino terminus. Immunofluorescence analysis with the anti-myc antibody revealed that the expressed protein was exclusively located in the nucleoplasm of the transfected cells (Figure 2, C and C′). From these results we conclude that we have cloned a novel, nuclear protein of X. laevis.
Given the differences in molecular mass and intracellular localization of the protein encoded by the pBT B2-Xen cDNA and the nucleolar antigen originally identified as the mAb B2 antigen, it is most likely that mAb B2 recognizes identical or overlapping epitopes in the 146-kDa nuclear and ∼70-kDa nucleolar proteins.
Biochemical Characterization of the Nuclear 146-kDa Protein
To study the intracellular distribution and localization of the endogenous 146-kDa protein, polyclonal antibodies against peptides deduced from the cDNA sequence of pBT B2-Xen (see Figure 1B) were raised. In immunoblots of total proteins isolated from cultured cells of different origin (Xenopus, chicken, rat kangaroo, mouse, rat, bovine, and human) antibody B2.4–1 reacted exclusively with a polypeptide of ∼140 kDa (Figure 3, A and A′), which is in good agreement with the results obtained by in vitro translation of pBT B2-Xen (cf. Figure 2, B and B′). Antibodies against other peptides of the 146-kDa protein gave identical results. The broad cross-reactivity observed again indicated that the 146-kDa protein has been highly conserved during evolution.
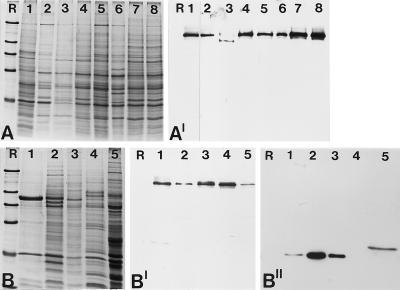
Figure 3.
Identification of the 146-kDa protein in different cell culture lines and nuclear fractions from Xenopus oocytes. (A) Coomassie blue-stained total cellular proteins. Cell lines shown are: X. laevis kidney epithelial cells, line A6 (lane 1); chicken embryonic fibroblasts line CEF (lane 2); rat kangaroo cells, PtK2 (lane 3); embryonal mouse cells of line 3T3-L1 (lane 4); rat vascular smooth muscle-derived cells of line RV (lane 5); bovine kidney epithelial cell line MDBK (lane 6); human primary liver carcinoma cells of line PLC (lane 7); and human cervical adenocarcinoma cells of line HeLa (lane 8). (A′) Corresponding autoradiogram showing immunochemiluminescence detection of the antigenic polypeptide using antibody B2.4–1. A shorter exposure of lane 1 is presented because of the very strong reaction of the antibodies generated against the Xenopus protein. The comparable weak reaction in lane 3 might be due to partial degradation of the protein. (B) Coomassie blue staining of various nuclear fractions of Xenopus oocytes separated by SDS-PAGE. Total mass-isolated nuclei (lane 1); proteins of the LSP, HSP, and HSS fractions of fractionated oocyte nuclei (lanes 2–4); and Xenopus egg extract (lane 5). (B′) Corresponding immunoblot probed with antibody B2.4–1, which specifically reacts with the 146-kDa protein present in all fractions analyzed. (B") Probing of a parallel immunoblot with mAB No-185 directed against the well characterized nucleolar protein NO38 to ascertain the fractionation procedure. Reference proteins (R) are the same as in Figure 2.
The transfection experiments shown above demonstrated the nuclear localization of the 146-kDa protein. To confirm this result on the biochemical level, we analyzed the distribution of the 146-kDa protein in different nuclear fractions of Xenopus oocytes (total oocyte nuclei, LSP, HSP, and HSS) and egg extract (Figure 3, B and B′). The 146-kDa protein was detected in both oocyte nuclei and egg extracts. In addition, its presence in the three fractions derived from oocyte nuclei indicated that under physiological buffer conditions a proportion of the 146-kDa protein remains bound to the nuclear remnant (LSP, HSP), whereas another part is in a soluble nucleoplasmic form (HSS). The identity of the fractions was ascertained by incubating a parallel blot with the nucleolar antigen NO38 (Figure 3B"), which was previously shown to be present in the LSP and HSP fractions, but absent from the HSS fraction, and a hyperphosphorylated form was found in egg extracts (Schmidt-Zachmann et al., 1987 and unpublished results).
The 146-kDa protein could also be immunoprecipitated in Coomassie blue-visible amounts from protein extracts of Xenopus eggs and XLKE-A6 cell lysates (Figure 4, A and B). Several weakly stained polypeptides in the range of 100–130 kDa could be detected in both immunoprecipitates. At present, it is not known whether these polypeptides are specifically associated with the 146-kDa protein or represent proteolytic breakdown products derived from the 146-kDa polypeptide. The use of an irrelevant antiserum directed against the nucleolar protein NO29 (Zirwes et al., 1997b) allowed verification of the specifity of antibody B2.4–1 (Figure 4A, lane 5).
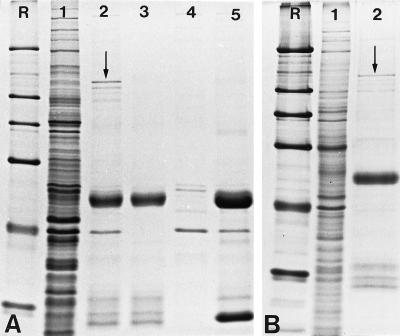
Figure 4.
Immunoprecipitation of the 146-kDa protein with antibody B2.4–1. The following fractions were separated by SDS-PAGE and stained with Coomassie blue. (A) Proteins of Xenopus egg extract before immunoprecipitation (lane 1); immunoprecipitate obtained from Xenopus egg extract (lane 2); immunoprecipitate after incubation with PBS as a negative control (lane 3); proteins of the egg extract that bind nonspecifically to protein G-Sepharose in the absence of antibodies (lane 4); immunoprecipitate obtained with antibodies against the nucleolar protein NO29. (B) Proteins of Xenopus A6 cell lysates before immunoprecipitation (lane 1); immunoprecipitate obtained from A6-lysates (lane 2). The 146-kDa protein (indicated by arrows) is immunoprecipitated in Coomassie blue-visible amounts (panel A, lane 2, and panel B, lane 2). The nature of the slightly smaller polypeptides (∼110–130 kDa) visible in the immunoprecipitates is unknown. R, reference proteins as shown in Figure 2.
To test for a possible association of the 146-kDa protein with other cellular constituents, the protein was analyzed by density gradient centrifugation of cellular lysates of XLKE-A6 cells (Figure 5, A and A′) and Xenopus egg extract (Figure 5, B and B′). Immunoblotting of the resulting fractions revealed that the protein sedimented at ∼12S (fraction 8 in Figure 5A′ and fraction 7 in Figure 5B′). In gel filtration experiments the bulk of the protein eluted in two distinct peaks corresponding to an apparent mass (Mapp) of 1,400,000 and 1,000,000, respectively (Figure 5, C and C′), and a smaller portion eluted at lower Mapp. The identity of smaller polypeptides that are decorated by antibody B2.4–1 in fractions 14–16 is unclear; they could represent proteolytic breakdown products or cross-reacting proteins. Taken together, the results from the gradient sedimentation and gel filtration suggest that the native 146-kDa protein is a constituent of a large particle.
Figure 5.
Analysis of the native state of the 146-kDa protein by sucrose gradient centrifugation and gel filtration. (A) Cell lysates of Xenopus A6 cells were fractionated after sucrose gradient centrifugation, separated by SDS-PAGE, and stained with Coomassie blue. Fraction numbers are indicated on top of the lanes (the top of the gradient is on the left). Bars indicate the peak positions of the reference proteins bovine serum albumin (4.3S; fraction 3), catalase (11.3S; fraction 7), and thyroglobulin (16.5S; fraction 11). R, reference proteins are the same as in Figure 2. (A′) Corresponding immunoblot using antibody B2.4–1. The 146-kDa protein is recovered in fraction 8 with a sedimentation coefficient of ∼12S. (B) A similar study was performed with a Xenopus egg extract. Coomassie blue staining of the resulting protein fractions after sucrose gradient centrifugation. (B′) Corresponding autoradiogram showing immunochemical detection of the 146-kDa protein in fractions 7–10. The bulk of the protein is recovered in fraction 8, which corresponds to a mean S value of 12.5. (C) Proteins from Xenopus A6 cell lysates were fractionated by gel filtration, separated by SDS-PAGE, and stained with Coomassie blue. Bars on top of the lanes indicate the peak positions of the cofractionated reference proteins: dextran blue (Mapp 2,000,000; fraction 2), thyroglobulin (Mapp 669,000; fraction 12), ferritin (Mapp 440,000; fraction 17), and catalase (Mapp 232,000; fraction 24). (C′) Corresponding immunoblot with antibody B2.4–1. The 146-kDa protein is detectable in fraction 5 and fractions 8–10 (main peaks are denoted by arrows), corresponding to Mapp 1,400,000 and 1,000,000, respectively.
Immunolocalization in Cultured Cells and Tissues
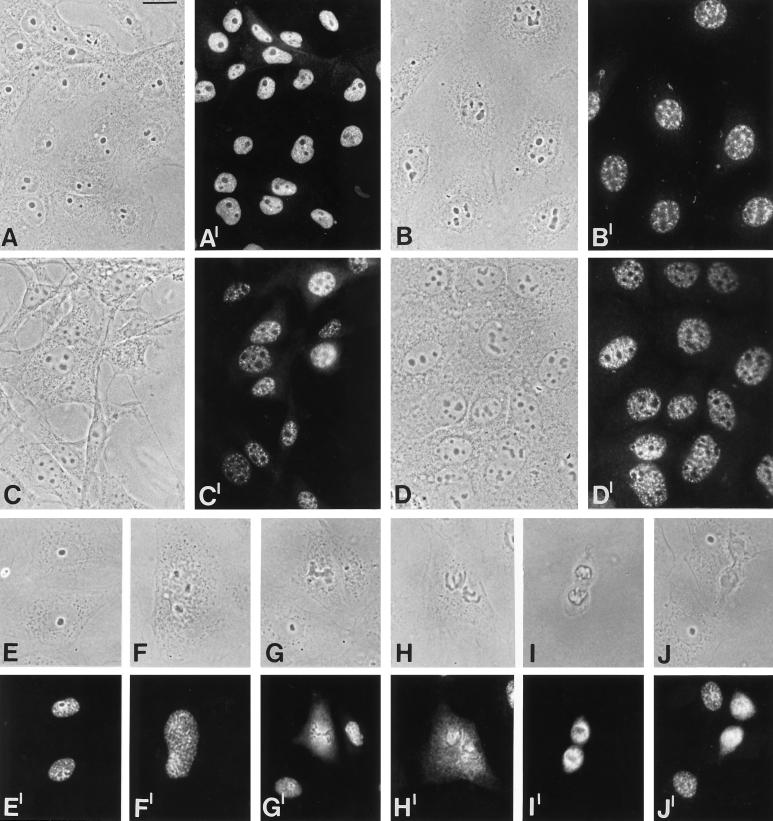
We next examined the intracellular location of the 146-kDa protein in cells from different vertebrate species. On monolayers of cultured cells including amphibian (Figure 6, A and A′), bovine (B and B′), mouse (C and C′), and human cells (D and D′), the antibodies stained the nucleoplasm in a finely punctate pattern. In some cells the protein was clearly enriched in larger, granular structures (B and B′). During mitosis, the protein displayed characteristic changes in its distribution. Various mitotic stages in rat kangaroo kidney epithelial cells (PtK2) are presented in Figure 6 (E and E′–J and J′). At the prophase–metaphase transition the protein is retained in the inner chromosomal regions, but also within the contours of the disintegrating nuclear envelope (F and F′). Subsequently, in metaphase and anaphase (G and G′–I and I′) the protein is transiently dispersed over most of the cytoplasm, leaving the chromosomes negative. In telophase, the protein is reconcentrated around the chromosomal masses in the forming daughter nuclei (J and J′).
Figure 6.
Immunolocalization studies in cultured cells from different species. Phase contrast micrographs are shown in A–J and the corresponding immunofluorescence micrographs in A′–J′. (A and A′) Xenopus kidney epithelial cells, line A6. (B, and B′) Bovine mammary gland-derived cells, line BMGE+H. (C and C′) Embryonal mouse line 3T3-L1. (D and D′) Human primary liver carcinoma line PLC. (E–J and E′–J′) Distribution of the 146-kDa protein during mitosis in rat kangaroo cells, PtK2; (E and E′) interphase; (F and F′) prophase; (G and G′) metaphase; (H and H′) late metaphase; (I and I′) anaphase; (J and J′) telophase. Bar, 20 μm
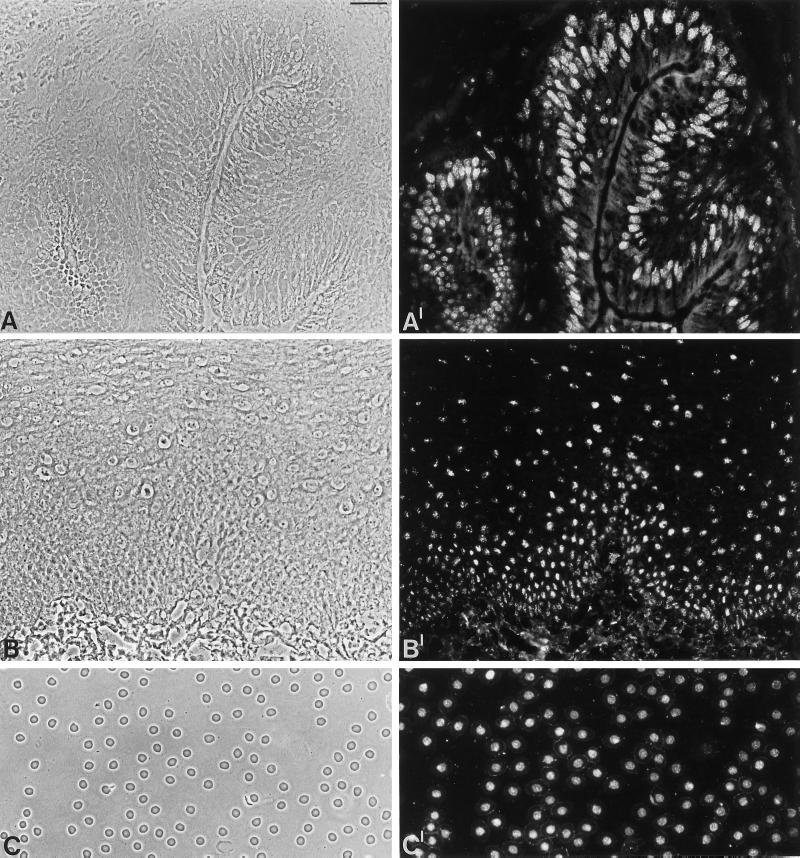
Immunofluorescence on frozen sections of tissues was seen in nuclei of all cell types examined, including epithelial cells as well as submucosal cells of Xenopus intestine (Figure 7, A and A′) and in human esophagus (B and B′). Moreover, an exclusive nuclear staining was observed in Xenopus oocytes, including the follicle epithelial cells surrounding the oocytes, in epithelial cells of glands and ducts, fibroblasts and other dermal cells in the skin, cardiomyocytes, endothelial cells, and erythrocytes (our unpublished observations). Interestingly, nuclei of Xenopus erythrocytes that are reported to be inactive in transcription and replication (Maclean et al., 1973; Gregory et al., 1977; Maclean and Gregory, 1981; Coppock et al., 1989) were intensely stained by the antibodies specific for the 146-kDa protein (C and C′), indicating that the protein is a general nuclear constituent and that its presence does not depend on RNA or DNA synthesis.
Figure 7.
Immunofluorescence microscopy of the 146-kDa protein in different tissues and X. laevis erythrocytes. (A and B) Phase contrast micrographs of frozen sections through Xenopus intestine and human esophagus, respectively. (A′ and B′) Corresponding immunofluorescence micrographs with antibody B2.4–1. (C) Phase contrast micrograph of X. laevis erythrocytes in a blood smear preparation. (C′) Corresponding immunofluorescence micrograph with antibody B2.4–1. Bar, 20 μm (in A and C); 40 μm (B)
Colocalization Studies with Well Characterized Nuclear Marker Proteins
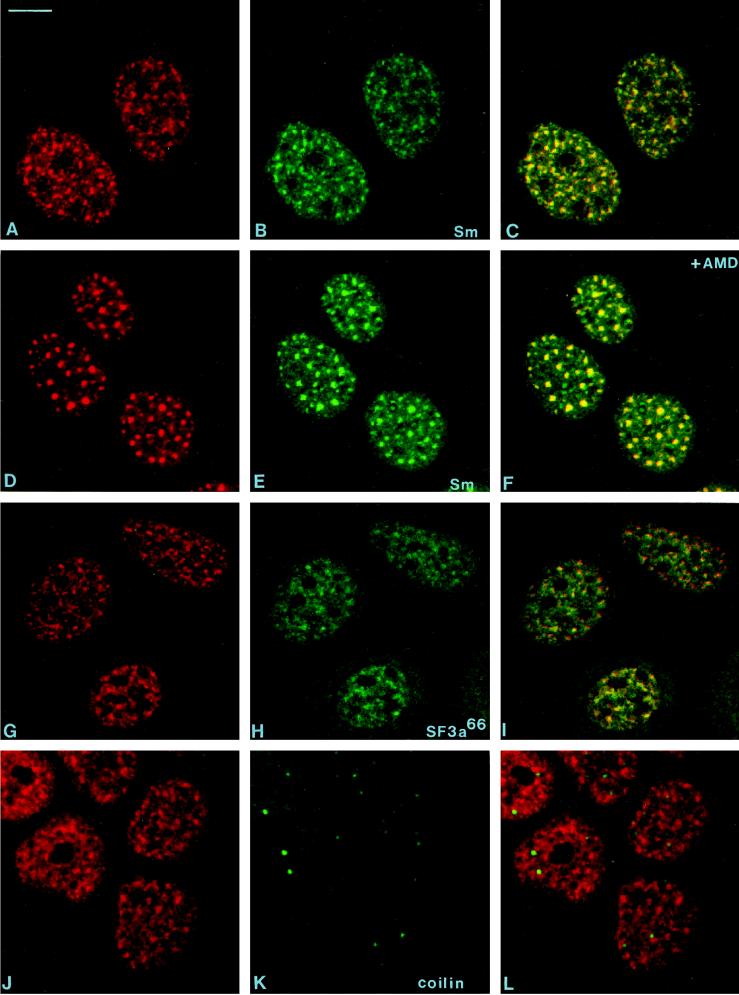
The observation that the 146-kDa protein was distributed diffusely in the nucleoplasm but also localized in nuclear speckles, which represent subnuclear compartments enriched in snRNPs and other splicing factors (reviewed by Spector, 1993), prompted us to compare the localization of the 146-kDa protein with those of known markers for these intranuclear structures.
By confocal laser scanning microscopy we directly compared the localization of the 146-kDa protein with that of Sm proteins, which represent general constituents of snRNPs and are located in speckles, coiled bodies, and the nucleoplasm. The experiments were performed in untreated cells (Figure 8, A–C) and upon treatment with the transcription inhibitor AMD (Figure 8, D–F). We further carried out colocalization studies with antibodies to the U2 snRNP-associated splicing factor SF3a66 (G–I) and coilin, a general constituent of coiled bodies (J–L). The results demonstrate that the 146-kDa protein does not colocalize with coilin and Sm proteins in coiled bodies, but clearly localizes in speckles, similar to the Sm proteins and the 66-kDa subunit of SF3a, although in both cases, the colocalization is not complete. The characteristic redistribution of the 146- kDa protein to enlarged nuclear speckles in response to AMD is noteworthy, a phenomenon earlier described for Sm proteins and various splicing factors (Carmo-Fonseca et al., 1991, 1992; Lamond and Carmo-Fonseca, 1993). Again, several of the structures decorated by the anti-Sm antibodies do not contain the 146-kDa protein. Notably, the distribution of the 146-kDa protein was not affected by treatment with DNase and RNase A, or heat shock (our unpublished observations) in contrast to snRNP antigens, which are known to become diffusely distributed after RNase digestion or heat shock (Spector et al., 1991). In this respect, the intracellular distribution of the 146 kDa protein resembles that of the splicing factor SC35 (Spector et al., 1991).
Figure 8.
Laser scanning confocal microscopy of double-labeling experiments. The distribution of the 146-kDa protein (A, D, G, and J; antibody B2.4–1) was compared with that of snRNP-specific (Sm) proteins (B and E; mAbY12), the protein splicing factor SF3a66 (panel H; mAb66), and the coiled body-specific protein coilin (panel K; anticoilin). The corresponding overlays are shown in panels C, F, I, and L. The cells shown in panels D–F were treated for 4 h with AMD (5 μg/ml). Bar, 10 μm
In summary, our localization studies revealed that the novel 146-kDa protein is distributed in a punctate pattern over a diffuse background throughout the nucleus but is excluded from coiled bodies and nucleoli. Moreover, it colocalizes with various components of the splicing machinery.
The 146-kDa Protein Cofractionates with the U2 snRNP-Associated Splicing Factor SF3b
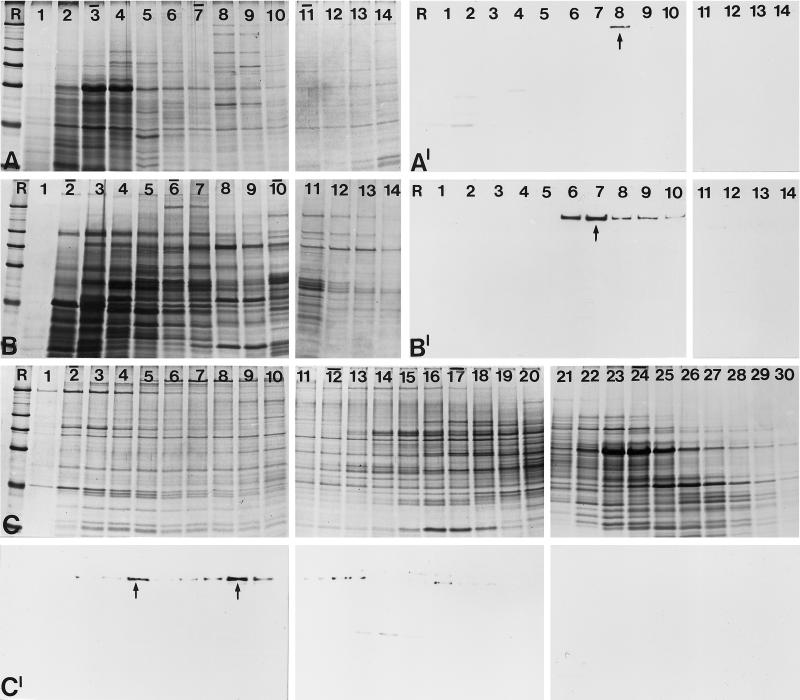
In view of the findings that the 146-kDa protein sedimented with an unexpectedly high S value and at least partially colocalized with marker proteins known to be involved in pre-mRNA splicing, we investigated whether it was present in chromatographic fractions enriched in U2 snRNP and the well characterized splicing factor SF3, which consists of two multimeric components (SF3a and SF3b) that were originally purified as non-snRNP–splicing factors (Brosi et al., 1993a,b). It is well established that both proteins are constituents of the 17S U2 snRNP, which functions in prespliceosome assembly, i.e., they can be regarded as loosely bound U2 snRNP-specific proteins (reviewed by Hodges and Beggs, 1994; Krämer, 1996; Will and Lührmann, 1997). It has been shown that SF3b binds first to the 12S U2 snRNP to form an intermediate complex of 15S, thereby facilitating the addition of SF3a to form the active 17S complex (Brosi et al., 1993a).
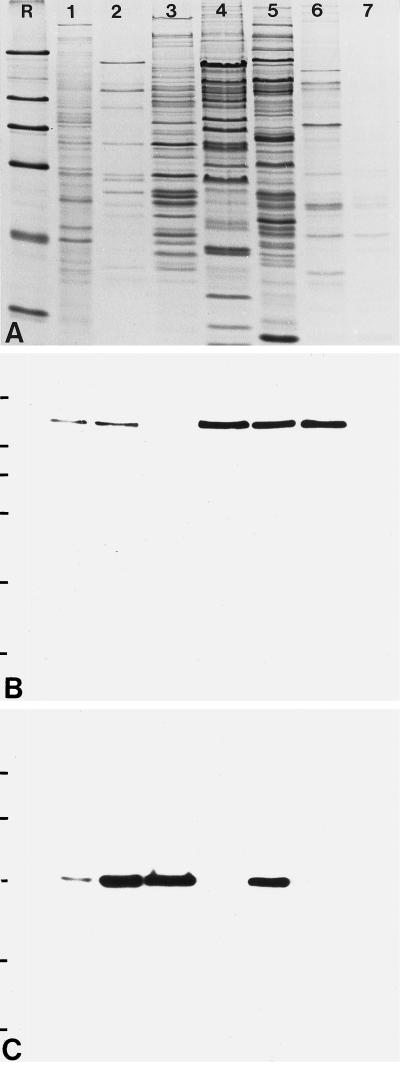
Chromatographic fractions from HeLa cell nuclei enriched in U2 snRNP particles of different size as well as snRNP-free fractions containing SF3a and SF3b were separated by SDS-PAGE (Figure 9A), transferred to nitrocellulose filters, and probed either with antibody B2.4–1 directed against the 146-kDa protein (Figure 9B) or with mAb66 specific for the 66-kDa subunit of SF3a (Brosi et al., 1993a,b; Figure 9C). The 146-kDa protein could be detected in all fractions that contain SF3b: in total nuclear extract (Figure 9B, lane 1), in a fraction containing SF3a and SF3b (lane 2), in a fraction enriched in SF3b (lane 4), and in fractions enriched in the 17S and 15S U2 snRNP particles (lanes 5 and 6), but it was completely absent from fractions enriched in SF3a (lane 3) and the 12S U2 snRNP (lane 7). In contrast, the 66-kDa subunit of SF3a was only detected in fractions containing SF3a (lanes 1, 2, 3, and 5).
Figure 9.
Identification of the 146-kDa protein in fractions of HeLa cell nuclei enriched in splicing factors and U2 snRNPs. (A) Coomassie blue-stained gel after SDS-PAGE of total nuclear extract (lane 1) and fractions enriched in splicing factors SF3a and SF3b (lane 2), SF3a (lane 3), SF3b (lane 4), and fractions enriched in U2 snRNPs of different composition: 17S, i.e., U2 snRNP associated with SF3a and SF3b (lane 5); 15S, containing U2 snRNP associated with SF3b (lane 6); and 12S, U2 snRNP lacking SF3a and SF3b (lane 7). R, reference proteins as shown in Figure 2. (B) Corresponding immunoblot with antibody B2.4–1 directed against the 146-kDa protein. (C) In parallel, a second filter was probed with mAb 66 directed against the 66-kDa component of SF3a.
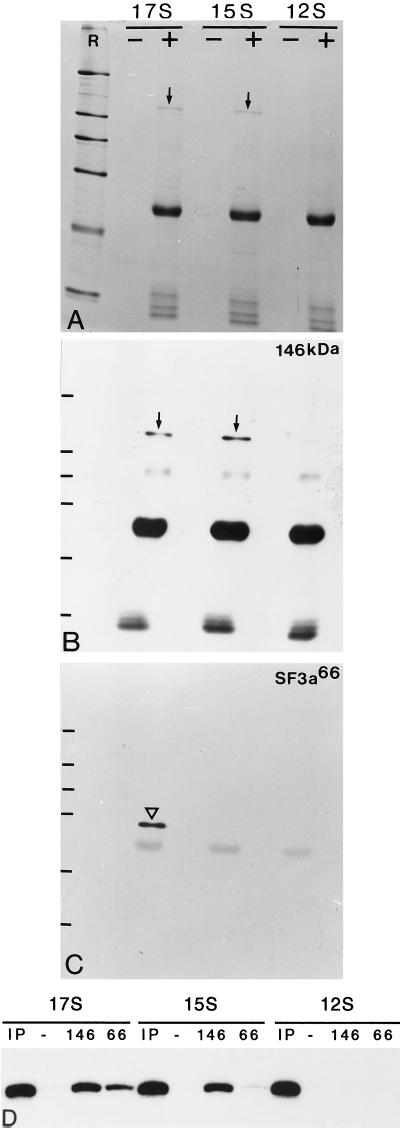
An association of the 146-kDa protein with the different forms of U2 snRNP was further investigated by immunoprecipitation. As shown in Figure 10A, antibody B2.4–1 precipitates a protein of 140–150 kDa from 17S and 15S U2 snRNP fractions, but not from fractions containing the 12S U2 snRNP. Several smaller proteins are precipitated in minor quantities. Western blotting of the immunoprecipitates confirmed the presence of the 146-kDa protein in fractions containing the 17S and 15S U2 snRNPs (Figure 10B). Moreover, Northern blot analysis demonstrated the specific precipitation of U2 snRNA from these fractions, whereas U2 snRNA was not precipitated from the 12S U2 snRNP fraction (Figure 10D). Such properties would be expected for a component of SF3b. The precipitate from the 17S, but not the 15S, U2 snRNP also contained SF3a66 (Figure 10C), suggesting that the 146-kDa protein is tightly associated not only with U2 snRNA but also with SF3a in the 17S U2 snRNP fraction. In a control experiments with mAb 66, U2 snRNA was only detected in immunoprecipitates of the 17S U2 snRNP, but not among the components precipitated from the 15S and 12S U2 snRNPs (Figure 10D), consistent with the previous observation that SF3a is only associated with the 17S U2 snRNP (Brosi et al., 1993a). Taken together, these results indicate that the 146-kDa protein is associated with U2 snRNPs. The specific precipitation with antibody B2.4–1 of the 146-kDa protein and U2 snRNA from the 17S and 15S U2 snRNPs, but not from the 12S particle, as well as the presence of the 146-kDa protein in fractions enriched in SF3b, but not SF3a, strongly suggest that this protein is a component of splicing factor SF3b.
Figure 10.
Immunoprecipitation of U2 snRNP particles with antibody B2.4–1. (A) Coomassie blue-stained SDS gel after immunoprecipitation of the 17S, 15S, and 12S U2 snRNP fractions in the absence (−) or presence (+) of antibody B2.4–1. Arrows indicate the 146-kDa protein. R, reference proteins as shown in Figure 2. (B) Immunoblot of a parallel SDS gel incubated with antibody B2.4–1. (C) After stripping of the membrane the same blot was incubated with mAb 66. The arrowhead indicates SF3a66. (D) A Northern blot of snRNAs immunoprecipitated from the 17S, 15S, and 12S U2 snRNP fractions (IP, input) in the absence of antibody (−) or in the presence of antibody B2.4–1 (146) or mAb 66 (66) was probed with a radiolabeled antisense U2 snRNA transcript. The input corresponds to the amount of fraction used for the immunoprecipitations.
DISCUSSION
In this study we have identified and characterized a novel nuclear protein that is present in various cell types of X. laevis to human. Both the high evolutionary conservation in aa sequence between the Xenopus protein and putative counterparts in human, C. elegans, and yeast, as well as the wide cross-reactivity of antibodies raised against the Xenopus protein, suggest that this protein participates in a fundamental cellular process. Immunolocalization studies revealed the presence of the 146-kDa protein in the cell nucleus, where it is found throughout the nucleoplasm, except in nucleoli, but particularly enriched in distinct nuclear domains, i.e., in nuclear speckles. This intranuclear localization is reminiscent of the pattern frequently observed with antibodies directed against proteins involved in the splicing of pre-mRNAs (Spector, 1993).
The localization of splicing factors in the nucleus is highly dynamic and influenced by a number of factors such as the rate of transcription and heat shock treatment (Spector et al., 1991; Carmo-Fonseca et al., 1992). Our results show that the intranuclear localization of the 146-kDa protein is also affected by these factors. The potent inhibitor of RNA transcription, AMD, has long been known to affect nuclear/nucleolar structure (Simard et al., 1974) and causes, for example, clumping of interchromatin granules and segregation of nucleolar components. The distribution of the 146-kDa protein varies from connected speckles in transcriptionally active cells to unconnected and enlarged speckles when cells are treated with AMD. At the same time, the diffuse nucleoplasmic staining outside of speckles, which is normally seen in untreated cells, is greatly reduced. In this respect, the distribution of the 146-kDa protein is very similar to that described earlier for various snRNP proteins and the splicing factor SC35 (Carmo-Fonseca et al., 1991, 1992; Spector et al., 1993). Recently, a sequence motif sufficient for targeting a protein to nuclear speckles has been described (Li and Bingham, 1991; Hedley et al., 1995). This sequence motif, originally identified in the primary sequence of the Drosophila Transformer (Tra) protein, but also found in a number of other splicing factors, including both subunits of U2AF (Zamore et al., 1992; Zhang et al., 1992), the U1 snRNP-specific 70-kDa protein (Mancebo et al., 1990), SC35 (Fu and Maniatis, 1992), and ASF/SF2 (Ge et al., 1991; Krainer et al., 1991), is characterized by a region rich in arginine/serine dipeptides (RS-domain, see Zahler et al., 1992). In fact, the RS-domain is the only known sequence that is shared by all of the splicing factors that localize to speckles. The 146-kDa protein presented here does not contain such an RS-domain, but clearly localizes to speckles. At the moment, we do not know whether the protein contains another sequence motif responsible for this specific intranuclear localization or whether it is directed to speckles via its interaction with other proteins carrying a RS-domain. Our observation that neither DNA nor RNA is needed for retention of the protein in the nucleus also suggests that its association with these nuclear structures takes place through protein–protein interactions, as has been described for splicing factor SC35 (Spector et al., 1991). Moreover, in the primary sequence of the 146-kDa protein no sequence motifs known to mediate binding to RNA and/or DNA are found (Burd and Dreyfuss, 1994). We are currently performing a detailed mutational analysis to identify sequence elements required for nuclear uptake and the characteristic intranuclear distribution of the 146-kDa protein.
Generally the snRNP proteins fall into two classes. The first class comprises the Sm proteins, B, B′, D1, D2, D3, E, F, and G. These are shared by the snRNP particles U1, U2, U5, and U4/U6 (Lehmeier et al., 1990). The second class comprises proteins that bind to the snRNPs in a particle-specific manner. For example, the U2-specific proteins can be divided into two groups, according to the conditions under which they bind. Under high-salt conditions, U2 snRNP is isolated in a 12S form; in addition to the common proteins, this particle contains two U2-specific proteins, A′ and B". However, in nuclear extracts active in splicing, U2 snRNP sediments at 17S (Behrens et al., 1993) and contains nine additional U2-specific proteins with molecular masses ranging from 35 to 160 kDa (reviewed by Krämer, 1996; Will and Lührmann, 1997). It has been shown that two protein complexes (SF3a and SF3b) interact specifically with the 12S U2 snRNP by converting it into the active 17S form (Brosi et al., 1993a,b). SF3a and SF3b can therefore be regarded as U2 snRNP-specific proteins (for recent reviews see Hodges and Beggs, 1994; Krämer, 1996).
SF3a consists of three polypeptides of 60, 66, and 120 kDa (Brosi et al., 1993b), which are also known as the spliceosome-associated proteins SAP61, SAP62, and SAP114 (Bennett and Reed, 1993). SF3b consists of four polypeptides of 50, 130, 145, and 155 kDa (SAP49, 130, 145 and 155; Staknis and Reed, 1994; reviewed by Krämer, 1996). cDNAs encoding the three subunits of SF3a and two SF3b subunits of 50 and 145 kDa have been isolated from human cells and homologous proteins have been identified in yeast (see Krämer, 1996). Moreover, functional studies have indicated that these factors play a critical role in spliceosome assembly (Brosi et al., 1993a,b; Gozani et al., 1996). By Western blotting we have shown that the 146-kDa protein is present in snRNP-free fractions enriched in SF3b but not in fractions enriched in SF3a. Moreover, the 146-kDa protein is precipitated from fractions containing the 15S and 17S U2 snRNPs but is absent from the 12S U2 snRNP fraction, a distribution that is reminiscent of the association of SF3b with the U2 snRNP (Brosi et al., 1993a).
The coprecipitation of U2 snRNA and SF3a66 with antibodies specific for the 146-kDa protein from U2 snRNP-containing fractions confirms the presence of this protein in the same complex with either U2 snRNP (in the 15S U2 particle) or with U2 snRNP and SF3a (in the 17S U2 snRNP). These results strongly suggest that the 146-kDa protein is a component of SF3b. It should be noted that in purified SF3b the four subunits are detected in nearly equimolar amounts (P. Grüter, K. Gröning, B. Kastner, and A. Krämer, manuscript in preparation). Antibodies directed against the 146-kDa protein precipitated the protein from Xenopus A6 cell lysates (Figure 4) or from the 17S and 15S U2 snRNP fractions (Figure 10) as the major polypeptide, whereas other polypeptides were present at lower concentration. Our failure to precipitate other SF3b subunits in equimolar amounts could be explained by a pool of free 146-kDa protein that is not part of the SF3b complex. On the other hand, the 145-kDa subunit of SF3b exhibits an aberrant migration in gels of different acrylamide concentrations and can comigrate with the 155-kDa subunit (Staknis and Reed, 1994; our unpublished observations). Thus, the apparent overrepresentation of the 146-kDa protein in the immunoprecipitates could result from a comigration of the two largest subunits of SF3b. cDNAs encoding SF3b50 and SF3b145 have been isolated (see Krämer, 1996), but cDNA sequences for SF3b130 and SF3b155 have not been reported. Given the calculated molecular mass (146.2 kDa) for the 146 kDa protein, we favor the notion that it corresponds to SF3b155. Further experiments are in progress to test the function of the 146-kDa protein in spliceosome assembly and splicing.
ACKNOWLEDGMENTS
We thank Dr. Bernadette Fouquet for providing hybridoma cell line B2, Sonja Reidenbach for expert technical assistance, Dr. Hans-Richard Rackwitz for preparing and KLH-coupling of synthetic peptides, Andreas Hunziker for competent sequencing work, Jutta Müller-Osterholt for excellent photographic work, and Dr. Harald Herrmann for reading the manuscript. We also gratefully acknowledge Dr. Herbert Spring for his expert cooperation in the laser scanning confocal microscopy. This study has been supported in part by the Deutsche Forschungsgemeinschaft (grant Schm 862/2–3 to M.S.-Z.).
REFERENCES
- Ankenbauer T, Kleinschmidt JA, Walsh MJ, Weiner OH, Franke WW. Identification of a widespread nuclear actin-binding protein. Nature. 1989;342:822–825. doi: 10.1038/342822a0. [DOI] [PubMed] [Google Scholar]
- Ascoli CA, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga SJ, Steitz JA. The diverse world of small ribonucleoproteins. In: Gesteland RF, Atkins JF, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 359–381. [Google Scholar]
- Behrens S-E, Tyc K, Kastner B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R, Gröning K, Behrens S-E, Lührmann R, Krämer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993a;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- Brosi R, Hauri H-P, Krämer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993b;268:17640–17646. [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino SML, Merdes A, Brunner C, Zamore PD, Green MR, Hurt E, Lamond AI. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J. 1991;10:195–206. doi: 10.1002/j.1460-2075.1991.tb07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho TT, Lammond AI. Transcription-dependent co-localization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Coppock DL, Lue RA, Wangh LJ. Replication of Xenopus erythrocyte nuclei in a homologous egg extract requires prior proteolytic treatment. Dev Biol. 1989;131:102–110. doi: 10.1016/s0012-1606(89)80041-7. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Waizenegger I, Krohne G. Nuclear pore complex glycoprotein p62 of Xenopus laevis and mouse: cDNA cloning and identification of its glycosylated region. Eur J Cell Biol. 1991;55:31–47. [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Köhler A, Stuurman N, van Driel R, Franke WW. Intranuclear filaments containing a nuclear pore complex protein. J Cell Biol. 1993;123:1333–1344. doi: 10.1083/jcb.123.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Franke WW. Cytoplasmic annulate lamellae in cultured cells: composition, distribution, and mitotic behavior. Cell Tissue Res. 1996;284:177–191. doi: 10.1007/s004410050578. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz H-R, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences — a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dumont JN. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory-maintained animals. J Morphol. 1972;136:153–164. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WH, Jr, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Kay BK. Biochemical fractionation of oocytes. Methods Cell Biol. 1991;36:133–148. doi: 10.1016/s0091-679x(08)60275-7. [DOI] [PubMed] [Google Scholar]
- Fouquet B. Expression von Intermediärfilament-Proteinen in Xenopus laevis. Ph.D. Thesis. Heidelberg, Germany: Faculty of Biology, University of Heidelberg; 1991. pp. 1–175. [Google Scholar]
- Franke WW, Schmid E, Winter S, Osborn M, Weber K. Widespread occurrence of intermediate-sized filaments of the vimentin-type in cultured cells from diverse vertebrates. Exp Cell Res. 1979;123:25–46. doi: 10.1016/0014-4827(79)90418-x. [DOI] [PubMed] [Google Scholar]
- Franke WW, Schmid E, Vandekerckhove J, Weber K. A permanently proliferating rat vascular smooth muscle cell with maintained expression of smooth muscle characteristics, including actin of the vascular smooth muscle type. J Cell Biol. 1980;87:594–600. doi: 10.1083/jcb.87.3.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW, Kleinschmidt JA, Spring H, Krohne G, Grund C, Trendelenburg MF, Stoehr M, Scheer U. A nucleolar skeleton of protein filaments demonstrated in amplified nucleoli of Xenopus laevis. J Cell Biol. 1981;90:289–299. doi: 10.1083/jcb.90.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- Gall JG, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M, Krauss RD, Chiang L, Valcárcel J, Green MR, Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Green MR. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Gregory SP, Hilder VA, Maclean N. Transcriptional reactivation of isolated Xenopus erythrocyte nuclei: patterns of RNA synthesis. J Cell Sci. 1977;28:49–60. doi: 10.1242/jcs.28.1.49. [DOI] [PubMed] [Google Scholar]
- Hedley ML, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid HW, Schmidt A, Zimbelmann R, Schäfer S, Winter-Simanowski S, Stumpp S, Keith M, Figge U, Schnölzer M, Franke WW. Cell type-specific desmosomal plaque proteins of the plakoglobin family: plakophilin 1 (band 6 protein) Differentiation. 1994;58:113–131. doi: 10.1046/j.1432-0436.1995.5820113.x. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Wiche G. Specific in situ phosphorylation of plectin in detergent-resistant cytoskeletons from cultured Chinese hamster ovary cells. J Biol Chem. 1983;258:14610–14618. [PubMed] [Google Scholar]
- Hodges PE, Beggs JD. U2 fulfils a commitment. Curr Biol. 1994;4:264–267. doi: 10.1016/s0960-9822(00)00061-0. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügle B, Scheer U, Franke WW. Ribocharin: a nuclear Mr 40,000 protein specific to precursor particles of the large ribosomal subunit. Cell. 1985;41:615–627. doi: 10.1016/s0092-8674(85)80034-9. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt JA, Franke WW. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell. 1982;29:799–809. doi: 10.1016/0092-8674(82)90442-1. [DOI] [PubMed] [Google Scholar]
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Korioth F, Gieffers C, Maul GG, Frey J. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J Cell Biol. 1995;130:1–13. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Krohne G, Wolin SL, McKeon FD, Franke WW, Kirschner MW. Nuclear lamin LI of Xenopus laevis: cDNA cloning, amino acid sequence and binding specificity of a member of the lamin B subfamily. EMBO J. 1987;6:3801–3808. doi: 10.1002/j.1460-2075.1987.tb02716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. Localisation of splicing snRNPs in mammalian cells. Mol Biol Rep. 1993;18:127–133. doi: 10.1007/BF00986767. [DOI] [PubMed] [Google Scholar]
- Lee KA, Green MR. Small-scale preparation of extracts from radiolabeled cells efficient in pre-mRNA splicing. Methods Enzymol. 1990;181:20–30. doi: 10.1016/0076-6879(90)81108-7. [DOI] [PubMed] [Google Scholar]
- Lehmeier T, Foulaki K, Lührmann R. Evidence for three distinct D proteins, which react differentially with anti-Sm autoantibodies, in the cores of the major snRNPs U1, U2, U4/U6 and U5. Nucleic Acids Res. 1990;18:6475–6484. doi: 10.1093/nar/18.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bingham PM. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- Maclean N, Gregory SP. The Cell Nucleus—Nuclear Particles. Vol. 3. H. Busch, New York: Academic Press; 1981. Transcription in isolated nuclei; pp. 139–191. [Google Scholar]
- Maclean N, Hilder VA, Baynes YA. RNA synthesis in Xenopus erythrocytes. Cell Differ. 1973;2:261–269. doi: 10.1016/0045-6039(73)90030-4. [DOI] [PubMed] [Google Scholar]
- Mancebo R, Lo PC, Mount SM. Structure and expression of the Drosophila melanogaster gene for the U1 small nuclear ribonucleoprotein particle 70K protein. Mol Cell Biol. 1990;10:2492–2502. doi: 10.1128/mcb.10.6.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens C, Kuhn C, Franke WW. Plakophilins 2a and 2b: Constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J Cell Biol. 1996;135:1009–1025. doi: 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to messenger RNAs by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Cooke CA, Earnshaw WC. Compartmentalization within the nucleus: discovery of a novel subnuclear regions. J Cell Biol. 1991;115:919–931. doi: 10.1083/jcb.115.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalenghe F, Buscaglia M, Steinheil C, Crippa M. Large scale isolation of nuclei and nucleoli from vitellogenic oocytes in Xenopus laevis. Chromosoma. 1978;66:299–308. doi: 10.1007/BF00328531. [DOI] [PubMed] [Google Scholar]
- Schmid E, Schiller DL, Grund C, Stadler J, Franke WW. Tissue type-specific expression of intermediate filament proteins in a cultured epithelial cell line from bovine mammary gland. J Cell Biol. 1983;96:37–50. doi: 10.1083/jcb.96.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Nigg EA. Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Hügle B, Scheer U, Franke WW. Identification and localization of a novel nucleolar protein of high molecular weight by a monoclonal antibody. Exp Cell Res. 1984;153:327–346. doi: 10.1016/0014-4827(84)90604-9. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Hügle-Dörr B, Franke WW. A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO J. 1987;6:1881–1890. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnölzer M, Alewood P, Jones A, Alewood D, Kent SB. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int J Pept Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- Simard R, Langelier Y, Mandeville R, Maestracci N, Royal A. The Cell Nucleus. Vol. 3. H. Busch, New York: Academic Press; 1974. Inhibitors as tools in elucidating the structure and function of the nucleus; pp. 447–487. [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, O’Keefe RT, Jiménez-García LF. Dynamics of transcription and pre-mRNA splicing within the mammalian cell nucleus. Cold Spring Harbor Symp Quant Biol. 1993;58:799–805. doi: 10.1101/sqb.1993.058.01.087. [DOI] [PubMed] [Google Scholar]
- Staknis D, Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994;14:2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO, Kornberg RD. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci USA. 1975;72:2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utans U, Behrens S-E, Lührmann R, Kole R, Krämer A. A splicing factor that is inactivated during in vivo heat shock is functionally equivalent to the [U4/U6. U5] triple snRNP-specific proteins. Genes Dev. 1992;6:631–641. doi: 10.1101/gad.6.4.631. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirwes RF, Kouzmenko AP, Peters J-M, Franke WW, Schmidt-Zachmann MS. Topogenesis of a nucleolar protein: determination of molecular segments directing nucleolar association. Mol Biol Cell. 1997a;8:231–248. doi: 10.1091/mbc.8.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirwes, R.F., Schmidt-Zachmann, M.S., and Franke, W.W. (1997b). Identification of a small, very acidic constitutive nucleolar protein (NO29) as a member of the nucleoplasmin family. Proc. Natl. Acad. Sci. USA (in press). [DOI] [PMC free article] [PubMed]