The Distribution of Polycomb-Group Proteins During Cell Division and Development in Drosophila Embryos: Impact on Models for Silencing (original) (raw)
Abstract
The subcellular three-dimensional distribution of three polycomb-group (PcG) proteins—polycomb, polyhomeotic and posterior sex combs—in fixed whole-mount Drosophila embryos was analyzed by multicolor confocal fluorescence microscopy. All three proteins are localized in complex patterns of 100 or more loci throughout most of the interphase nuclear volume. The rather narrow distribution of the protein intensities in the vast majority of loci argues against a PcG-mediated sequestration of repressed target genes by aggregation into subnuclear domains. In contrast to the case for PEV repression (Csink, A.K., and S. Henikoff. 1996. Nature. 381:529–531), there is a lack of correlation between the occurrence of PcG proteins and high concentrations of DNA, demonstrating that the silenced genes are not targeted to heterochromatic regions within the nucleus. There is a clear distinction between sites of transcription in the nucleus and sites of PcG binding, supporting the assumption that most PcG binding loci are sites of repressive complexes. Although the PcG proteins maintain tissue-specific repression for up to 14 cell generations, the proteins studied here visibly dissociate from the chromatin during mitosis, and disperse into the cytoplasm in a differential manner. Quantitation of the fluorescence intensities in the whole mount embryos demonstrate that the dissociated proteins are present in the cytoplasm. We determined that <2% of PH remains attached to late metaphase and anaphase chromosomes. Each of the three proteins that were studied has a different rate and extent of dissociation at prophase and reassociation at telophase. These observations have important implications for models of the mechanism and maintenance of PcG- mediated gene repression.
Early in Drosophila embryogenesis, both larval and adult body segment identities are determined by the expression patterns of the homeotic genes. The expression domains in the embryo are established by products of the segmentation genes, which decay by 5–7 h of development. The spatial distribution of repression and expression of the homeotic genes, however, is maintained for up to ten cell divisions before terminal cell differentiation. As first deduced by Lewis (1978), the class of genes responsible for maintenance of repression is the polycomb group (PcG; for reviews see Orlando and Paro, 1995; Paro, 1990; Pirrotta, 1995).1 Animals mutant for any of the approximately 15 members of the PcG identified so far show multiple homeotic transformations resulting from ectopic expression of homeogenes outside their normally restricted domains (Simon et al., 1992). Maintaining the determined state is a process required in every multicellular organism undergoing development. Similar proteins presumably fix homeotic gene repression in higher mammals, as has been deduced by the isolation of genes homologous to polycomb (Pc), polyhomeotic (PH), posterior sex combs (Psc), and enhancer of zeste (_E_[_z_]; Brunk et al., 1991; Gecz et al., 1995; Kanno et al., 1995; Mueller et al., 1995; Pearce et al., 1992; van der Lugt et al., 1994; van Lohuizen et al., 1991). It may be assumed that a better understanding of the mechanism of PcG-mediated repression in Drosophila will have broad ramifications with respect to the problem of developmental control in higher eukaryotes. However, neither a description of the molecular interactions by which PcG repression is established nor how this is maintained through cell division has been determined.
Six of the proteins encoded by the PcG genes—polycomb (PC), polyhomeotic (PH), polycomb-like, posterior sex combs (PSC), suppressor of zeste-2, and enhancer of zeste—were shown to be present in largely overlapping patterns at more than 60 discrete sites on polytene chromosomes of larval salivary glands, including the homeotic gene clusters (Carrington and Jones, 1996; Franke et al., 1992; Lonie et al., 1994; Martin and Adler, 1993; Rastelli et al., 1993; Zink and Paro, 1989). Indeed, large multiprotein particles are immunoprecipitated from embryonic nuclear extracts by antibodies against PcG proteins (Alkema et al., 1997; Franke et al., 1992). These data suggest that the proteins exert their function by forming multiprotein complexes that bind DNA. However, neither purified PC nor PH have been shown to have DNA-binding activity in vitro. The genes of the PcG show a number of similarities, such as dosage sensitivity and synergism, to the modifiers of position effect variegation (PEV) in Drosophila. The finding of a homologous protein domain in PC (the chromodomain; Paro and Hogness, 1991) and the heterochromatin-specific protein HP1 (Eissenberg et al., 1990; James and Elgin, 1986) links the PcG and the modifiers of PEV at the molecular level. Such observations have led to the proposal that the PcG gene products function in a manner similar to the heterochromatin proteins, i.e., by acting as multimeric complexes and packaging the DNA into a repressed chromatin structure (Messmer et al., 1992; Paro, 1990; Reuter and Spierer, 1992).
Alternative models for the mechanism of PcG-mediated repression have been proposed, such as (a) the sequestration of target genes into specific subnuclear compartments that restrict access to transcription factors and RNA polymerase (Felsenfeld, 1996; Paro, 1993; Schloßherr et al., 1994; Strouboulis and Wolffe, 1996; Wakimoto and Hearn, 1990); (b) nucleosome positioning at promoter sequences (Pirrotta and Rastelli, 1994); and (c) the remodeling of DNA-looping interactions from promoter–enhancer to promoter–PcG interactions (Bienz and Müller, 1995; Pirrotta and Rastelli, 1994).
Earlier immunochemical fluorescence and enzyme-linked assays in situ concluded that the PcG proteins are localized constitutively in the nucleus throughout embryogenesis (DeCamillis and Brock, 1994; Paro and Zink, 1992), and appear to remain attached to the chromatin through the cell cycle (DeCamillis and Brock, 1994; Martin and Adler, 1993). Studies addressing the intranuclear patterns of PcG protein distribution at high resolution could provide data allowing one to distinguish between some of the proposed repression mechanisms. The focus of this study has been to determine the qualitative and semiquantitative distribution of the PcG proteins (PC, PH, and PSC) in whole mount embryos throughout embryogenesis and across the cell cycle using high-resolution confocal laser scanning microscopy.
Materials and Methods
Flies and Egg Collection
Drosophila wild-type strain Oregon R-P2 (Allis et al., 1977) was used throughout. Larvae were raised in plastic bottles on a medium of cornmeal, agar, soy bean meal, malt extract, molasses, yeast, 0.5% (vol/vol) propionic acid, and the mold inhibitor methyl p-hydroxybenzoate (Nipagin™; Caesar and Lorentz, Hilden, Germany). Flies were held in population cages at room temperature and at ∼70% humidity. Eggs were collected from these cages on gauze nets containing fresh yeast.
Antibodies
PC antibodies were produced and affinity-purified as described (Zink and Paro, 1989). PH antibodies were either produced as described previously by immunization with a β-gal/Polyhomeotic (aa753–922) fusion protein (DeCamillis et al., 1992), or with a fusion protein containing an NH2-terminal 6-histidine tag purified by nickel chelate chromatography (Petty, 1987). In this latter case, a 1,304-bp Xho I-Sal I fragment (bases 750–2054) of the ph cDNA (DeCamillis et al., 1992) was cloned into the Sal I site of pQE-31 (QIAGEN Inc., Chatsworth, CA) and expressed in E. coli strain M15. PSC antibodies were raised from an NH2-terminal 6-histidine tag fusion protein (a 1,044-bp Hind III fragment [bases 2566–3610]) of the Psc cDNA (Brunk et al., 1991; van Lohuizen et al., 1991) cloned into the Hind III site of pRSETB (Invitrogen Corp., Carlsbad, CA) and expressed in strain BL21 (DE3; Studier and Moffatt, 1986).
Antibodies were raised by immunizing rabbits with 250 μg fusion protein at four weekly intervals until a strong reaction was obtained. Antibodies were affinity-purified by chromatography on an Affigel 10 (Bio-Rad Laboratories, Hercules, CA) column to which the fusion protein was covalently coupled.
NonA mAb Bj6 and Hrb57A mAb Q18 were generated as described (Frasch and Saumweber, 1989; Saumweber et al., 1980) and purified from hybridoma cell supernatants by chromatography on a protein G Sepharose column (Pharmacia Biotech, Inc., Piscataway, NJ).
Secondary fluorescently labeled antibodies, Cy3-conjugated F(ab′)2 fragment goat anti–rabbit IgG, and Cy3-conjugated F(ab′)2 fragment goat anti–mouse IgG, were purchased (Jackson Laboratories, Bar Harbor, ME).
Coupling Fluorescent Dyes to Primary Antibodies
The mAbs were labeled as previously described (Buchenau et al., 1993; Buchenau et al., 1997) with fluorescein-isothiocyanate (Molecular Probes, Inc., Eugene, Oregon) or the succinimidyl ester Cy-3-OSu (Cy3; Nycomed Amersham Inc., Buckinghamshire, United Kingdom). The spectroscopically determined molar dye/protein ratio was 4. The anti-PH antibody was labeled at a concentration of 2 mg/ml in 100 μl bicine buffer (100 mM, pH 9.0) by adding a sixfold molar excess of succinimidyl ester Cy-5.18-OSu (Cy5; Nycomed Amersham Inc.). After 1 h at room temperature, the reaction was stopped by adding Tris-HCl buffer (pH 7.2) to a final concentration of 2 mM, and nonbound dye molecules were removed from the labeled antibody by gel filtration on a Sephadex G-25 column (Pharmacia Biotech, Inc.). The molar dye/protein ratio was 2.9.
Immunostaining and DNA Staining of Embryos
Immunocytochemistry was carried out according to the method of Mitchison and Sedat (1983) with minor modifications. In brief, embryos were dechorionated in a 3% Na hypochlorite solution for 90 s, fixed by shaking in a mixture of heptane, buffer A (60 mM KCl, 15 mM NaCl, 0.5 mM spermidine, 0.15 mM spermine, 2 mM EDTA, 0.5 mM EGTA, and 15 mM Pipes, pH 7.4) and 37% paraformaldehyde (9:0.9:0.1, by vol) for 20–40 min, and devitellinized by vigorous shaking in a 1:1 mixture of heptane and methanol. The embryos were then transferred into buffer A (or in some cases PBS + 0.05% Tween 20) via a graded solvent series (methanol/buffer: 90:10, 75:25, 50:50, 25:75) and washed extensively in buffer.
Incubations in 200–500 μl of diluted solutions of primary and secondary antibodies were routinely performed on a rotation wheel at room temperature for 2–4 h. Embryos were washed three times for 20 min in 1 ml of buffer A or PBS, and mounted for analysis (see below) or processed further for staining of the DNA.
DNA in the embryos was stained with 5 μM DAPI or 3 μM Hoechst 33342 (Molecular Probes, Inc.) without further processing, or after RNAse A digestion (100–500 μg/ml, 2–4 h) with YOYO®1, BOBO®1 at 100 nM, or TO-PRO™3 at 1 μM (Molecular Probes, Inc.). Thereafter the embryos were washed briefly and then transferred to the mounting medium Mowiol 4-88 (Hoechst AG, Frankfurt, Germany; prepared as described by Heimer and Taylor, 1974) through increasing concentration of the Mowiol/buffer series.
Double antibody staining (for example, PH and Q18) was accomplished by simultaneous incubation with directly labeled primary antibodies (see above). In other experiments, one antibody was indirectly labeled (for example, Cy3-coupled goat anti–mouse antibody with the mAb and Cy5-coupled anti-PH).
Confocal Microscopy and Image Processing
Digital optical sections from whole mount embryos were recorded with a confocal laser scanning microscope (model LSM310; Carl Zeiss, Oberkochen, Germany) using a 63× NA 1.4 Plan-apochromat oil immersion objective, a 63× NA 1.2 C-apochromat water immersion objective, or a 25× NA 0.8 immersion objective with a refractive index correction collar (Carl Zeiss). The microscope was equipped with two internal lasers (HeNe, 633 nm; and argon ion, 488 and 514 nm) and three additional external lasers (argon ion, tunable; HeNe, 543 nm; HeNe, 594 nm). The external lasers were brought into the microscope via fiber optics coaxially aligned to the internal ones.
The different dyes were excited with the following laser lines: DAPI or Hoechst, 365 nm; YOYO1, BOBO1, and fluorescein, 488 nm; Cy3, 543 or 514 nm; Cy5 or TOPRO3, 633 nm. The following emission filters were used: DAPI and Hoechst, LP 418; YOYO1, BOBO1, and fluorescein, BP530; Cy3, LP550, or BP 565 (when excited at 514 nm); LP575 or LP610 (when excited at 543 nm); Cy5 or TOPRO3, LP665. For each double staining the single labeled controls were prepared to establish imaging conditions without cross talk between channels.
Images (eight bit) were acquired with an appropriate scanning time and frame averaging. For doubly stained embryos, the images of the two fluorophore distributions were recorded separately and saved to separate channels of an RGB image. Reconstructions of stereo images were performed using the projection functions of the LSM310 software, NIH- Image (National Institutes of Health, Bethesda, Maryland), or SCILImage (Delft Technical University, Delft, The Netherlands). Image-processing tasks such as grey level ratioing or determining the number of binding loci in image stacks at high resolution (a combination of thresholding and binary labeling functions) were carried out with SCILImage. Figures were composed with Photoshop 3.0 and Illustrator 5.5 (Adobe Systems, Mountain View, CA).
Quantitation of Polyhomeotic Protein
Fixed embryos (aged 0–15 h) were stained for PH or PSC (using affinity-purified antibodies) and DNA (with YOYO1, 100 nM, and TO-PRO3, 1 μM; or Hoechst 33342, 5 μM) and mounted in Mowiol as described above. Serial confocal sections in two channels were recorded from mitotic regions of embryos using a 63×, NA 1.4 Plan-apochromat oil or a 63×, NA 1.2 C-apochromat water objective. The voxel dimensions (x, y, z) in such image stacks were 0.1 × 0.1 × 0.3 μm.
Data were transferred to a Silicon Graphics workstation and quantified with the depth analyzer module of Imaris 2.5 (Bitplane AG, Zürich, Switzerland) or DeltaVision 2.0 (Applied Precision, Inc., Issaquah, WA). The programs open the red and green channels into a single RGB stack, allow the interactive definition of polygons in subsequent sections, and calculate volume, mean, and integrated grey values of the two channels in three dimensions. In these stacks, the cells were grouped into the five categories: inter-, pro-, meta-, ana- and telophase. Within the cellular or nuclear/chromosomal polygon masks, we measured the integrated fluorescence. Mean PH ratios were then determined for each category of the cell cycle by averaging over all nuclei of a class. Image restoration was performed by deconvolution of the fluorescence data using the Huygens module of the Imaris software that uses a maximum likelihood estimation algorithm (van der Voort and Straster, 1995). The average intensity/pixel of the protein fluorescence was calculated inside and outside of the DNA bitmask for the z-axis section of metaphase and anaphase cells with the highest DNA intensity. The total intensities of the protein in the DNA and cytoplasmic two-dimensional areas/three-dimensional vol were then calculated by multiplying by their respective pixel/voxel sums.
Results
The Three-dimensional Distribution of PcG Proteins, Polycomb, Polyhomeotic, and Posterior Sex Combs in Embryonic Interphase Nuclei
The developmental profile of the distribution of the proteins PC, PH, and PSC in fixed whole mount embryos has been analyzed at low resolution by immunocytochemistry with enzymatically developed color precipitates (DeCamillis and Brock, 1994; Franke et al., 1992; Martin and Adler, 1993; Paro and Zink, 1992). In the present study we have taken advantage of the higher spatial resolution that is possible when using fluorescence signals and confocal laser scanning microscopy to describe the subnuclear distribution patterns of the three proteins in detail. Wild-type embryos were fixed with 3.7% formaldehyde in the presence of a buffer containing polyamines (see Materials and Methods). These conditions have been shown to be optimal for the preservation of nuclear structure (Belmont et al., 1989). The PcG proteins were detected by incubation with affinity-purified fluorescently labeled polyclonal antibodies, or by indirect immunofluorescence with Cy3-coupled Fab secondary antibody fragments. DNA was visualized by DNA-specific dyes (DAPI or Hoechst 33342) or with one of the nucleic acid–specific cyanine dyes YOYO1, BOBO1, or TO-PRO3 after RNAse digestion of the embryos. Images were taken with a confocal microscope (Carl Zeiss). All of the antibody preparations were checked for specificity by Western blotting. In addition, antibody specificity in the whole mount embryos was further verified by staining mutant embryos carrying deletions for the individual PcG proteins. Antibodies to topoisomerase II and to core histones were used as controls for chromatin accessibility throughout the cell cycle in whole mount embryos.
In the syncytial blastoderm, the three proteins localize predominantly in the nuclei. They stain all tissues throughout embryogenesis. The conclusions from this study have been drawn from data on the distributions of the proteins in stages 8–14 embryos, developmental times when homeotic gene regulation is established and maintained in embryogenesis. By ratio imaging of PC, PH, or PSC and the DNA on the scale of whole embryos, we confirmed quantitatively the earlier qualitative observation that the PcG proteins are not distributed in concentration gradients along the anterior-posterior axis, and show a very similar distribution in individual nuclei through stage 14 embryos (Fig. 1) after which stage the staining predominates in nuclei of the neural system. In all interphase nuclei, the PcG proteins localize to a large number of discrete spots as depicted in the field of PH-stained interphase nuclei from a gastrula embryo (Fig. 1 D).
Figure 1.
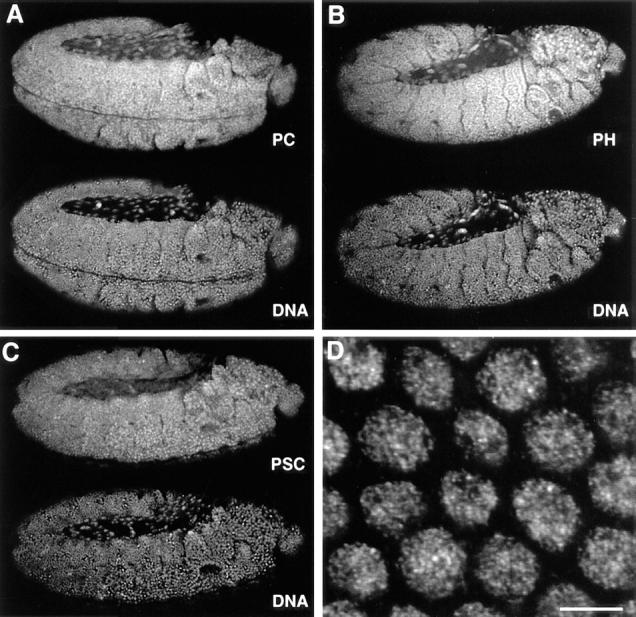
Ubiquitous expression of PcG proteins in developing Drosophila embryos. (A–C) Antibody staining to (A) PC, (B) PH, and (C) PSC (top) and DNA (bottom) in whole mount stage 11 (extended germ band) embryos with anterior right and posterior left. The images are single confocal sections. Objective, Neofluar 25×, NA 0.8 immersion with refractive index correction collar. Field widths are 590 μm. (D) Higher magnification image of a field of 2n nuclei from a whole mount embryo stained for PH. Bar, 5 μm.
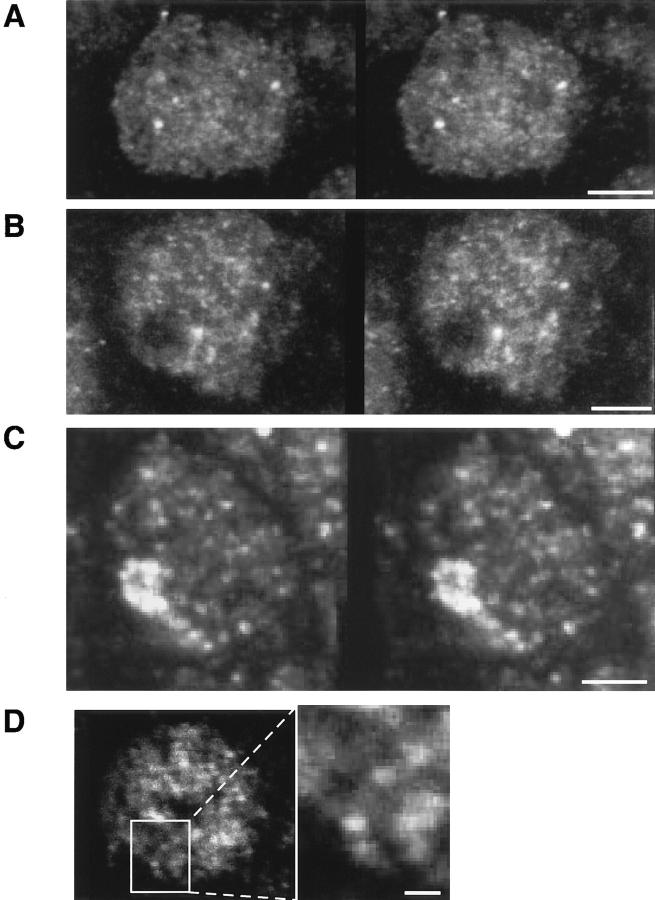
Fig. 2 shows representative examples of high-resolution stereo images of the PH, PC, and PSC protein patterns in embryonic interphase nuclei. These images have been reconstructed from series of optical sections recorded at intervals of 0.2–0.3 μm by projection of the maximal intensities of the stack. The patterns of the PcG proteins are characterized by 100 or more discrete spots distributed throughout most of the nuclear volume. In some parts of the nuclei, threadlike less intense fluorescent signals appeared to connect the spotted pattern (Fig. 2 D). None of the three proteins were found in the nucleoli, which were usually discerned as dark subvolumes within the nuclei (Fig. 2 B). Some of the PSC was present in higher concentrations in a cluster of spots in addition to the more discrete and less intense signals characteristic for PC and PH (Fig. 2 C). These PSC clusters appeared in almost all interphase nuclei from the late blastoderm to about the time of dorsal closure, and appear in different parts of the nucleus, often very near to or partially coincident with the centromeric heterochromatin (not shown).
Figure 2.
Three-dimensional distribution of PcG proteins in embryonic interphase nuclei. (A–C) The stereo images show the distribution of PH (A), PC (B), and PSC (C) in representative interphase nuclei. The images were reconstructed from (A) 30 confocal sections separated by 0.2 μm, (B) 21 sections, 0.2 μm, (C) 15 sections, 0.3 μm, by projection of the maximal pixel values along two viewing lines of the image stack. (D) This overlay of two adjacent confocal sections through a PH-stained interphase nucleus demonstrates one example in which a faint fluorescence between the PH spots suggests the location of the loci on a chromatin fiber. Bars, 2 μm (A–C), 0.5 μm (D).
The fractional volume of PcG complexes in 2n nuclei compared with the nuclear volume was estimated from the voxels of fluorescence intensity in PcG loci. These estimations gave a range of sizes from 0.005% to as much as 0.1% of the nuclear volume for a PcG complex (mean, 0.016%; n = 43). These values are comparable to the ratios of occupied to unoccupied bands measured on images of polytene chromosomes (0.008% for a small and 0.05% for a large PcG-positive band). Therefore, we interpret the spots discerned by the high-resolution imaging in 2n nuclei as accumulations of PC, PH, and PSC on individual interphase chromosomal loci. The order of magnitude of the number of loci also supports the interpretation that we are looking predominantly at individual gene loci. These data provide no evidence for a favored region for PcG accumulation in the nucleus, or for aggregation of PcG loci into larger compartments except for the PSC-specific cluster mentioned above which, however, does not colocalize with the other Polycomb group proteins.
The size and distribution of the PcG loci in embryonic nuclei are different from those discerned in Drosophila tissue culture cells (our unpublished data and Messmer et al., 1992) or the homologous PcG protein distributions in transformed mammalian tissue culture cells (Alkema et al., 1997). In these cases, large domains of PcG accumulation are seen within the nuclei. All of the Drosophila and mammalian cells lines that show such a distribution are heteroploid or aneuploid. Thus, the accumulations may be the result of overproduction of some of the PcG proteins that cannot form competent repression complexes. We have not observed similar PcG distributions in embryonic tissues or primary cell preparations.
The Spatial Relationship between PcG Proteins and DNA in Fixed Embryonic Interphase Nuclei
A number of the proposed models for repression by the PcG proteins invoke restructuring chromatin by a multiprotein complex (see Discussion). In some versions of the models, this restructuring would lead to a local condensation of chromatin, or a relocalization of the repressed sites into heterochromatic DNA domains within the nucleus. In mammalian cells there are conflicting data as to whether repressed genes are relocalized to heterochromatic nuclear domains (Xing et al., 1995), or whether they occupy the same territory as when they are transcriptionally active (Kurz et al., 1996). To determine whether PcG complexes associate preferentially with heterochromatin, we used multiwavelength confocal microscopy to investigate the correlation of PcG protein distribution and DNA concentration.
We imaged whole-mount embryos stained with antibodies against the three proteins and a variety of DNA dyes. In most cases the protein binding appeared to be excluded from the condensed heterochromatin (with the exception of the PSC cluster mentioned above). Two-dimensional correlation plots (Demandolx and Davoust, 1997a, b) of the intensities of antibody staining compared with the intensities of stained DNA in many nuclei in a number of embryos showed no propensity of the PH or PC to be associated with regions of high DNA intensity. Fig. 3 A shows a representative example of such a double staining for PH and DNA in a single optical section through an interphase nucleus. A similar lack of correlation was found between DNA concentration and both PC (Fig. 3 B) and PSC binding (Fig. 3 C). In the merged images in the far right column of Fig. 3, pixels in which both signals have intensities above half the maximum value appear yellow. In fact, few discrete protein spots were yellow (white arrowheads in Fig. 3), that is, most of the high protein intensity and condensed DNA pixels were uncorrelated. We conclude that the PcG proteins appear less frequently in regions of very high DNA concentration than in regions of less condensed DNA. A careful evaluation of the PcG-binding loci on polytene chromosomes of third instar larvae revealed a nonuniform ratio of protein/DNA concentration (data not shown). These data taken together imply that PcG protein binding does not relocalize the repressed genes to areas of condensed heterochromatin.
Figure 3.
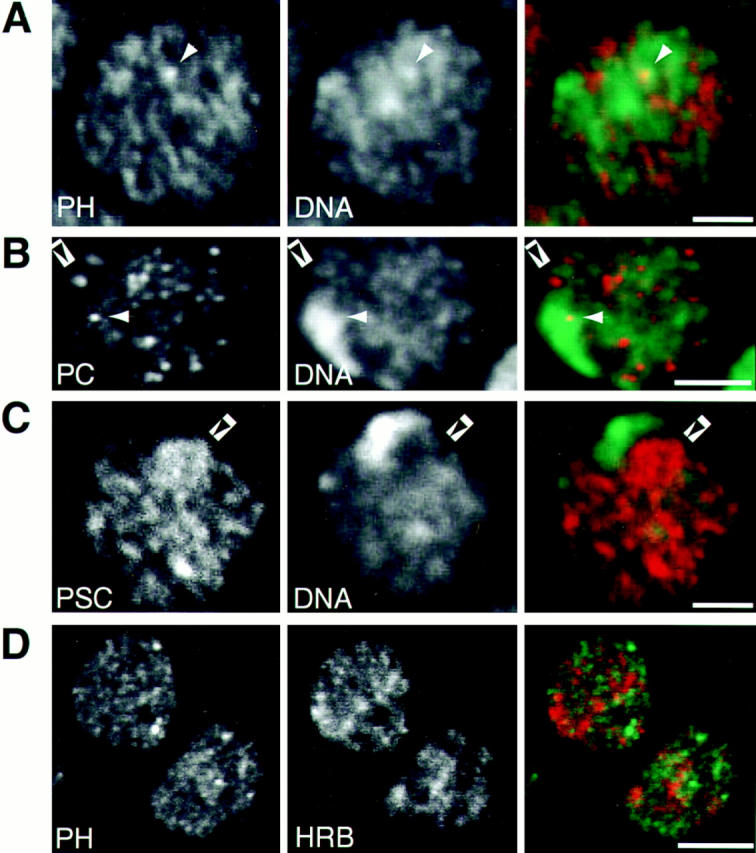
Interphase localization of PcG proteins in relation to DNA distribution and transcription sites. (A–C) The distribution of PH (A), PC (B), and PSC (C) compared with the DNA concentration as seen in single confocal sections through embryonic interphase nuclei from stage 9 whole-mount embryos. The protein concentration visualized by Cy-3 labeled antibody staining (left, red channel), and the DNA concentration visualized by YOYO-1 staining (middle, green channel) are seen to be relatively uncorrected as demonstrated by the lack of many strongly overlapping intensities (white arrowheads) that appear yellow in the merged images (right). Black arrowheads indicate examples for loci lying within regions of decondensed DNA. (D) The distribution of PH (left, red channel) compared with the distribution of the hnRNPK protein, Hrb57A (middle, green channel) and their merged images (right). Bars, 2 μm (A–C), 5 μm (D).
The Spatial Relationship between PcG Proteins and Transcriptional Complexes
Genetic experiments first implicated the PcG genes as repressors of the homeotic gene complexes (e.g., bithorax complex [BX-C], antennepedia complex [ANT-C]) in specific segments and tissues. The PcG proteins PC, PH, and PSC were mapped to ∼60–100 common loci on polytene chromosomes from salivary glands. These sites include the cut, Distalless, even-skipped, engrailed, ANT-C, and BX-C genes (DeCamillis et al., 1992; Rastelli et al., 1993; Zink et al., 1991). Not all DNA binding sites necessarily form repression complexes since both PH and PSC show self-regulatory binding (Fauvarque et al., 1995; Paro and Zink, 1992). Additionally, the GAGA transcription factor has been shown to bind to both active and inactive promotors (O'Brien et al., 1995), suggesting that transcription factors and repressor proteins may not be mutually and exclusively distributed in the nucleus. For these reasons, we were interested in determining to what extent the PcG loci overlap transcriptionally active loci. Several transcription-associated proteins shown to colocalize with active RNA polymerase II (Saumweber et al., 1980; Buchenau et al., 1993; 1997; Buchenau, 1996) were used in combination with one or another of the PcG probes. Fig. 3 D shows representative nuclei in which PH and Hrb57A, an hnRNPK homolog associated strongly with active transcription loci (Buchenau et al., 1997), have been localized. Similar results were obtained for simultaneous staining for PH or PC and other transcription site probes (data not shown). The transcription sites and the PcG loci were distributed in predominantly nonoverlapping distributions as would be expected if PcG antibodies localize repression complexes. Thus, although PcG proteins do not direct loci to inactive heterochromatin domains, neither are they coincident with transcriptionally active chromatin loci.
The Subcellular Distribution of PC, PH, and PSC During Mitosis
Chromatin structure is drastically reorganized at mitosis, whereby selective dissociation of nonhistone chromatin proteins occurs, accompanied by the inhibition of transcription (Martinez-Balbas et al., 1995; Segil et al., 1996). In mammals, terminally differentiated cells express a differential spectrum of genes that are distinguished from their repressed counterparts by unique DNA methylation patterns, a modification unavailable in Drosophila. The function of the PcG proteins as the molecular memory of the transcriptionally repressed state of certain selector genes in Drosophila implies a continuous presence of these proteins on the chromatin (Bienz, 1992). In earlier experiments with Drosophila tissue culture cells, a native PC protein could not be detected on mitotic chromosomes, but a transfected PC-βGal fusion protein expressed in the cells was bound (Messmer et al., 1992). In syncytial blastoderm Drosophila embryos, PSC protein appeared associated with mitotic figures by immunoenzymatic detection (Martin and Adler, 1993), whereas a PSC mammalian homolog was not detected on anaphase mouse chromosomes (Alkema et al., 1997). Analysis of the behavior of the PcG proteins throughout mitosis in embryos could help resolve whether these differences are species or cell type–specific, and lead us to a plausible model for molecular memory.
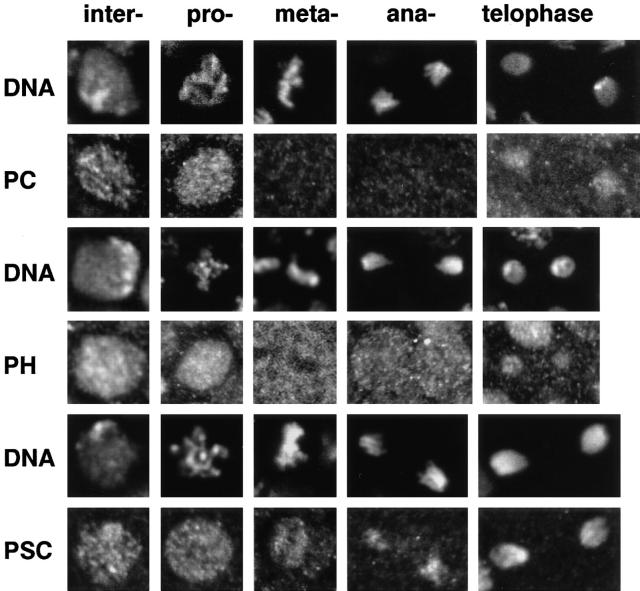
Confocal microscopy of the whole mount embryos allowed us to closely observe mitotic domains in middle and late embryonic developmental stages when the PcG proteins should be exerting their maintenance function. Compared with interphase nuclei, the signal intensities of all three proteins on mitotic chromosomes were drastically reduced or completely absent. Fig. 4, A and B shows an overview of the distribution of PH protein in the anterior half of an embryo during stage 8, when the embryonic ectoderm contains a number of mitotically active regions designated by their order of mitotic cleavage, as determined by Foe (1989). Fig. 4, C and D show stereo image pairs of a region of an older embryo at a higher magnification. At the highest resolution we discerned that while the interphase and prophase nuclei clearly contained PH, the metaphase and anaphase chromosomes of the mitotic fields appeared free of staining. At the same time the signal intensity of PH in the cytoplasm of mitotic cells increased. A complete analysis of the protein distribution with regard to cell cycle was performed in a number of embryos for each of the PcG proteins. Representative raw images of nuclei across the cell cycle from late-stage embryos showing DNA and protein staining for these three PcG proteins are composed into a gallery in Fig. 5. We can summarize the behavior of the proteins as follows: (a) PC dissociates completely from the chromatin and disperses into the cytoplasm after prophase. Clear labeling of the chromatin does not occur until the next interphase. (b) PH shows a similar behavior as PC whereby it disperses into the cytoplasm after prophase, remaining visible outside the condensed chromatin volume until telophase. During telophase there is preferential rebinding of the protein to the DNA, which is still in a condensed state. This binding increases dramatically with formation of the interphase nucleus. (c) PSC also dissociates in prophase, but in contrast to the other two proteins becomes strongly reassociated with the chromatin during anaphase B, rebinding already being complete by telophase when the other two group members are in the process of reassociation.
Figure 4.
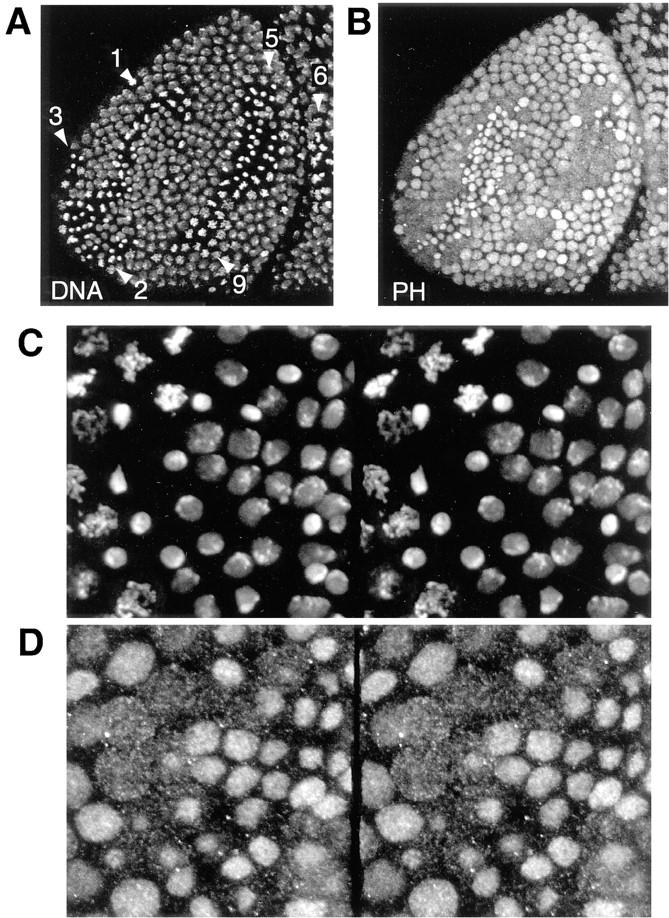
Distribution of PH in an embryo with various cell cycle phases. (A and B) The distribution of DNA and PH in the anterior region of an embryo of stage 8. The DNA staining (A) allows the identification of the mitotic regions 1–3, 5, 6, and 9 in nuclear cycles 15 and 14, respectively (Foe, 1989). The PH antibody staining (B) reveals that the chromosomal protein concentration is drastically reduced during mitosis. Both images are maximum projection overlays of four confocal sections. Field width, 138 μm. (C and D) Stereo image pairs of a field of cells from an embryo at a later developmental stage. (C) DNA staining; (D) PH staining. Field width, 50 μm.
Figure 5.
Distribution of PcG proteins across the cell cycle. A gallery of single confocal sections showing the distribution of PC, PH, and PSC during all distinguishable phases of the cell cycle for 2n nuclei. The images were extracted from whole mount embryos older than stage 8. From left to right: interphase, prophase, metaphase, anaphase, and telophase. The corresponding DNA image is shown above each protein image. Rows from top to bottom: PC, PH, PSC (see text).
The observable fluorescence of the three proteins is lost from the metaphase chromosomes. These observations cannot be attributed to inaccessibility of the antibodies to the antigens in condensed mitotic chromosomes. We clearly see PSC staining on anaphase and telophase chromosomes that have the same level of condensation as metaphase chromosomes. Furthermore, in control experiments with antibodies to topoisomerase II and to core histones, we can access these antigens throughout all phases of mitosis (data not shown; Buchenau, 1996).
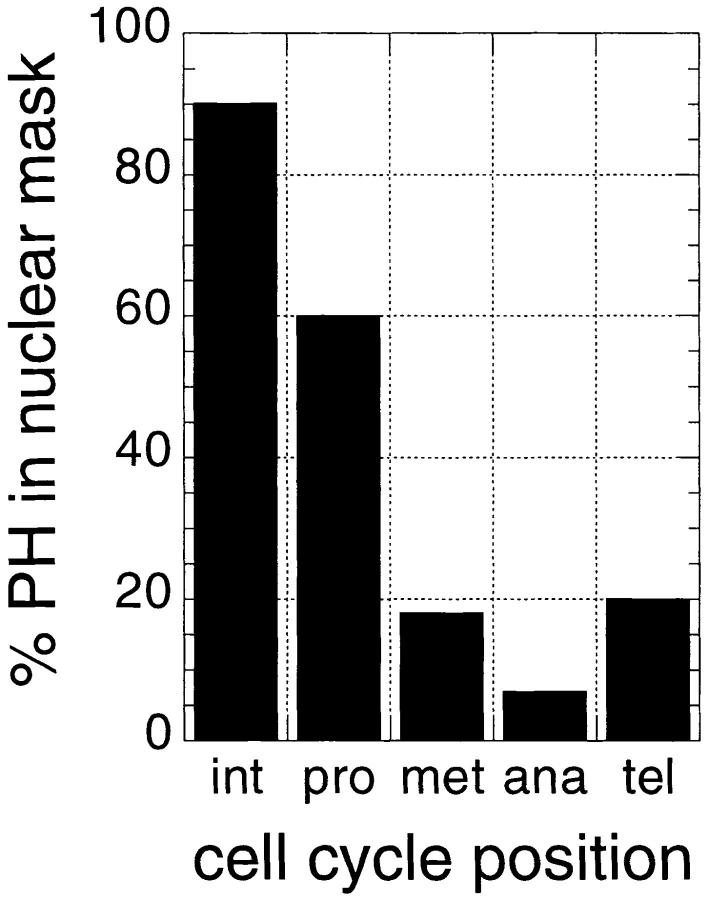
The dissociation and concordant dissolution of the PcG repression complexes in metaphase requires a reassessment of present models about how repression through multiple cell cycles is maintained. We verified our qualitative observations concerning the behavior of PH by quantification of the total protein fluorescence intensity on the chromatin and in the cytoplasm in different phases of the cell cycle. First, we found that the ratio of the total PH fluorescence signal in the whole cell to the total DNA signal remains constant, confirming that the dissociated PH is recognized by the antibody, and thus that which is lost from the chromatin can be accounted for in the cytoplasm. The three-dimensional distributions of PH and DNA in embryonic mitotic fields were quantified in several experiments from stacks of confocal images for 30 or more cells. Segmentation of the volumes for the cells and their nuclei/ chromosomes was performed by interactively defining labeled polygons in three dimensions and generating binary masks (see Materials and Methods). Within these volumes the integrated signal intensities for PH was measured. The data derived directly from the confocal laser scanning microscopy (CLSM) images without correction for z-axis cross talk between optical sections shows only 5–10% of the PH signal remaining in the mask, defining the late metaphase and anaphase chromosomes as seen in the bar graph (Fig. 6). Much of this signal may in fact arise from contamination of signal from adjacent optical sections due to the lower resolution of the CLSM in the axial direction. We can deconvolve the CLSM data to improve the resolution (van der Voort and Straster, 1995; van Kempen et al., 1997), as is commonly done for reconstruction of confocal images from wide-field microscope images (Agard et al., 1989). After these corrections, the optical midsection through the metaphase or anaphase chromosomes is essentially free of intensities arising outside the chromatin domain. The intensity of the PH signal in the pixels within the chromatin mask shows only 1.3 ± 0.2% of the signal determined for pixels in the cell outside the DNA mask in these sections. Thus, only a very small amount of the PcG protein complex present on the interphase chromatin may be necessary to mark the repressed loci through mitosis.
Figure 6.
Quantitation of the cell cycle–dependent PH fluorescence. The total PH fluorescence intensity within the volume of 53 nuclei (or chromosome sets) and 35 cells was measured as described in Materials and Methods. By inspecting DNA staining, the cells were grouped into a phase of the cell cycle. The bars show the percentage of PH intensity associated with the nuclear mask.
These data confirm that (a) the total amount of PH staining in the cell is approximately constant throughout the cell cycle; (b) the vast majority of the PH proteins disperse into the cytoplasm after condensation of the chromosomes in prophase; (c) only 1–2% of the protein remains attached to the metaphase chromosomes, and (d) beginning in telophase, the PH protein reassociates strongly with the reforming nuclei and disappears from the cytoplasm. While the bulk of PH is nuclear during interphase, and cytoplasmic during meta- and anaphase prophase nuclei are special cases. Most of the PcG proteins are already extrachromosomal by this time point of mitosis (Figs. 5 and 6), but are still located within the nuclear volume, presumably retained by the as yet intact nuclear membrane.
PSC protein dissociated from the chromatin for a much shorter period during mitosis than the other two proteins, making quantitative measurements less precise due to fewer cells in the depleted mitotic phase. While metaphase chromosomes are visibly depleted of PSC fluorescence, a significant proportion of the protein reassociated already with anaphase chromatin, and essentially all of the premitotic protein reassociated with telophase chromatin (Fig. 5). We calculate that retention of the protein on metaphase chromosomes does not exceed 5–7%. The results from all three proteins indicates that there is a hierarchy in the assembly of the PcG protein complexes on interphase chromatin.
Discussion
The cytological study of the embryonic distribution of three proteins of the Polycomb group presented here challenges some of the existing models for PcG repression. The new information about the dissociation of the proteins during mitosis sets restrictions on the possible modes of PcG action.
The fact that the same DNA sequence may be transcribed in one cellular, developmental, or genetic context and repressed in another has led geneticists and molecular biologists to postulate that the activity of any given sequence is dependent on the local chromatin structure. However, there is no clear molecular picture of the composition of a repressed chromatin structure. PEV, the phenomenon by which a gene displaced near a telomeric or a heterochromatic region becomes clonally repressed, presumably by taking on the chromatin structure of the neighboring DNA, has served as a model for repression. It must be said, however, that only in the case of the yeast telomere or the silent mating type locus is there molecular evidence for particular interactions that can be extrapolated to a specific structure (Aparicio et al., 1991; Hecht et al., 1995; Johnson et al., 1990; Moretti et al., 1994; Thompson et al., 1994).
A number of arguments have been advanced in support of a higher order heterochromatin-like structure for PcG repression loci: (a) products of the PcG genes show many common polytene chromosome–binding loci (DeCamillis et al., 1992; Martin and Adler, 1993; Rastelli et al., 1993), implying the existence of multimeric protein complexes at the repressed loci; (b) PC and PH have been isolated in a high–molecular weight complex from cell and embryo extracts (Alkema et al., 1997; Franke et al., 1992); (c) double and triple mutants of PcG genes demonstrate synergistic effects in homeotic transformations compared with single mutations; and (d) there is a region of sequence homology, the chromodomain, between PC (Messmer et al., 1992; Paro, 1993) and the heterochromatin-associated protein HP1 (Eissenberg et al., 1995; Eissenberg et al., 1990; James and Elgin, 1986).
Since in the case of PEV there is evidence of association of repressed sequences with condensed heterochromatin (Csink and Henikoff, 1996), we asked in our experiments if there is a positive correlation between PcG protein binding and association with heterochromatin regions of the interphase nucleus. We used the non-sequence-specific DNA binding dyes YOYO1 or TO-PRO 3 (Hirons et al., 1994) for quantitative determination of DNA concentration, and compared the intensity (DNA compaction) with the intensity distribution of antibody staining for the three PcG proteins studied here. The PcG proteins were largely excluded from the most condensed and heterochromatic regions within interphase nuclei as seen in Fig. 3. Two- dimensional correlation plots of the intensity distributions for the protein and DNA revealed no obvious correlation except for the avoidance of highly condensed DNA regions. In polytene chromosomes, the protein is associated with DNA regions of all different levels of condensation and intensity. Thus, PcG complexes do not seem to favor areas in the nucleus where there is a visible compaction of chromatin, as is the case for repression of HP1-associated or Y chromosome heterochromatic sequences. Models for PcG repression that involve remodeling entire genes (Orlando and Paro, 1993) imply changes in structure of hundreds of kilobases of DNA. A reorganization of such an extensive sequence as the BX-C is within the resolution of the light microscope. However, accepting that our data show a number and intensity of PcG-binding loci compatible with repression at individual genes throughout the nuclear volume (see discussion below), a change in compaction or condensation of short sequences of only a few kilobases involving only promoters and control elements would not be detectable at the resolution of the light microscope with the present techniques. Thus, our data do not rule out the local remodeling of the chromatin by the PcG protein complex within a restricted gene locus, but argue against association into heterochromatin compartments as discussed below.
A number of models have been proposed for PcG-mediated repression that assume some kind of compartmentalization of the repressed loci away from transcription factors and sites of transcription. These models range from functionally (and physically) distinct domains (Franke et al., 1995; Orlando and Paro, 1993) to local association of repressed genes (Orlando and Paro, 1995) to molecular exclusion of other proteins (Müller, 1995), or exclusion of only active RNA polymerase II complexes (Bienz and Müller, 1995; McCall and Bender, 1996; Pirrotta, 1995).
Our results of the three-dimensional imaging of PcG protein patterns in interphase nuclei of whole-mount embryos demonstrate a distribution of PcG complexes throughout the nuclear volume as discrete loci (Fig. 2). The number of fluorescent spots found in PcG protein–stained interphase nuclei is on the order of 100 or higher. This is in the same order of magnitude found for the number of loci on polytene chromosomes (Franke et al., 1992; Lonie et al., 1994; Rastelli, et al., 1993; Zink and Paro, 1989). A few of the protein binding sites are brighter than the vast majority, one of which we recently identified in repressed regions of the embryo as the linked genes of the BC-X by simultaneous FISH and PcG labeling (Gemkow and Arndt-Jovin, unpublished observations). However, there is no physical evidence for a distinct compartmentalization of PcG target genes by association into microscopically visible subdomains of the nucleus (Figs. 2 and 3, A–C). These patterns of staining are not similar to the large accumulations of PcGs in the nuclei, as have been described in aneuploid and heteroploid cells (Alkema et al., 1997; Messmer et al., 1992; and unpublished observations of the authors). Such large accumulations may not reflect the distribution of functional PcG complexes since they are not observed in primary tissue or embryos, and may be due to overproduction of some of the PcG proteins in transformed cells.
Our data are in clear contrast to two recent studies of the PEV brownDominant (bwD) mutation. Inserting a large block of AAGAG satellite leads to physical association of the bw locus with the centromeric heterochromatin; i.e., compartmentalization, as shown for larval neuroblasts (Csink and Henikoff, 1996) and imaginal disk cells (Dernburg et al., 1996). Thus, although the phenomena of PEV and PcG-mediated repression show some similarities, their molecular mechanisms must be quite different.
Although a number of people have proposed functionally distinct domains for active and inactive euchromatic genes within the nucleus (Jackson and Cook, 1995; Strouboulis and Wolffe, 1996), the few data that actually have been acquired to test these hypotheses suggest that the domains are not distinct. In the case of the Drosophila salivary gland nucleus, transcription sites are not favored either towards the center or towards the periphery of the nucleus (Mathog et al., 1984). A recent examination of several mammalian genes in three cell types in their transcriptionally active or inactive states by fluorescence in situ hybridization (Kurz et al., 1996) demonstrated that the genes, regardless of their activity, were preferentially located on the periphery of chromosomal domains that themselves were randomly distributed in the nucleus. Furthermore, transcription sites appear to be very mobile as observed by in vivo experiments in Drosophila embryos (Buchenau, 1996; Buchenau et al., 1993; Buchenau et al, 1997).
The model of functionally and physically restricted domains of repression has been challenged by experiments testing the accessibility of BX-C genes in chromatin isolated from repressed or nonrepressed tissue to bacterial restriction enzymes (Schloßherr et al., 1994). In the embryo, McCall and Bender (1996) found that T7 RNA polymerase was able to transcribe from a T7 promoter inserted into an intron in the control region of the Ultrabithorax (Ubx) gene, albeit after heat shock treatment, whereas accessibility to RNApol II, assayed by Gal4 transcription at the same locus in another construct, was inhibited if Ubx was repressed by PC. These observations imply that the final transcriptional state of the chromatin is probably determined by a complex interplay between activating and repressing chromatin proteins rather than by a simple compaction of the structure or association of repressed regions into a large complex that limits accessibility of all other proteins. The trithorax group genes, trx-G, are the positive effectors of homeotic genes (for reviews see Tamkun, 1995; and Simon, 1995). The Trithorax-like GAGA protein has been shown to bind to regions in the ANT-C and BX-C, even when these genes are repressed as in the polytene chromosomes or SL-2 cells (Chinwalla et al., 1995; Strutt et al., 1997). Rastelli et al. (1993) have also documented both repressor and activator proteins binding on the same or neighboring polytene chromosomal sites.
These data taken together argue against restricted nuclear domains for repressive complexes. However, there seems to be a clear distinction within the interphase 2n nucleus between the distribution of the PcG protein complexes and the distribution of factors associated with active transcriptional complexes since we visualize mutually exclusive staining patterns (Fig. 3 D).
Gross displacement of PcG-binding loci to heterochromatic domains in the nucleus or compartmentalization of repressed loci are ruled out by our data. However, gene-by-gene–regulated repression by differential protein exclusion or an inhibition of protein–protein interactions necessary for transcription initiation would be consistent with our results. A number of other models for the mechanism of PcG action have been proposed, and should be explored further. One proposal is that PcG proteins modify and/or rephase nucleosome structure or position (Pirrotta and Rastelli, 1994), thus blocking the transcription complex. Histone modification has been shown to influence gene expression, and a direct role in PEV has been inferred from the effects of histone mutations on PEV. However, similar effects have not been observed in histone mutant backgrounds for the PcG (Reuter and Spierer, 1992), and a definitive statement concerning the interplay of PcG repression and histone modification is lacking. In fact, histone modification could be important in assisting the fixation of repression signals through mitosis (see discussion below).
In vertebrates, silencing is often associated with a hypermethylation of the DNA in the repressed region. This imprinting on the DNA level provides a possible basis for a memory mechanism used for maintenance of the inactive state over many cell generations in these organisms. However, since there is no methylation of the Drosophila genome, the maintenance of silencing or gene repression must proceed by another mechanism in the fly. The important question of the mitotic distribution of PcG proteins has been addressed in earlier studies, with conflicting results (Alkema et al., 1997; DeCamillis and Brock, 1994; Martin and Adler, 1993; Messmer et al., 1992; Müller, 1995). We have now carried out a more detailed qualitative and quantitative analysis of this question. Our results show the dissociation of the proteins from metaphase chromosomes. In contrast to the results shown here, fusion proteins of PC and heterologous proteins have been detected as a general part of mitotic chromosomes (Messmer et al., 1992; Müller, 1995). We have found that while a PC-specific antiserum could not detect endogenous PC on mitotic chromosomes of SL2-cells or in embryos, a PC-βgal fusion protein expressed in embryos was clearly part of condensed mitotic chromosomes using a βgal-specific antibody (unpublished data of the authors). This PC-βgal fusion protein is not capable of rescuing the mutant PC phenotypes, and presumably cannot form a competent PcG repression complex. Similarly, it has been reported that a GAL4-PC fusion protein is localized in high amounts in the mitotic chromosomes of embryos from transformed lines (Müller, 1995). Although such fusion proteins are, like the endogenous PC, primarily localized in the nucleus during most of the cell cycle, they may not form productive chromatin associations through multiprotein complexes as the endogenous proteins do, and may be uncoupled from normal dynamic regulation in mitosis. This result is not surprising in light of the fact that none of the purified PcG proteins so far isolated can be shown to bind specifically to DNA, and thus are inferred to achieve their DNA recognition through the formation of a multiprotein complex. Additional COOH-terminal or NH2-terminal protein fragments could easily disrupt these normal and essential interactions. Our studies on embryos show strong depletion of PC or PH on mitotic chromosomes after embryonic blastoderm formation using affinity-purified antibodies. Therefore, we caution that the extrapolation of results from the study of fusion proteins may not be justified in all cases.
The apparent lack of fluorescence staining by antibodies to the PcG proteins on metaphase chromosomes for PH, PC, and PSC does not absolutely eliminate the possibility that small amounts of these proteins or larger amounts of some other PcG proteins remain attached to the mitotic chromosomes. We believe that our results cannot be explained by antibody inaccessibility since PSC clearly binds to anaphase chromosomes that have the same compaction ratio as metaphase chromosomes. In addition, we have observed topoisomerase II and core histones in the condensed metaphase chromosomes of similarly staged embryos by antibody staining (data not shown and Buchenau, 1996). The fact that several polyclonal antibodies raised against several different sequences within the PcG proteins all show dissociation at metaphase also argues against loss of staining by masking of epitopes. Additionally, we made quantitative measurements of the total PH fluorescence signal across the cell cycle, and can account for the dissociated protein in the cytoplasmic fraction during mitosis. By integrating the fluorescence within the volume of the cell occupied by the metaphase plate compared with that in the cytoplasmic volume, we can put an upper limit on the amount of PH protein that might remain associated with the chromatin. The results described here show that <2% of PH protein that was originally present on the interphase chromatin remains associated during metaphase (Figs. 4–6, and data not shown). If the continuous presence of these proteins on DNA is necessary to maintain the repressed state, then we must assume that only a very minor fraction of the total protein is sufficient for this role, or that other PcG members are responsible for maintenance.
The minor fraction of the PcG proteins that remains bound to mitotic chromosomes may be associated with specific nucleation sites within the repressed genes. Repression could then be initiated in telophase (Fig. 6) from these sites via cooperative binding of previously dispersed PcG protein complexes, insuring that promoters are blocked before reassembly of functional transcription complexes. A similar marking mechanism has been proposed for active genes by residual transcription factors on mitotic chromosomes (Segil et al., 1996). In the BX-C, possible candidates for repression nucleation sites are regulatory elements such as Mcp and bxd, which have been shown recently by immunoprecipitation experiments with formaldehyde cross-linked chromatin to be preferentially associated with PC (Strutt et al., 1997).
It is possible that the repression complex only need be present during transcriptional states of the cell, and thus may be disassembled during mitosis. In this case, only active gene loci would have to be marked in metaphase (Michelotti et al., 1997). Reassembly of the repression complex in telophase would occur by default on the unmarked PREs.
Interestingly, of the three PcG proteins studied, PSC reassociates significantly earlier with anaphase chromatin than do either PC or PH (Fig. 6). Sequence studies on this and analogous proteins in mammals show the presence of putative zinc fingers that predict a DNA-binding capability (Brunk et al., 1991; Martin and Adler, 1993). It is clear from the analysis of the PcG-binding sites on polytene chromosomes that the multimeric PcG protein complex does not have an identical composition at all target loci during interphase. Although PC and PH overlap to more than 90% in their binding on polytene chromosomes, Rastelli et al. (1993) have shown that PSC has somewhat more than half of its binding sites in common with PC. The mutant phenotypes of several of the PcG group genes demonstrate that there are tissue-specific differences in their effects, implying that the proteins are unequal in their timing and locus of action (Campbell et al., 1995; Cheng et al., 1994; Fauvarque et al., 1995). Our data are the first indication that different components of the complex might have different levels of importance in the maintenance function and organization of the repression complex. The reduced dispersion of the PSC protein during mitosis after dissociation from the chromatin may indicate that PSC remains in association with other proteins in a higher-order complex, even after dissociation. Indeed, Martin and Adler (1993) have shown that PSC extracted from embryos for Western blots required addition of urea to the gels to band the protein, an observation consistent with the formation of a very stable complex with itself or other proteins. Intragenic complementation in Psc alleles are consistent with a multidomain structure for PSC, which would support interaction with several different chromatin components (Wu and Howe, 1995). A temperature-sensitive enhancer of zeste E(z)S2 mutation was shown to cause gradual dissociation of PSC from polytene chromosomes, suggesting again an association with other proteins (Jones and Gelbart, 1990; Rastelli et al., 1993).
Clearly, the molecular nature of the PcG repression complex is still to be fully elucidated. Our results show that PcG proteins are associated with genetic loci that show repression in 2n interphase nuclei, and suggest a variety of experimental approaches for further investigating the intriguing phenomenon of how developmentally regulated gene repression is maintained over many cell generations.
Acknowledgments
We thank R. Paro and H. Brock for a rewarding collaboration with their laboratories, stimulating discussions, and constructive criticisms to the manuscript. We thank P. Verveer for assistance with and advice on image processing problems. Part of this work was presented in a doctoral thesis to the Humboldt University, Berlin by P. Buchenau, 1996.
Abbreviations used in this paper
ANT-C
antennepedia complex
BX-C
bithorax complex
CLSM
confocal laser scanning microscopy
PC
polycomb
PcG
Polycomb-group
PEV
position effect variegation
PH
polyhomeotic
PSC
Posterior sex combs
Footnotes
Address all correspondence to Donna J. Arndt-Jovin, Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, 37070 Göttingen, Germany. Phone: 49-551-201-1381; FAX: 49-551-201-1467; E-mail: djovin@mpc186.mpibpc.gwdg.de
The present address of Helen Strutt is Department of Molecular Biology and Biotechnology, University of Sheffield, Firth Court, Western Bank, Sheffield S10 2TN, England. The present address of P. Buchenau is Botanical Institute, University of Bonn, Kirschallee 1, 53115 Bonn, Germany.
References
- Agard D, Hiraoka Y, Shaw P, Sedat J. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Alkema M, Bronk M, Verhoeven E, Otte A, van't Verr L, Berns A, von Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- Allis CD, Waring GL, Mahowald AP. Mass isolation of pole cells from Drosophila melanogaster. . Dev Biol. 1977;56:372–381. doi: 10.1016/0012-1606(77)90277-9. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Billington BI, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–143. doi: 10.1007/BF00291049. [DOI] [PubMed] [Google Scholar]
- Bienz M. Molecular mechanisms of determination in Drosophila. . Curr Opin Cell Biol. 1992;4:955–961. doi: 10.1016/0955-0674(92)90124-u. [DOI] [PubMed] [Google Scholar]
- Bienz M, Müller J. Transcriptional silencing of homeotic genes in Drosophila. . BioEssays. 1995;17:775–784. doi: 10.1002/bies.950170907. [DOI] [PubMed] [Google Scholar]
- Brunk BP, Martin EC, Adler PN. Drosophilagenes posterior sex combs and suppressor two of zeste encode proteins with homology to the murine bmi-1 oncogene. Nature. 1991;353:351–353. doi: 10.1038/353351a0. [DOI] [PubMed] [Google Scholar]
- Buchenau, P. 1996. Verteilung, funktion und in vivo-dynamik chromosomaler proteine in Drosophila melanogaster. Ph.D. thesis. Humboldt University of Berlin, Berlin, Germany. pp. 105.
- Buchenau P, Arndt-Jovin DJ, Saumweber H. In vivo observation of the puff-specific protein no-on transient A (NONA) in nuclei of Drosophilaembryos. J Cell Sci. 1993;106:189–199. doi: 10.1242/jcs.106.1.189. [DOI] [PubMed] [Google Scholar]
- Buchenau P, Saumweber H, Arndt-Jovin DJ. The dynamic nuclear redistribution of an hnRNP-K homologous protein during Drosophilaembryo development and heat shock. Flexibility of transcription sites in vivo. J Cell Biol. 1997;137:291–303. doi: 10.1083/jcb.137.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RB, Sinclair DA, Couling M, Brock HW. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. . Mol Gen Genet. 1995;246:291–300. doi: 10.1007/BF00288601. [DOI] [PubMed] [Google Scholar]
- Carrington EA, Jones RS. The Drosophilaenhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development. 1996;122:4073–4083. doi: 10.1242/dev.122.12.4073. [DOI] [PubMed] [Google Scholar]
- Cheng NN, Sinclair DA, Campbell RB, Brock HW. Interactions of polyhomeotic with Polycomb group genes of Drosophila melanogaster. . Genetics. 1994;138:1151–1162. doi: 10.1093/genetics/138.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinwalla V, Jane EP, Harte PJ. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. . Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- DeCamillis M, Brock HW. Expression of the polyhomeotic locus in development of Drosophila melanogaster. Roux's . Arch Dev Biol. 1994;203:429–438. doi: 10.1007/BF00188692. [DOI] [PubMed] [Google Scholar]
- DeCamillis M, Cheng NS, Pierre D, Brock HW. The polyhomeotic gene of Drosophilaencodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev. 1992;6:223–232. doi: 10.1101/gad.6.2.223. [DOI] [PubMed] [Google Scholar]
- Demandolx D, Davoust J. Multicolour analysis and local image correlation in confocal microscopy. J Microsc. 1997a;185:21–36. [Google Scholar]
- Demandolx D, Davoust J. Multiparameter image cytometry: from confocal micrographs to subcellular fluorograms. Bioimaging. 1997b;5:159–169. [Google Scholar]
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J.C., S.C.R. Elgin, and R. Paro. 1995. Epigenetic regulation in Drosophila: a conspiracy of silence. In Chromatin Structure and Gene Expression. S.C.R. Egin, editor. IRL Press, Oxford, United Kingdom. 147–171.
- Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SCR. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophilamelanogaster. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvarque MO, Zuber V, Dura JM. Regulation of polyhomeotic transcription may involve local changes in chromatin activity in Drosophila. . Mech Dev. 1995;52:343–355. doi: 10.1016/0925-4773(95)00412-t. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- Franke A, DeCamillis M, Zink D, Cheng N, Brock HW, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Messmer S, Paro R. Mapping functional domains of the Polycomb protein of Drosophila melanogaster. . Chromosome Res. 1995;3:351–360. doi: 10.1007/BF00710016. [DOI] [PubMed] [Google Scholar]
- Frasch M, Saumweber H. Two proteins from Drosophila nuclei are bound to chromatin and are detected in a series of puffs on polytene chromosomes. Chromosoma. 1989;97:272–281. doi: 10.1007/BF00371966. [DOI] [PubMed] [Google Scholar]
- Gecz J, Gaunt SJ, Passage E, Burton RD, Cudrey C, Pearce JJ, Fontes M. Assignment of a Polycomb-like chromobox gene (CBX2) to human chromosome 17q25. Genomics. 1995;26:130–133. doi: 10.1016/0888-7543(95)80091-y. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Heimer GV, Taylor CED. Improved mountant for immunofluorescence preparations. J Clin Pathol. 1974;27:254–256. doi: 10.1136/jcp.27.3.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirons GT, Fawcett JJ, Crissman HA. TOTO and YOYO: new very bright fluorochromes for DNA content analyses by flow cytometry. Cytometry. 1994;15:129–140. doi: 10.1002/cyto.990150206. [DOI] [PubMed] [Google Scholar]
- Jackson, D.A., and P.R. Cook. 1995. The structural basis of nuclear function. Int. Rev. Cytol. 162A:125–149. [DOI] [PubMed]
- James TC, Elgin SCR. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogasterand its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of silent mating loci in Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. . Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno M, Hasegawa M, Ishida A, Isono K, Taniguchi M. Mel-18, A polycomb group related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. EMBO J. 1995;14:5672–5678. doi: 10.1002/j.1460-2075.1995.tb00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz A, Lampel S, Nickolenko H, Bradl J, Benner A, Zierbel R, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. . Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lonie A, D'Andrea R, Paro R, Saint R. Molecular characterisation of the Polycomb like gene of Drosophila melanogaster, a trans-acting negative regulator of homeotic gene expression. Development. 1994;120:2629–2636. doi: 10.1242/dev.120.9.2629. [DOI] [PubMed] [Google Scholar]
- Martin EC, Adler PN. The polycomb group gene posterior sex combs encodes a chromosomal protein. Development. 1993;117:641–655. doi: 10.1242/dev.117.2.641. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Mathog D, Hochstrasser M, Gruenbaum Y, Saumweber H, Sedat J. Characteristic folding pattern of polytene chromosomes in Drosophilasalivary gland nuclei. Nature. 1984;308:414–421. doi: 10.1038/308414a0. [DOI] [PubMed] [Google Scholar]
- McCall K, Bender W. Probes for chromatin accessibility in the Drosophilabithorax complex respond differently to Polycomb-mediated repression. EMBO J. 1996;15:569–580. [PMC free article] [PubMed] [Google Scholar]
- Messmer S, Franke A, Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. . Genes Dev. 1992;6:1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Sedat JW. Localization of antigenic determinants in whole Drosophilaembryos. Dev Biol. 1983;99:261–264. doi: 10.1016/0012-1606(83)90275-0. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Mueller J, Gaunt S, Lawrence PA. Function of the Polycomb protein is conserved in mice and flies. Development. 1995;121:2847–2852. doi: 10.1242/dev.121.9.2847. [DOI] [PubMed] [Google Scholar]
- Müller J. Transcriptional silencing by the Polycomb protein in Drosophilaembryos. EMBO J. 1995;14:1209–1220. doi: 10.1002/j.1460-2075.1995.tb07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Wilkins RC, Giardina C, Lis JT. Distribution of GAGA protein on Drosophilagenes in vivo. Genes Dev. 1995;9:1098–1110. doi: 10.1101/gad.9.9.1098. [DOI] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Orlando V, Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev. 1995;5:174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- Paro R. Imprinting a determined state into the chromatin of Drosophila. . Trends Genet. 1990;6:416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- Paro R. Mechanisms of heritable gene repression during development of Drosophila. . Curr Opin Cell Biol. 1993;5:999–1005. doi: 10.1016/0955-0674(93)90084-4. [DOI] [PubMed] [Google Scholar]
- Paro R, Hogness DS. The polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. . Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R, Zink B. The polycomb gene is differentially regulated during oogenesis and embryogenesis of Drosophila melanogaster. . Mech Dev. 1992;40:37–46. doi: 10.1016/0925-4773(93)90086-d. [DOI] [PubMed] [Google Scholar]
- Pearce JJ, Singh PB, Gaunt SJ. The mouse has a polycomb-like chromobox gene. Development. 1992;114:921–929. doi: 10.1242/dev.114.4.921. [DOI] [PubMed] [Google Scholar]
- Petty, K.J. 1987. Metal chelate affinity chromatography. In Current Protocols in Molecular Biology. K. Janssen, editor. John Wiley & Sons, Inc. New York. 10.11.8–10.11.22.
- Pirrotta V. Chromatin complexes regulating gene expression in Drosophila. . Curr Opin Genet Dev. 1995;5:466–472. doi: 10.1016/0959-437x(95)90050-q. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Rastelli L. White gene expression, repressive chromatin domains and homeotic gene regulation in Drosophila. . BioEssays. 1994;16:549–556. doi: 10.1002/bies.950160808. [DOI] [PubMed] [Google Scholar]
- Rastelli L, Chan CS, Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and polycomb group proteins in Drosophilaand their dependence on enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Spierer P. Position effect variegation and chromatin proteins. BioEssays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Saumweber H, Symmons P, Kabisch R, Will H, Bonhoeffer F. Monoclonal antibodies against chromosomal proteins of Drosophila melanogaster. . Chromosoma. 1980;80:253–275. doi: 10.1007/BF00292684. [DOI] [PubMed] [Google Scholar]
- Schloßherr J, Eggert H, Paro R, Cremer S, Jack RS. Gene inactivation in Drosophilamediated by the polycomb gene product or by position-effect variegation does not involve major changes in the accessibility of the chromatin fibre. Mol Gen Genet. 1994;243:453–462. doi: 10.1007/BF00280476. [DOI] [PubMed] [Google Scholar]
- Segil N, Guermah M, Hoffmann A, Roeder R, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- Simon J. Locking in stable states of gene expression: transcriptional control during Drosophiladevelopment. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development. 1992;114:493–505. doi: 10.1242/dev.114.2.493. [DOI] [PubMed] [Google Scholar]
- Strouboulis J, Wolffe A. Functional compartmentalization of the nucleus. J Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Strutt H, Cavalli G, Paro R. Co-localisation of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tamkun J. The role of brahma and related proteins in transcription and development. Curr Opin Genet Dev. 1995;5:473–477. doi: 10.1016/0959-437x(95)90051-h. [DOI] [PubMed] [Google Scholar]
- Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- van der Lugt NMT, Domen J, Linders K, van Roon M, Robanus-Maandag E, Te H, Riele, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, Berns A. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- van der Voort H, Straster K. Restoration of confocal images for quantitative image analysis. J Microsc. 1995;178:165–181. [Google Scholar]
- van Kempen GMP, van Vliet LJ, Verveer PJ, van der Voort HTM. A quantitative comparison of image restoration methods for confocal microscopy. J Microsc. 1997;185:354–365. [Google Scholar]
- van Lohuizen M, Frasch M, Wientjens E, Berns A. Sequence similarity between the mammalian bmi-1 proto-oncogene and the Drosophilaregulatory genes Psc and Su(z)2. Nature. 1991;353:353–355. doi: 10.1038/353353a0. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT, Hearn MG. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. . Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CT, Howe M. A genetic analysis of the suppressor 2 of zeste complex of Drosophila melanogaster. . Genetics. 1995;140:139–181. doi: 10.1093/genetics/140.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Johnson C, Moen P, McNeil J, Lawrence JL. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink B, Engstrom Y, Gehring WJ, Paro R. Direct interaction of the polycomb protein with antennapedia regulatory sequences in polytene chromosomes of Drosophila melanogaster. . EMBO J. 1991;10:153–162. doi: 10.1002/j.1460-2075.1991.tb07931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink B, Paro R. In vivo binding pattern of a trans-regulator of homeotic genes in Drosophila melanogaster. . Nature. 1989;337:468–471. doi: 10.1038/337468a0. [DOI] [PubMed] [Google Scholar]