Short-Insert Libraries as a Method of Problem Solving in Genome Sequencing (original) (raw)
Abstract
As the Human Genome Project moves into its sequencing phase, a serious problem has arisen. The same problem has been increasingly vexing in the closing phase of the Caenorhabditis elegans project. The difficulty lies in sequencing efficiently through certain regions in which the templates (DNA substrates for the sequencing process) form complex folded secondary structures that are inaccessible to the enzymes. The solution, however, is simply to break them up. Specifically, the offending fragments are sonicated heavily and recloned, as much smaller fragments, into pUC vector. The sequences obtained from the resulting library can subsequently be assembled, free from the effects of secondary structure, to produce high-quality, complete sequence. Because of the success and simplicity of this procedure, we have begun to use it for the sequencing of all regions in which standard primer walking has been at all difficult.
[The sequence data described in this paper have been submitted to the EMBL data library under accession nos. Z93392, Z92540, and Z81558.]
Genome sequencing projects are releasing a wealth of genetic information, with the complete human DNA sequence and that of the nematode Caenorhabditis elegans expected in 2005 and 1998, respectively. Sequencing projects deal with chromosomes as a series of mapped and ordered large inserts cloned into such vectors as cosmids, YACs, BACs, or PACs. The sequence of each of these large clones is then most often derived via a shotgun strategy followed by directed walking (Berks 1995). For a variety of reasons, sections of the genomes remain unsequenced after this procedure and to complete them, new strategies must be developed.
First, either by accident or design, gaps may remain in the large insert map. Some large or problematic gaps require new large insert clones to be found, but many gaps can be closed by PCR.
Second, unsequenced gaps may remain within large insert clones. These fall broadly into three classes: (1) those that are bridged by subclones, but for which sequencing reactions fail; (2) those for which no bridging subclone can be found but that can be bridged by PCR; and (3) cases in which neither subcloning nor PCR succeeds so the region must be recovered as a restriction fragment. Because of these difficulties some early proposals for the sequencing of the human genome suggested that gaps of this type might be left unclosed. More recently, however, there has been international agreement that these should be filled to the same accuracy as the rest of the genome (Second International Strategy Meeting on Human Genome Sequencing 1997). It is therefore urgent to find efficient ways of doing this.
A number of methods are currently used to close sequence gaps within clones. It is possible to sequence a gap PCR product by primer walking; this is, however, an inefficient and time-consuming process, often producing sequence data of low quality in that particular bases are reproducibly unreadable. The PCR amplification method has, however, been extended to give products of up to ∼35 kb (Barnes 1994), enabling some gaps between large insert clones to be isolated by “long distance” PCR. Hybridization has been used successfully to select subclones specific to a region of interest within a clone (Beck and Alderton 1993; Alderton et al. 1994), a method that has been extended by taking advantage of unidirectional hybridization to single-stranded M13 clones (Flint et al. 1998). Recently, a transposon insertion method (Devine et al. 1997) has been used with some success to generate new reads within a subclone. Although this is a powerful method that may be of use in solving large, perfect tandem repeat sequences, it is labor intensive and time consuming.
Here we report a simpler method for obtaining the sequence of gaps within or between clones that have either been amplified by PCR, isolated as a restriction fragment, or are subcloned but not sequenceable using standard techniques. We have found that these regions can be sequenced as a series of small, random shotgun fragments producing sequence of high quality, even from regions of complex secondary structure in both C. elegans and Homo sapiens. Furthermore, because this method is purely mechanical it is simple, cheap, robust, and highly successful.
Sequencing by shotgun of DNA fragments is not new (Sanger et al. 1978; Roe et al. 1996); however, by reducing the size of the sonicated fragments in our approach, we overcome problems of secondary structure. Large-scale sequencing has been accelerated during the 1990s by an increase of insert size and of read lengths but has thereby become more vulnerable to the effects of secondary structure. By selectively fragmenting these large inserts in problematic areas, we combine the advantages of old and new. However, it is not simply a matter of using short inserts. The method provides a heavy shotgun of new ends across the problem area, the effect being to break up inverted repeats and G–C runs into small fragments, free from the original structural problems.
RESULTS
Y48E1–C. elegans Chromosome II
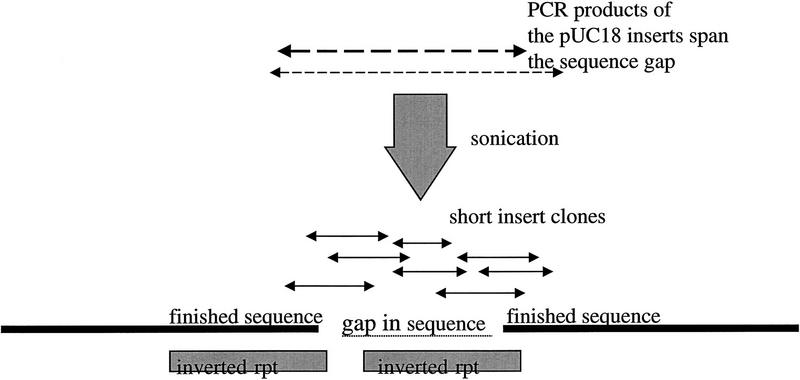
Y48E1 contains an inverted repeat of 1 kb that was spanned by three pUC18 subclones from the shotgun assembly. However, the sequence obtained from the three subclones was unreadable in the inverted repeat region, and restriction digest data confirmed that a region of 800 bases was missing from that region. The inserts of two of the spanning pUC18 subclones were isolated by PCR and sonicated to 200–300 bases and 300–500 bases, respectively. A total of 100 reads were prepped and sequenced, to a depth of a least three reads, to allow for errors caused by single-base changes in the PCR process, enabling the gap in the sequence to be closed, again with high-quality data (Fig. 1).
Figure 1.
Y48E1–C. elegans chromosome II. Restriction digest analysis showed that a fragment of 800 bases was missing from the assembly, and although three pUC18 shotgun subclones spanned the gap, they were unsequenceable in that region. The small-insert clones obtained from the inserts of two of the shotgun pUC18 subclones provided complete and unequivocal contiguation of the gap, which could then be identified as containing one arm of a 1-kb inverted repeat. EMBL accession no. Z93392, bases 263250–264250. Sequence starts, ATCATGGTTGATAACGTAAATTCCCAGAC; sequence ends, CGCTGCGTATCGATTTTTATGAAACTGTG.
The 179I15–BRCA2 Region of Human Chromosome 13
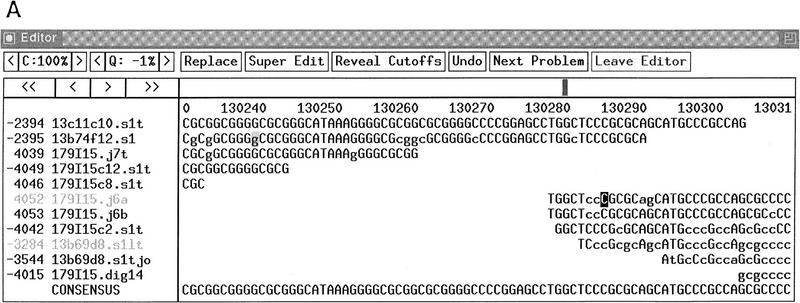
The 179I15 region contains a large CpG in which we found 11 bp to be unreadable using standard sequencing techniques on m13 subclones, although the sequence subsequently recovered sufficiently for a join to be made in adjacent reads. A restriction fragment spanning the region was isolated and sonicated to fragments of ∼100 bases. One of these fragments, subcloned into pUC18 as described, gave clear, high-quality sequence to enable the missing 11 bases to be assigned (Fig. 2).
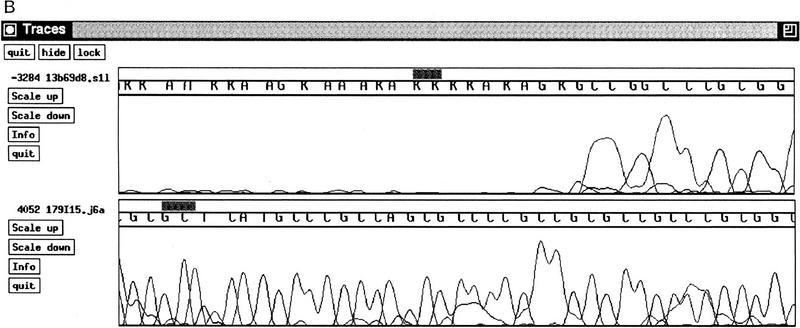
Figure 2.
179I15–H. sapiens chromosome 13 BRCA2 region. (A) After finishing, 179I15 contained a region of 11 bp within a CpG island in which the sequence was unreadable using standard dye primer or dye terminator sequencing. (B) An example of a reverse direction dye primer terminator sequencing reaction over the region (read no. 3284); the sequence obtained from a small insert clone across the same region (read no. 4052). EMBL accession no. Z92540, bases 13000–131140. Sequence starts, CCTGCACGGCTCCCGGGAGCTGGGAGAAA; sequence ends, GTGAGTGCGAGGGGCCAGGCGGAGGGCCA.
F59D12–C. elegans Chromosome II
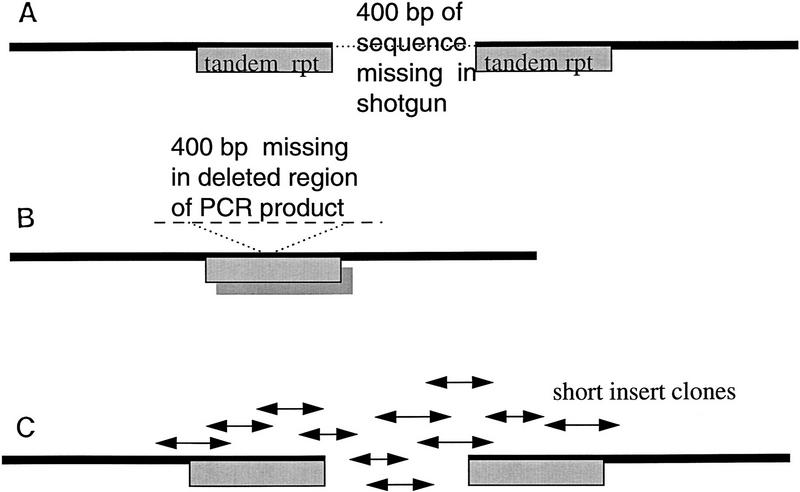
F59D12 was apparently contiguous after shotgun assembly and finishing. However, restriction digest data showed that there was a region of 400 bases missing with two identical copies of a tandem repeat flanking the missing region (Fig. 3A). This suggested that the region, although present in the cosmid, had become deleted from all subclones during the shotgun process. PCR on cosmid DNA across the missing region also gave a deleted product caused by the PCR reaction “skipping” from one repeat motif to the other (Fig. 3B), but a short-insert pUC18 library of a restriction fragment of the region revealed the missing sequence (Fig. 3C). A DOTTER diagram (Sonnhammer et al. 1995) showing the secondary structures that produced this problem is shown in Figure 4.
Figure 3.
F59D12–C. elegans chromosome II. (A) Restriction digest revealed that although the assembly appeared contiguous, there was a 400-bp fragment missing between two identical repeat motifs. This was present in the cosmid but had become deleted from all shotgun subclones. (B) A PCR product was obtained across the region, but this concurred with the original deleted assembly. The PCR reaction had “skipped” between the two repeat regions giving a product that was also missing the 400-bp fragment. (C) A restriction fragment containing the missing sequence was isolated and sonicated to give a small insert library which, when sequenced, revealed the missing 400 bp. EMBL accession no. Z81558, bases 18910–19550. Sequence starts, GTCCACTTACGGGAAAAGGCAAAAATTTA; sequence ends, TTCCCATGACTTTCCGAAAAAAAGGCGGG.
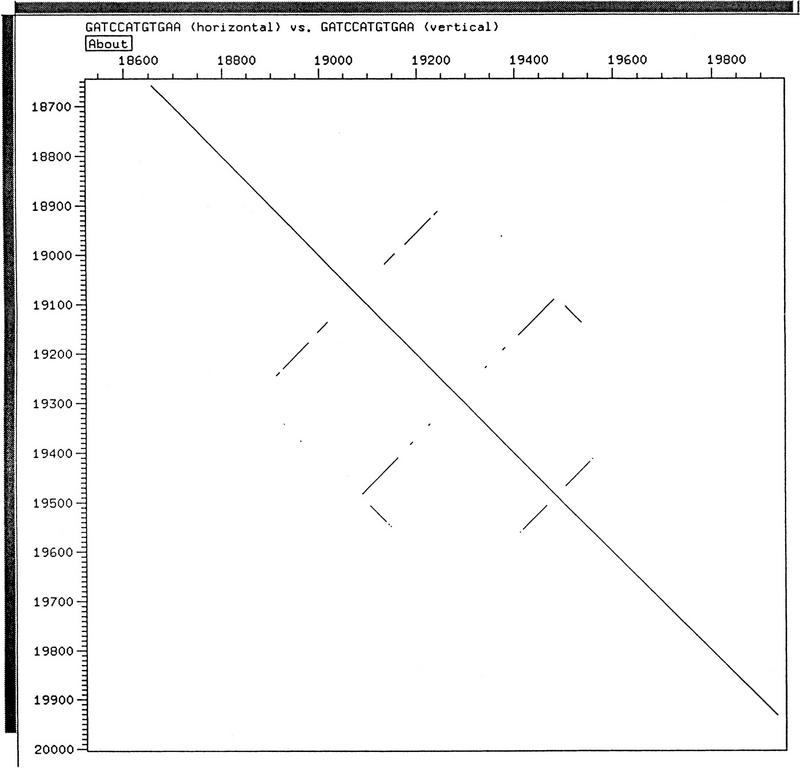
Figure 4.
View of the finished region of F59D12 in DOTTER (Sonnhammer et al. 1994) showing comparison of the sequence with itself. The main diagonal from top left to bottom right shows the in-phase identity. The three broken lines perpendicular to the main diagonal represent the three inverted repeats that caused the problem. The short lines parallel to the main diagonal are the tandem repeats that allowed 400 bp to delete.
CONCLUSION
Subsequently, we have closed in excess of 150 gaps with a success rate of 100% using short-insert libraries, both from fragments isolated by restriction digest or PCR and from plasmid subclones containing DNA that proved impossible to sequence otherwise—many containing elements of potential secondary structure. Typically, libraries of 300–500 bp were found to be adequate for gap closure, although 100- to 300-bp libraries have been used for products of <1 kb and 500- to 1000-bp libraries proved adequate for gaps of 8–12 kb that did not contain any secondary structure. Regions of secondary structure such as those described are particularly prone to deletion during PCR or cloning so it is essential that all regions finished in this way are checked for size accuracy by restriction digest. In addition, because of the inherent possibility of inaccuracy caused by single-base mutation in the PCR process, all regions closed by PCR must have at least a threefold coverage of sequenced fragments, ideally from more than one PCR reaction.
In summary, we have described a novel yet simple technique for the closure of gaps in sequencing projects that is invaluable for sequencing regions of DNA that contain elements that interfere with the progression of sequencing reactions. We envisage that this technique will make a major contribution to genome sequencing projects, both large and small.
METHODS
Gap DNA in the form of a restriction fragment or PCR product, from a clone or unsequenceable subclone, was first sonicated into smaller fragments. To the DNA, which was contained in a maximum volume of 54 μl, was added 6 μl of 10× MB buffer (50% glycerol, 0.5 m sodium chloride, 10 mm zinc chloride, 300 mm sodium acetate at pH 5.0) and water to a final volume of 60 μl. The resulting mixture was sonicated for 2 × 10 sec at full power when placed 1 mm from the face of a cup–horn probe (Heat Systems, Inc., Farmingdale, NY), covered with distilled water, pre-equilibrated to 4°C. The efficiency of sonication was then analyzed by gel electrophoresis on a 1-μl aliquot. This process was repeated until the bulk of the DNA had been sonicated into fragments of between 100 bp and 1 kb. These fragments were then made blunt-ended by incubation with 0.3 μl of mung bean nuclease (156 U/μl, Pharmacia Biotech), at 30°C for 10 min. Surprisingly, the termini of PCR fragments subcloned in this manner were found to be present in most cases. However, where sequence information close to the end of PCR fragments was required, a brief incubation with T4 polynucleotide kinase was performed prior to sonication, but only on 20% of the total volume, to prevent over-representation of the ends. Subsequently, the DNA was concentrated by ethanol precipitation before being fractionated by electrophoresis through a low-melting-point agarose gel. After staining, gels were viewed on a long-wavelength UV transilluminator, agarose slices containing DNA of the required size range (typically 100–300, 300–500, or 500–1000 bp) were excised, and the DNA eluted by using phenol extraction, the Prep-A-Gene DNA purification kit (Bio-Rad), or the Qiaquick gel extraction kit (Qiagen). Recovered DNA was ligated with _Sma_I-digested and phosphatased pUC18 vector DNA, and transformed into Escherichia coli TG1 cells by electroporation. After growth on TYE agar plates containing 100 μg/ml ampicillin, 50 μg/ml X-gal, and 50 μg/ml IPTG, white colonies were picked and grown up, and plasmid DNA was prepared from them by alkaline lysis. Under the conditions described, inserts were of the expected size with <3% contamination by sequencing vector. Sequencing reactions were performed using the ABI Prism dye terminator cycle sequencing kit prior to sequencing on ABI model 373/377 automated sequencers. Data were transferred to a Unix workstation, and complete sequences were assembled automatically using Xgap (Staden 1994). The data were edited to our normal accuracy of <1 error in 10,000 bases of DNA sequence. Other methods were as described by Sambrook et al. (1989).
Acknowledgments
We thank Nicola Lennard, Julia Lightning, Christine Lloyd, Lucy Matthews, Sarah White and other members of staff at the Sanger Centre for the supply and finishing of problem regions from C. elegans. We also thank our colleagues at The Genome Sequencing Center, Washington University, for discussion and support. This work is supported by the Medical Research Council of Great Britain and The Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jes@sanger.ac.uk; FAX 1223 494919.
REFERENCES
- Alderton R, Kitau J, Beck S. Automated DNA hybridization. Analy Biochem. 1994;218:98–102. doi: 10.1006/abio.1994.1146. [DOI] [PubMed] [Google Scholar]
- Barnes W. PCR amplification of up to 35kB DNA with high fidelity and high yield from lambda-bacteriophage templates. Proc Natl Acad Sci. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S, Alderton R. A strategy for the amplification, purification, and selection of M13 templates for large-scale DNA-sequencing. Analy Biochem. 1993;212:498–505. doi: 10.1006/abio.1993.1359. [DOI] [PubMed] [Google Scholar]
- Berks M. The C. elegans genome sequencing project. Genome Res. 1995;5:99–104. doi: 10.1101/gr.5.2.99. [DOI] [PubMed] [Google Scholar]
- Devine SE, Chissoe SL, Eby Y, Wilson RK, Boeke JD. A transposon-based strategy for sequencing repetitive DNA in eukaryotic genomes. Genome Res. 1997;7:551–563. doi: 10.1101/gr.7.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, J., M. Sims, R. Staden, and K. Thomas. 1998. An oligo-screening strategy to fill gaps found during shotgun sequencing projects. DNA Sequence (in press). [DOI] [PubMed]
- Roe, B., J. Crabtree, and A. Khan. 1996. DNA isolation and sequencing: Essential techniques series (ed. D. Rickwood), pp. 85–86, 116–117. John Wiley and Sons, New York, NY.
- Sanger F, Coulson AR, Friedmann T, Air GM, Barrell BG, Brown NL, Fiddes JC, Hutchison III CA, Slocombe PM, Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978;125:225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sonnhammer ELL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Staden R. Methods in molecular biology. Totowa, NJ: Human Press; 1994. The Staden package. [Google Scholar]