Hydrolysis of sequenced beta-casein peptides provides new insight into peptidase activity from thermophilic lactic acid bacteria and highlights intrinsic resistance of phosphopeptides - PubMed (original) (raw)
Hydrolysis of sequenced beta-casein peptides provides new insight into peptidase activity from thermophilic lactic acid bacteria and highlights intrinsic resistance of phosphopeptides
S M Deutsch et al. Appl Environ Microbiol. 2000 Dec.
Free PMC article
Abstract
The peptidases of thermophilic lactic acid bacteria have a key role in the proteolysis of Swiss cheeses during warm room ripening. To compare their peptidase activities toward a dairy substrate, a tryptic/chymotryptic hydrolysate of purified beta-casein was used. Thirty-four peptides from 3 to 35 amino acids, including three phosphorylated peptides, constitute the beta-casein hydrolysate, as shown by tandem mass spectrometry. Cell extracts prepared from Lactobacillus helveticus ITG LH1, ITG LH77, and CNRZ 32, Lactobacillus delbrueckii subsp. lactis ITG LL14 and ITG LL51, L. delbrueckii subsp. bulgaricus CNRZ 397 and NCDO 1489, and Streptococcus thermophilus CNRZ 385, CIP 102303, and TA 060 were standardized in protein. The peptidase activities were assessed with the beta-casein hydrolysate as the substrate at pH 5.5 and 24 degrees C (conditions of warm room ripening) by (i) free amino acid release, (ii) reverse-phase chromatography, and (iii) identification of undigested peptides by mass spectrometry. Regardless of strain, L. helveticus was the most efficient in hydrolyzing beta-casein peptides. Interestingly, cell extracts of S. thermophilus were not able to release a significant level of free proline from the beta-casein hydrolysate, which was consistent with the identification of numerous dipeptides containing proline. With the three lactic acid bacteria tested, the phosphorylated peptides remained undigested or weakly hydrolyzed indicating their high intrinsic resistance to peptidase activities. Finally, several sets of peptides differing by a single amino acid in a C-terminal position revealed the presence of at least one carboxypeptidase in the cell extracts of these species.
Figures
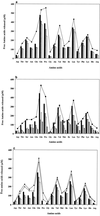
FIG. 1
β-Casein sequence (209 amino acids; one-letter amino acid code). Phosphorylated serines are indicated by the letter “U” and highlighted in grey; every 10th amino acid is framed in bold. The 34 initial peptides of the hydrolysate are represented in black rectangles above the β-casein sequence. After 72 h of hydrolysis, nonhydrolyzed peptides were identified by MS-MS and are presented below the sequence with different patterns, depending on the cell extract used: ▥ for L. helveticus ITG LH1, ▧ for L. delbrueckii subsp. lactis ITG LL14, and  for S. thermophilus TA 060.
for S. thermophilus TA 060.
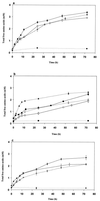
FIG. 2
Time course of hydrolysis of the β-casein hydrolysate by cell extracts of the following strains: (a) _L. helveticus_ITG LH1 (—●—), ITG LH77 (○), and CNRZ 32 (--- ---), (b)L. delbrueckii subsp. lactis ITG LL14 (—■—) and ITG LL51 (□), and L. delbrueckii subsp.bulgaricus NCDO 1489 (▴) and CNRZ 397 (▵); and (c)S. thermophilus CNRZ 385 (—⧫—), TA 060 (◊), and CIP (Pasteur Institut Collection, Paris, France) 102303 (---
---), (b)L. delbrueckii subsp. lactis ITG LL14 (—■—) and ITG LL51 (□), and L. delbrueckii subsp.bulgaricus NCDO 1489 (▴) and CNRZ 397 (▵); and (c)S. thermophilus CNRZ 385 (—⧫—), TA 060 (◊), and CIP (Pasteur Institut Collection, Paris, France) 102303 (--- ---). The enzyme reactions were performed for 72 h at 24°C in 20 mM acetate ammonium buffer (pH 5.5) with 137.5 μg of cell extract proteins per ml for 0.7 mg of lyophilized β-casein hydrolysate per ml. Blanks (⋯●⋯, ⋯⧫⋯, and ⋯▴⋯) were assayed with the same conditions but with boiled cell extracts.
---). The enzyme reactions were performed for 72 h at 24°C in 20 mM acetate ammonium buffer (pH 5.5) with 137.5 μg of cell extract proteins per ml for 0.7 mg of lyophilized β-casein hydrolysate per ml. Blanks (⋯●⋯, ⋯⧫⋯, and ⋯▴⋯) were assayed with the same conditions but with boiled cell extracts.
FIG. 3
Hydrolysis of the β-casein hydrolysate by cell extracts (▴) of L. helveticus ITG LH77 (a) and_S. thermophilus_ CIP 102303 (b): effect of the addition at 72 h of fresh β-casein hydrolysate (■) or fresh cell extracts (●).
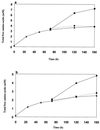
FIG. 4
Free amino acids released after 24 h by hydrolysis of β-casein peptides by cell extracts of the following strains: (a)L. helveticus ITG LH1 (&cjs3744;), CNRZ 32 ( ), and ITG LH77 (▯); (b) L. delbrueckii subsp. bulgaricus NCDO 1489 (&cjs3744;), and CNRZ 397 (
), and ITG LH77 (▯); (b) L. delbrueckii subsp. bulgaricus NCDO 1489 (&cjs3744;), and CNRZ 397 ( and L. delbrueckii subsp. lactis ITG LL14 (▯), and ITG LL51 (
and L. delbrueckii subsp. lactis ITG LL14 (▯), and ITG LL51 ( ); and; (c)S. thermophilus CNRZ 385 (&cjs3744;), TA 060 (
); and; (c)S. thermophilus CNRZ 385 (&cjs3744;), TA 060 ( ), and CIP 102303 (▯). Curves indicate values obtained after 72 h of incubation for L. helveticus ITG LH1 (a; ●), L. delbrueckii subsp.lactis ITG LL14 (b; ▪), and S. thermophilus CNRZ 385 (♦), and TA 060 (
), and CIP 102303 (▯). Curves indicate values obtained after 72 h of incubation for L. helveticus ITG LH1 (a; ●), L. delbrueckii subsp.lactis ITG LL14 (b; ▪), and S. thermophilus CNRZ 385 (♦), and TA 060 ( ) (c).
) (c).
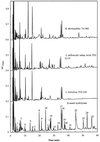
FIG. 5
RP-HPLC profiles of β-casein peptides at time zero and after incubation for 24 h at 24°C with cell extracts of_L. helveticus_ ITG LH1, L. delbrueckii subsp.lactis ITG LL14, and S. thermophilus TA 060.
Similar articles
- Characterization of the pattern of alphas1- and beta-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581.
Hebert EM, Mamone G, Picariello G, Raya RR, Savoy G, Ferranti P, Addeo F. Hebert EM, et al. Appl Environ Microbiol. 2008 Jun;74(12):3682-9. doi: 10.1128/AEM.00247-08. Epub 2008 Apr 18. Appl Environ Microbiol. 2008. PMID: 18424544 Free PMC article. - Identification of endopeptidase genes from the genomic sequence of Lactobacillus helveticus CNRZ32 and the role of these genes in hydrolysis of model bitter peptides.
Sridhar VR, Hughes JE, Welker DL, Broadbent JR, Steele JL. Sridhar VR, et al. Appl Environ Microbiol. 2005 Jun;71(6):3025-32. doi: 10.1128/AEM.71.6.3025-3032.2005. Appl Environ Microbiol. 2005. PMID: 15932998 Free PMC article. - Hydrolysis of casein-derived peptides alpha(S1)-casein(f1-9) and beta-casein(f193-209) by Lactobacillus helveticus peptidase deletion mutants indicates the presence of a previously undetected endopeptidase.
Christensen JE, Broadbent JR, Steele JL. Christensen JE, et al. Appl Environ Microbiol. 2003 Feb;69(2):1283-6. doi: 10.1128/AEM.69.2.1283-1286.2003. Appl Environ Microbiol. 2003. PMID: 12571058 Free PMC article. - Original features of cell-envelope proteinases of Lactobacillus helveticus. A review.
Sadat-Mekmene L, Genay M, Atlan D, Lortal S, Gagnaire V. Sadat-Mekmene L, et al. Int J Food Microbiol. 2011 Mar 15;146(1):1-13. doi: 10.1016/j.ijfoodmicro.2011.01.039. Epub 2011 Feb 2. Int J Food Microbiol. 2011. PMID: 21354644 Review. - The Microfloras and Sensory Profiles of Selected Protected Designation of Origin Italian Cheeses.
Licitra G, Carpino S. Licitra G, et al. Microbiol Spectr. 2014 Feb;2(1):CM-0007-2012. doi: 10.1128/microbiolspec.CM-0007-2012. Microbiol Spectr. 2014. PMID: 26082116 Review.
Cited by
- Lactobacillus helveticus: the proteolytic system.
Griffiths MW, Tellez AM. Griffiths MW, et al. Front Microbiol. 2013 Mar 5;4:30. doi: 10.3389/fmicb.2013.00030. eCollection 2013. Front Microbiol. 2013. PMID: 23467265 Free PMC article. - Phylogenetic comparative analysis: Chemical and biological features of caseins (alpha-S-1, alpha-S-2, beta- and kappa-) in domestic dairy animals.
Hassanin AA, Osman A, Atallah OO, El-Saadony MT, Abdelnour SA, Taha HSA, Awad MF, Elkashef H, Ahmed AE, Abd El-Rahim I, Mohamed A, Eldomiaty AS. Hassanin AA, et al. Front Vet Sci. 2022 Sep 15;9:952319. doi: 10.3389/fvets.2022.952319. eCollection 2022. Front Vet Sci. 2022. PMID: 36187819 Free PMC article. - Small-scale analysis of exopolysaccharides from Streptococcus thermophilus grown in a semi-defined medium.
Levander F, Svensson M, Rådström P. Levander F, et al. BMC Microbiol. 2001;1:23. doi: 10.1186/1471-2180-1-23. Epub 2001 Sep 26. BMC Microbiol. 2001. PMID: 11602017 Free PMC article. - Little Impact of NaCl Reduction in Swiss-Type Cheese.
Gagnaire V, Lecomte X, Richoux R, Genay M, Jardin J, Briard-Bion V, Kerjean JR, Thierry A. Gagnaire V, et al. Front Nutr. 2022 Jun 16;9:888179. doi: 10.3389/fnut.2022.888179. eCollection 2022. Front Nutr. 2022. PMID: 35782920 Free PMC article. - Simultaneous presence of PrtH and PrtH2 proteinases in Lactobacillus helveticus Strains improves breakdown of the pure alphas1-casein.
Sadat-Mekmene L, Jardin J, Corre C, Mollé D, Richoux R, Delage MM, Lortal S, Gagnaire V. Sadat-Mekmene L, et al. Appl Environ Microbiol. 2011 Jan;77(1):179-86. doi: 10.1128/AEM.01466-10. Epub 2010 Oct 29. Appl Environ Microbiol. 2011. PMID: 21037305 Free PMC article.
References
- Atlan D, Gilbert C, Blanc B, Portalier R. Cloning, sequencing and characterization of a PepIP gene encoding a proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricusCNRZ 397. Microbiology. 1994;140:527–535. - PubMed
- Baer A, Riba I, Meyer J, Bütikofer U. Microplate assay of free amino acids in Swiss cheeses. Lebensm Wiss Technol. 1996;29:58–62.
- Bockelman W, Hoppe-Seyler T, Heller K. Purification and characterization of an endopeptidase from Lactobacillus delbrueckii subsp. bulgaricusB14. Int Dairy J. 1996;6:1167–1180.
- Booth M, Jennings V, Fhaolain I, O'Cuinn G. Prolidase activity of Lactococcus lactis ssp. cremorisAM2: partial purification and characterization. J Dairy Res. 1990;57:245–254.
- Christensen J E, Dudley E G, Pederson J A, Steele J L. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:217–246. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources