SIFT: Predicting amino acid changes that affect protein function - PubMed (original) (raw)
SIFT: Predicting amino acid changes that affect protein function
Pauline C Ng et al. Nucleic Acids Res. 2003.
Abstract
Single nucleotide polymorphism (SNP) studies and random mutagenesis projects identify amino acid substitutions in protein-coding regions. Each substitution has the potential to affect protein function. SIFT (Sorting Intolerant From Tolerant) is a program that predicts whether an amino acid substitution affects protein function so that users can prioritize substitutions for further study. We have shown that SIFT can distinguish between functionally neutral and deleterious amino acid changes in mutagenesis studies and on human polymorphisms. SIFT is available at http://blocks.fhcrc.org/sift/SIFT.html.
Figures
Figure 1
An example of SIFT prediction on amino acid changes in a protein. Substitutions with score less than 0.05 are predicted to affect protein function. In the last prediction, the median conservation of the sequences does not meet the threshold so a warning is issued.
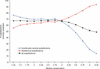
Figure 2
Prediction depends on the diversity of the sequences used in the alignment. Percentage of substitutions correctly predicted is based on over 4000 substitutions that were assayed throughout the LacI protein of Escherichia coli (2,12). When the sequences in the alignment used for prediction are closely related (high median conservation) then many positions appear conserved and important for function. In this situation, prediction accuracy on deleterious substitutions is high but many functionally neutral substitutions are erroneously predicted to be deleterious. To obtain an alignment with a specified median conservation, the LacI protein sequence of E.coli was submitted to the SIFT website and the median conservation setting adjusted. Because the homologous sequences available are distantly related to E.coli LacI, alignments with higher median conservation values could not be obtained. In order to obtain alignments with median conservation values more than 3.25, closely related sequences were simulated by starting with an alignment of identical E.coli LacI sequences. A position and a sequence were randomly selected from the LacI alignment with median conservation 2.75. The amino acid corresponding to this location was substituted in the starting alignment. Amino acids continued to be randomly selected and substituted until the desired median conservation was met. The simulated alignment was then evaluated for its performance as previously described (2) and the plotted value is the average performance of 100 simulated alignments.
Similar articles
- Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm.
Kumar P, Henikoff S, Ng PC. Kumar P, et al. Nat Protoc. 2009;4(7):1073-81. doi: 10.1038/nprot.2009.86. Epub 2009 Jun 25. Nat Protoc. 2009. PMID: 19561590 - SIFT web server: predicting effects of amino acid substitutions on proteins.
Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. Sim NL, et al. Nucleic Acids Res. 2012 Jul;40(Web Server issue):W452-7. doi: 10.1093/nar/gks539. Epub 2012 Jun 11. Nucleic Acids Res. 2012. PMID: 22689647 Free PMC article. - Predicting deleterious amino acid substitutions.
Ng PC, Henikoff S. Ng PC, et al. Genome Res. 2001 May;11(5):863-74. doi: 10.1101/gr.176601. Genome Res. 2001. PMID: 11337480 Free PMC article. - Predicting the effects of amino acid substitutions on protein function.
Ng PC, Henikoff S. Ng PC, et al. Annu Rev Genomics Hum Genet. 2006;7:61-80. doi: 10.1146/annurev.genom.7.080505.115630. Annu Rev Genomics Hum Genet. 2006. PMID: 16824020 Review. - State-of-the-art bioinformatics protein structure prediction tools (Review).
Pavlopoulou A, Michalopoulos I. Pavlopoulou A, et al. Int J Mol Med. 2011 Sep;28(3):295-310. doi: 10.3892/ijmm.2011.705. Epub 2011 May 23. Int J Mol Med. 2011. PMID: 21617841 Review.
Cited by
- BRCAIndica: a resource for ACMG/AMP classified BRCA1 and BRCA2 variants.
Vatsyayan A, Anu RI, Mathur P, Uchil D, Joshi A, Dwivedi A, Sirohi B, Mathew A, Damodaran D, Panda SS, Kolluri S, Ayillath SK, Amalnath D, Shankar G, Pandhare K, Scaria V. Vatsyayan A, et al. Fam Cancer. 2024 Nov 15;24(1):4. doi: 10.1007/s10689-024-00429-5. Fam Cancer. 2024. PMID: 39546087 - Revealing SARS-CoV-2 Mpro mutation cold and hot spots: Dynamic residue network analysis meets machine learning.
Barozi V, Chakraborty S, Govender S, Morgan E, Ramahala R, Graham SC, Bishop NT, Tastan Bishop Ö. Barozi V, et al. Comput Struct Biotechnol J. 2024 Oct 22;23:3800-3816. doi: 10.1016/j.csbj.2024.10.031. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39525081 Free PMC article. - Unraveling the protective genetic architecture of COVID-19 in the Brazilian Amazon.
Barros MC, de Souza JES, Gomes DHF, Pinho CT, Silva CS, Braga-da-Silva C, Cavalcante GC, Magalhães L, Azevedo-Pinheiro J, Quaresma JAS, Falcão LFM, Costa PF, Salgado CG, Carneiro TX, Burbano RR, Dos Santos Vieira JR, Santos S, Soares-Souza GB, de Souza SJ, Ribeiro-Dos-Santos Â. Barros MC, et al. Sci Rep. 2024 Nov 9;14(1):27332. doi: 10.1038/s41598-024-78170-3. Sci Rep. 2024. PMID: 39521879 Free PMC article. - Prenatal diagnosis of fetal skeletal anomalies via whole-exome sequencing in a tertiary referral center.
Xue H, Yu A, Zhao W, Chen L, Fang R, Ling W, Zhang L, Guo Q, Lin N, Xu L, Huang H. Xue H, et al. Sci Rep. 2024 Nov 9;14(1):27371. doi: 10.1038/s41598-024-75738-x. Sci Rep. 2024. PMID: 39521787 Free PMC article. - Molecular Modeling and In Vitro Functional Analysis of the RGS12 PDZ Domain Variant Associated with High-Penetrance Familial Bipolar Disorder.
Agogo-Mawuli PS, Mendez J, Oestreich EA, Bosch DE, Siderovski DP. Agogo-Mawuli PS, et al. Int J Mol Sci. 2024 Oct 24;25(21):11431. doi: 10.3390/ijms252111431. Int J Mol Sci. 2024. PMID: 39518985 Free PMC article.
References
- Krawczak M., Ball,E.V., Fenton,I., Stenson,P.D., Abeysinghe,S., Thomas,N. and Cooper,D.N. (2000) Human gene mutation database-a biomedical information and research resource. Hum. Mutat., 15, 45–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources