Adenovirus protein VI mediates membrane disruption following capsid disassembly - PubMed (original) (raw)
Adenovirus protein VI mediates membrane disruption following capsid disassembly
Christopher M Wiethoff et al. J Virol. 2005 Feb.
Abstract
In contrast to enveloped viruses, the mechanisms involved in membrane penetration by nonenveloped viruses are not as well understood. In these studies, we determined the relationship between adenovirus (Ad) capsid disassembly and the development of membrane lytic activity. Exposure to low pH or heating induced conformational changes in wild-type Ad but not in temperature-sensitive Ad (ts1) particles that fail to escape the early endosome. Wild-type Ad but not ts1 particles permeabilized model membranes (liposomes) and facilitated the cytosolic delivery of a ribotoxin. Alterations in wild-type Ad capsids were associated with the exposure of a pH-independent membrane lytic factor. Unexpectedly, this factor was identified as protein VI, a 22-kDa cement protein located beneath the peripentonal hexons in the viral capsid. Recombinant protein VI and preprotein VI, but not a deletion mutant lacking an N-terminal amphipathic alpha-helix, possessed membrane lytic activity similar to partially disassembled virions. A new model of Ad entry is proposed based on our present observations of capsid disassembly and membrane penetration.
Figures
FIG. 1.
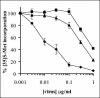
Cytosolic translocation of the ribotoxin α-sarcin by Ad5 or _ts_1 particles. Sarcin entry into A549 cells was performed in the presence of various amounts of Ad5 (♦), Ad5 plus 0.3 uM bafilomycin (▪) or _ts_1 particles (▴). Toxin activity was assessed by inhibition of [35S]methionine incorporation into cellular proteins. Less than 10% inhibition of protein synthesis was observed with the highest amount of Ad5 in the absence of α-sarcin.
FIG. 2.
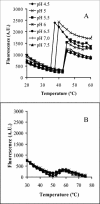
Analysis of Ad capsid stability as a function of pH and temperature. The stability of Ad5 (A) or _ts_1 particles (B) was assessed at various pH levels and temperatures by monitoring the accessibility of the viral DNA to TOTO-1, a bis-intercalating fluorophore.
FIG. 3.
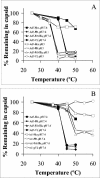
Examination of thermally induced capsid disassembly. Ad5 at pH 7.4 (A and B, solid symbols) or pH 5.0 (A, open symbols) or _ts_1 particles at pH 7.4 (B, open symbols) were heated to various temperatures and then subjected to density gradient centrifugation to separate partially disassembled virions from soluble capsomers. Individual capsid proteins associated with the treated virions were analyzed by SDS-PAGE and Simply Blue staining and then quantitated by scanning densitometry. Densities were normalized to that of the major core protein, protein VII.
FIG. 4.
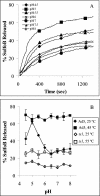
Ad-mediated disruption of model membranes (liposomes) containing an entrapped fluorophore. (A) Ad5 particles (75 μg/ml) were incubated with liposomes (2 μg/ml) at different pH levels, and the release of SulfoB was monitored over time as described in Materials and Methods. (B) The amount of SulfoB released from liposomes incubated with Ad5 or _ts_1 particles was measured at different pHs, following preincubation of virions at 25 or 45°C (for WT) or 55°C (for _ts_1).
FIG. 5.
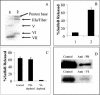
Identification of the Ad capsid protein associated with pH-independent membrane disruption. (A) Immunoblot of partially disassembled Ad5 capsids (lane 1) and the soluble capsomer fraction (lane 2). Individual Ad proteins were detected with a polyclonal anti-Ad antibody. (B) Liposome disruption by virion (lane 1) and supernatant fraction (lane 2) was analyzed by the SulfoB release assay. (C) The supernatant fraction was immunodepleted with a control or anti-penton base (PB) or protein VI antibodies, followed by immunoblot analysis. (D) The supernatant- or PB- or protein VI-depleted fractions were analyzed for liposome disruption at pH 7.4 by the SulfoB release assay.
FIG. 6.
Schematic representation of Ad protein VI. Sites for cleavage by the 23K cysteine protease, hexon binding, and nuclear export (NES1) and import (nuclear localization signal 2 [NLS2]) are indicated. A predicted amphipathic α-helical domain is located between residues 36 to 53. A helical wheel diagram of this region is shown with the hydrophobic residues in black and the hydrophilic residues in white. An alignment of protein VI sequences corresponding to the amphipathic helical domain of human and nonhuman Ads is shown. Predicted Δ_G_interfacial values for the partitioning of these domains into lipid membranes are provided for comparison.
FIG. 7.
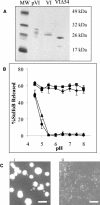
Analysis of the membrane lytic activity recombinant protein VI molecules. (A) SDS-PAGE of recombinant preprotein VI, protein VI, and a truncated protein VI (VIΔ54), following purification on nitriloacetic acid-Ni2+ agarose columns. (B) Liposome disruption by equivalent amount of pVI ♦, VI ▪, VIΔ54 ▴, or BSA • as a control was assessed at various pH levels by SulfoB release. (C) EMs of untreated liposomes (i) or liposomes treated with an equivalent weight of protein VI at pH 7.4 (ii) obtained at a magnification of ×21,000 after being negatively stained with 2% phosphotungstate. Bar, 500 nm.
FIG. 8.
Hypothetical model of Ad-mediated endosomal membrane disruption. (Top) Penton base binding to αv integrins triggers Ad uptake into endosomes, coincident with the loss of the fiber protein. (Middle) Endosome acidification induces partial disassembly of the capsid, resulting in the removal of the peripentonal hexons, penton base, IIIa, and VI. (Bottom) Protein VI dissociates from peripentonal hexons and associates with the endosome, resulting in membrane disruption and release of partially disassembled particles into the cytoplasm.
Similar articles
- A Single Maturation Cleavage Site in Adenovirus Impacts Cell Entry and Capsid Assembly.
Moyer CL, Besser ES, Nemerow GR. Moyer CL, et al. J Virol. 2015 Oct 21;90(1):521-32. doi: 10.1128/JVI.02014-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26491163 Free PMC article. - Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption.
Chandran K, Farsetta DL, Nibert ML. Chandran K, et al. J Virol. 2002 Oct;76(19):9920-33. doi: 10.1128/jvi.76.19.9920-9933.2002. J Virol. 2002. PMID: 12208969 Free PMC article. - Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection.
Reddy VS, Nemerow GR. Reddy VS, et al. Proc Natl Acad Sci U S A. 2014 Aug 12;111(32):11715-20. doi: 10.1073/pnas.1408462111. Epub 2014 Jul 28. Proc Natl Acad Sci U S A. 2014. PMID: 25071205 Free PMC article. - Breach: Host Membrane Penetration and Entry by Nonenveloped Viruses.
Kumar CS, Dey D, Ghosh S, Banerjee M. Kumar CS, et al. Trends Microbiol. 2018 Jun;26(6):525-537. doi: 10.1016/j.tim.2017.09.010. Epub 2017 Oct 25. Trends Microbiol. 2018. PMID: 29079499 Review. - Adenovirus membrane penetration: Tickling the tail of a sleeping dragon.
Wiethoff CM, Nemerow GR. Wiethoff CM, et al. Virology. 2015 May;479-480:591-9. doi: 10.1016/j.virol.2015.03.006. Epub 2015 Mar 19. Virology. 2015. PMID: 25798531 Free PMC article. Review.
Cited by
- Complex regulation of mitochondrial signaling by human adenovirus minor capsid protein VI.
Schubert E, Mun K, Larsson M, Panagiotou S, Idevall-Hagren O, Svensson C, Punga T. Schubert E, et al. J Virol. 2024 Jul 23;98(7):e0035624. doi: 10.1128/jvi.00356-24. Epub 2024 Jun 5. J Virol. 2024. PMID: 38837380 - Integrin-Targeting Strategies for Adenovirus Gene Therapy.
Nemerow GR. Nemerow GR. Viruses. 2024 May 13;16(5):770. doi: 10.3390/v16050770. Viruses. 2024. PMID: 38793651 Free PMC article. Review. - The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
de Sautu M, Herrmann T, Scanavachi G, Jenni S, Harrison SC. de Sautu M, et al. PLoS Pathog. 2024 Apr 4;20(4):e1011750. doi: 10.1371/journal.ppat.1011750. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38574119 Free PMC article. - Advances of Recombinant Adenoviral Vectors in Preclinical and Clinical Applications.
Scarsella L, Ehrke-Schulz E, Paulussen M, Thal SC, Ehrhardt A, Aydin M. Scarsella L, et al. Viruses. 2024 Feb 28;16(3):377. doi: 10.3390/v16030377. Viruses. 2024. PMID: 38543743 Free PMC article. Review. - Cholesterol-Dependent Membrane Deformation by Metastable Viral Capsids Facilitates Entry.
Jiao M, Danthi P, Yu Y. Jiao M, et al. bioRxiv [Preprint]. 2024 Feb 1:2024.01.10.575085. doi: 10.1101/2024.01.10.575085. bioRxiv. 2024. PMID: 38260524 Free PMC article. Updated. Preprint.
References
- Anderson, C. W. 1990. The proteinase polypeptide of adenovirus serotype 2 virions. Virology 177:259-272. - PubMed
- Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. - PubMed
- Blumenthal, R., P. Seth, M. C. Willingham, and I. Pastan. 1986. pH-dependent lysis of liposomes by adenovirus. Biochemistry 25:2231-2237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007354/AI/NIAID NIH HHS/United States
- AI055729/AI/NIAID NIH HHS/United States
- R01 HL054352/HL/NHLBI NIH HHS/United States
- R01 AI055729/AI/NIAID NIH HHS/United States
- AI007354/AI/NIAID NIH HHS/United States
- EY011431/EY/NEI NIH HHS/United States
- R01 EY011431/EY/NEI NIH HHS/United States
- HL054352/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases