Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations - PubMed (original) (raw)
Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations
Heidi G Sutherland et al. Mol Cell Biol. 2006 Oct.
Abstract
PC4- and SF2-interacting protein 1 (Psip1)-also known as lens epithelium-derived growth factor (Ledgf)-is a chromatin-associated protein that has been implicated in transcriptional regulation, mRNA splicing, and cell survival in vitro, but its biological function in vivo is unknown. We identified an embryonic stem cell clone with disrupted Psip1 in a gene trap screen. The resulting Psip1-betageo fusion protein retains chromatin-binding activity and the PWWP and AT hook domains of the wild-type protein but is missing the highly conserved C terminus. The majority of mice homozygous for the disrupted Psip1 gene died perinatally, but some survived to adulthood and displayed a range of phenotypic abnormalities, including low fertility, an absence of epididymal fat pads, and a tendency to develop blepharitis. However, contrary to expectations, the lens epithelium was normal. The mutant mice also exhibited motor and/or behavioral defects such as hind limb clenching, reduced grip strength, and reduced locomotor activity. Finally, both Psip1(-/-) neonates and surviving adults had craniofacial and skeletal abnormalities. They had brachycephaly, small rib cages, and homeotic skeletal transformations with incomplete penetrance. The latter phenotypes suggest a role for Psip1 in the control of Hox expression and may also explain why PSIP1 (LEDGF) is found as a fusion partner with NUP98 in myeloid leukemias.
Figures
FIG. 1.
Gene trap disruption of the Psip1 locus. (A) Predicted intron-exon structure of mouse Psip1 and antisense Snapc3 with integration sites of the F9/3G5 and ES149 gene trap lines indicated. (B) Diagram of the Psip1 p52 and p75 protein isoforms and the predicted gene trap fusion proteins in F9/3G5 and ES149 gene trap lines. Known protein motifs predicted by SMART/PFAM and the experimentally determined human immunodeficiency virus IBD are depicted.
FIG. 2.
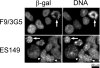
Subcellular localization of Psip1-βgeo fusion proteins. Shown is immunostaining of F9/3G5 and ES149 cells with an anti-β-gal antibody showing localization of the Psip1-β-geo fusion proteins in the nuclei and on chromosomes in mitotic cells (arrows). In some ES149 nuclei, the fusion protein is concentrated at the DAPI-bright heterochromatic foci (arrowheads).
FIG. 3.
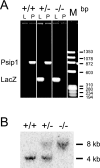
Genotyping of ES149 mice for gene trap disruption of Psip1. (A) PCR analysis of WT (+/+) and heterozygous (+/−) and homozygous (−/−) Psip1 mutant mice with primers for lacZ (L) (432-bp band) and endogenous Psip1 (P) (860-bp band). (B) Southern blot analysis of HindIII-digested genomic DNA from +/+, +/−, and −/− mice probed with a PCR product for exon 9 of Psip1. This results in a 4-kb band for the endogenous Psip1 locus and an 8-kb band for the gene-trapped locus.
FIG. 4.
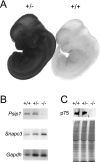
Psip1 expression in WT and mutant mice. (A) X-Gal staining of WT and Psip1+/− embryos at 11.5 dpc. (B) RT-PCR analysis of Psip1 and Snapc3 mRNAs in Psip1+/+, Psip1+/−, and _Psip1_−/− mice. Gapdh was used as a control. (C) Western blot analysis of p75/Psip1 in protein extracts form +/+, +/−, and −/− embryos (top). The Coomassie-stained polyacrylamide gel at the bottom shows protein loading.
FIG. 5.
Phenotypic analysis of surviving adult _Psip1_−/− mutant mice in the MF1 background. (A) _Psip1_−/− mutant mice have a reduction of intraabdominal fat, in particular, an absence of epididymal fat pads (fp), compared to their +/+ littermates. (B) H&E staining of seminiferous tubules from heterozygous (+/−) and _Psip1_−/− mutant mice. Both show normal-appearing basilar spermatogonia, several layers of differentiating spermatocytes, and maturing spermatids at the lumen. Bars, 50 μm. (C) H&E staining of eyes from WT (+/+) and _Psip1_−/− mutant mice showing a normal lens epithelium (le). (D) Chronic ulcerative blepharitis in _Psip1_−/− mutant mice. The mouse on the left is 2 months old; the right eye is asymptomatic, and the left eye shows mild symptoms. The mouse on the right is 4 months old; symptoms in the right eye are mild, and those in the left eye are severe. (E) H&E staining of the eyelid of a _Psip1_−/− mutant mouse showing ulcerated epidermis covered with adherent fibrin plaque. The adjacent intact epidermis is hyperplastic, and chronic mixed inflammatory cells have infiltrated the dermis. Bar, 100 μm. (F) _Psip1_−/− mutant mice clench their hind limbs, in contrast to the hind limb extension of +/+ mice. (G) _Psip1_−/− mutant mice have brachycephaly with broad, shortened faces and jaws. (H) Alizarin red-alcian blue staining of skulls shows that the cranial bones and nasal process are broader and shorter in _Psip1_−/− mutant mice compared to those of +/− (and +/+) mice.
FIG. 6.
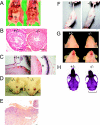
Alizarin red-alcian blue staining reveals craniofacial and skeletal abnormalities in _Psip1_−/− mutant mice. (A) Staining of +/+ and _Psip1_−/− mutant MF1 newborn skeletons. −/− pups have domed skulls (arrowheads) and reduced interparietal, exoccipital, and supraoccipital bones (double arrowheads). (B) _Psip1_−/− mutant pups lack physiological curvature of the spine and have a small rib cage and a short sternum, compared to +/+ and +/− littermates. (C) Dissected rib cages from C57BL/6 +/+, +/−, and −/− newborn pups. (D) Cervical region, showing misshapen C1 and C2 and an incomplete ectopic rib (arrow) from C7 that is fused with the cartilage of T1. (E) Adult skeletons showing an incomplete ectopic rib (arrowed) from C7 unilaterally fused with T1 and a prominent spinous process, characteristic for T2, incorrectly associated with T3 in a _Psip1_−/− mutant animal. (F) +/+ and −/− MF1 newborn skeletons showing a reduced number of lumbar vertebrae and transformation of L6 into S1.
Similar articles
- Embryonic Lethality Due to Arrested Cardiac Development in Psip1/Hdgfrp2 Double-Deficient Mice.
Wang H, Shun MC, Dickson AK, Engelman AN. Wang H, et al. PLoS One. 2015 Sep 14;10(9):e0137797. doi: 10.1371/journal.pone.0137797. eCollection 2015. PLoS One. 2015. PMID: 26367869 Free PMC article. - Psip1/Ledgf p75 restrains Hox gene expression by recruiting both trithorax and polycomb group proteins.
Pradeepa MM, Grimes GR, Taylor GC, Sutherland HG, Bickmore WA. Pradeepa MM, et al. Nucleic Acids Res. 2014 Aug;42(14):9021-32. doi: 10.1093/nar/gku647. Epub 2014 Jul 23. Nucleic Acids Res. 2014. PMID: 25056311 Free PMC article. - Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing.
Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Pradeepa MM, et al. PLoS Genet. 2012;8(5):e1002717. doi: 10.1371/journal.pgen.1002717. Epub 2012 May 17. PLoS Genet. 2012. PMID: 22615581 Free PMC article. - Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: evidence for unique and redundant function.
Horan GS, Kovàcs EN, Behringer RR, Featherstone MS. Horan GS, et al. Dev Biol. 1995 May;169(1):359-72. doi: 10.1006/dbio.1995.1150. Dev Biol. 1995. PMID: 7750651 - PSIP1/p75 promotes tumorigenicity in breast cancer cells by promoting the transcription of cell cycle genes.
Singh DK, Gholamalamdari O, Jadaliha M, Ling Li X, Lin YC, Zhang Y, Guang S, Hashemikhabir S, Tiwari S, Zhu YJ, Khan A, Thomas A, Chakraborty A, Macias V, Balla AK, Bhargava R, Janga SC, Ma J, Prasanth SG, Lal A, Prasanth KV. Singh DK, et al. Carcinogenesis. 2017 Oct 1;38(10):966-975. doi: 10.1093/carcin/bgx062. Carcinogenesis. 2017. PMID: 28633434 Free PMC article.
Cited by
- Multivalency of nucleosome recognition by LEDGF.
Koutná E, Lux V, Kouba T, Škerlová J, Nováček J, Srb P, Hexnerová R, Šváchová H, Kukačka Z, Novák P, Fábry M, Poepsel S, Veverka V. Koutná E, et al. Nucleic Acids Res. 2023 Oct 13;51(18):10011-10025. doi: 10.1093/nar/gkad674. Nucleic Acids Res. 2023. PMID: 37615563 Free PMC article. - Psip1/p52 regulates posterior Hoxa genes through activation of lncRNA Hottip.
Pradeepa MM, McKenna F, Taylor GC, Bengani H, Grimes GR, Wood AJ, Bhatia S, Bickmore WA. Pradeepa MM, et al. PLoS Genet. 2017 Apr 6;13(4):e1006677. doi: 10.1371/journal.pgen.1006677. eCollection 2017 Apr. PLoS Genet. 2017. PMID: 28384324 Free PMC article. - Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting.
Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. Marshall HM, et al. PLoS One. 2007 Dec 19;2(12):e1340. doi: 10.1371/journal.pone.0001340. PLoS One. 2007. PMID: 18092005 Free PMC article. - Integration of transcriptomics, proteomics, and microRNA analyses reveals novel microRNA regulation of targets in the mammalian inner ear.
Elkan-Miller T, Ulitsky I, Hertzano R, Rudnicki A, Dror AA, Lenz DR, Elkon R, Irmler M, Beckers J, Shamir R, Avraham KB. Elkan-Miller T, et al. PLoS One. 2011 Apr 5;6(4):e18195. doi: 10.1371/journal.pone.0018195. PLoS One. 2011. PMID: 21483685 Free PMC article. - LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors.
Meehan AM, Saenz DT, Morrison JH, Garcia-Rivera JA, Peretz M, Llano M, Poeschla EM. Meehan AM, et al. PLoS Pathog. 2009 Jul;5(7):e1000522. doi: 10.1371/journal.ppat.1000522. Epub 2009 Jul 17. PLoS Pathog. 2009. PMID: 19609362 Free PMC article.
References
- Abramovich, C., N. Pineault, H. Ohta, and R. K. Humphries. 2005. Hox genes: from leukemia to hematopoietic stem cell expansion. Ann. N. Y. Acad. Sci. 1044:109-116. - PubMed
- Cherepanov, P., E. Devroe, P. A. Silver, and A. Engelman. 2004. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279:48883-48892. - PubMed
- Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials