FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors - PubMed (original) (raw)
. 2007 Aug 6;204(8):1813-24.
doi: 10.1084/jem.20070876. Epub 2007 Jul 23.
Jonathan Grim, Peter Strack, Sudhir Rao, Deanne Tibbitts, Christopher Winter, James Hardwick, Markus Welcker, Jules P Meijerink, Rob Pieters, Giulio Draetta, Rosalie Sears, Bruce E Clurman, A Thomas Look
Affiliations
- PMID: 17646409
- PMCID: PMC2118656
- DOI: 10.1084/jem.20070876
FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors
Jennifer O'Neil et al. J Exp Med. 2007.
Abstract
gamma-secretase inhibitors (GSIs) can block NOTCH receptor signaling in vitro and therefore offer an attractive targeted therapy for tumors dependent on deregulated NOTCH activity. To clarify the basis for GSI resistance in T cell acute lymphoblastic leukemia (T-ALL), we studied T-ALL cell lines with constitutive expression of the NOTCH intracellular domain (NICD), but that lacked C-terminal truncating mutations in NOTCH1. Each of the seven cell lines examined and 7 of 81 (8.6%) primary T-ALL samples harbored either a mutation or homozygous deletion of the gene FBW7, a ubiquitin ligase implicated in NICD turnover. Indeed, we show that FBW7 mutants cannot bind to the NICD and define the phosphodegron region of the NICD required for FBW7 binding. Although the mutant forms of FBW7 were still able to bind to MYC, they do not target it for degradation, suggesting that stabilization of both NICD and its principle downstream target, MYC, may contribute to transformation in leukemias with FBW7 mutations. In addition, we show that all seven leukemic cell lines with FBW7 mutations were resistant to the MRK-003 GSI. Most of these resistant lines also failed to down-regulate the mRNA levels of the NOTCH targets MYC and DELTEX1 after treatment with MRK-003, implying that residual NOTCH signaling in T-ALLs with FBW7 mutations contributes to GSI resistance.
Figures
Figure 1.
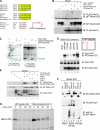
Treatment with MRK-003 results in cell cycle arrest, apoptosis, and inhibition of NICD production in human T-ALL cell lines. (A) Cell cycle analysis of the GSI-sensitive HPB-ALL cell line. Cells were treated with DMSO (vehicle) or 1 μM MRK-003 for 7 d, stained with propidium iodide, and analyzed by flow cytometry. (B) Annexin V–FITC/propidium iodide staining of the DND41 cell line after 7 d in DMSO or 1 μM MRK-003. Numbers represent the percentage of cells in each quadrant. Similar results were observed in other GSI-sensitive cell lines. (C) Cell cycle analysis of the GSI-resistant Molt13 cell line after 7 d in DMSO (vehicle) or MRK-003. Similar results were observed for all other GSI-resistant cell lines listed in Table I. (D) Annexin V–APC/propidium iodide staining of the Molt4 cell line after 7 d in DMSO or 1 μM MRK-003. Similar results were observed in all other GSI-resistant cell lines. (E) Activated NOTCH1 Western blot analysis. T-ALL cell lines were treated with 1 μM MRK-003 for 3 d. Whole cell lysates were subjected to SDS-PAGE electrophoresis and immunoblotting with the NOTCH1 (V1744) antibody.
Figure 2.
NOTCH T2512A mutant shows increased stability and decreased binding to FBW7. (A) Alignment of the four human NOTCH proteins, as well as Notch proteins from various species, shows strong conservation of the putative NOTCH CPD. Known CPDs are aligned with NOTCH for comparison. (B) The T2512A NOTCH mutant is deficient in binding to WT FBW7, and the tumor-derived FBW7 arginine mutant (R465C) can no longer bind WT NOTCH. Cells were transfected as indicated, and FLAG-FBW7 was immunoprecipitated. Samples were subjected to SDS-PAGE electrophoresis and immuno blotting with anti-MYC tag (9E10) to detect transfected Notch proteins. Whole cell lysates were analyzed as indicated to verify expression of transfected constructs. (C) Co-immunoprecipitation assays in the K562 erythroleukemia cell line show that T2512A is deficient in binding to FBW7 in hematopoietic cells. Whole cell lysates were analyzed as indicated to verify expression of transfected constructs. (D) Phosphorylation of S2514 is not required for the FBW7–Notch ICD interaction. 293a cells were transfected as indicated and analyzed as in B above. (E) NICD preferentially associates with nuclear and nucleolar FBW7 isoforms. 293a cells were transfected with the indicated plasmids and analyzed as in B above. (F) In vivo ubiquitination assays show that the NOTCH T2512A mutant is resistant to FBW7-mediated ubiquitination. 293a cells were transfected as indicated. Cell lysates were prepared and immuno precipitated with anti-HA antibody to pull down ubiquitinated proteins. Samples were subjected to SDS-PAGE electrophoresis and immunoblotted with 9E10 to detect MYC-tagged NOTCH proteins. Whole cell lysates were analyzed as indicated to verify expression of transfected constructs. (G) The NOTCH T2512A mutant has an extended half-life in 293a cells compared with WT NOTCH. 293a cells were transfected with either WT NICD or the T2512A mutant. 48 h later, cells were pulse labeled in vivo with 35S-methionine/cysteine and chased in medium with excess unlabeled methionine and cysteine for the indicated times. Transfected NOTCH proteins were immunoprecipitated with MN-1 antisera, and samples were subjected to SDS-PAGE electrophoresis. Gels were then exposed to x-ray film.
Figure 3.
GSI treatment results in decreased MYC protein levels in a subset of T-ALL cell lines. T-ALL cell lines were treated with 1 μM MRK-003 or DMSO for 3 d, and whole cell lysates were prepared using RIPA lysis buffer. Western blot analysis was performed using antibodies against MYC and β-actin as a loading control.
Figure 4.
T-ALL cell lines have increased MYC half-life. (A) T-ALL cell lines with WT FBW7 (Molt4, DND41 and KOPTK1) were pulse-labeled in vivo with 35S-methionine/cysteine and chased in medium with excess unlabeled methionine and cysteine for the indicated times. Endogenous MYC was immunoprecipitated from an equal number of cells for each time point and analyzed by gel electrophoresis. 35S-labeled MYC from each sample was quantitated by phosphoimager. The rate of degradation of MYC for each cell line is represented in the graph by best-fit exponential lines. Half-lives of MYC were calculated from exponential line equations. (B) Pulse-chase experiments were similarly performed on T-ALL cell lines with mutant FBW7 (CEM and Jurkat). All T-ALL cell lines analyzed have increased MYC half-life compared with JY cells, a B cell lymphoblastoid cell line.
Figure 5.
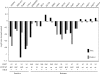
NOTCH target genes are not decreased upon GSI treatment in T-ALL cell lines with mutant FBW7. MYC and DELTEX RNA levels in 18 T-ALL cell lines after 3 d of 1 μM MRK-003 GSI treatment (compared with DMSO-treated cells) as determined by microarray gene expression analysis. Values are a log10 ratio of the expression level of MYC and DELTEX in MRK-003–treated cells compared with DMSO-treated cells.
Figure 6.
Mutant FBW7 cannot bind to NICD and acts in a dominant-negative manner to prevent MYC degradation. (A) FBW7 arginine point mutants efficiently co-precipitate with MYC but not with NOTCH. 293a cells were transfected with the indicated plasmids. Whole cell lysates were subjected to immunoprecipitation with anti-FLAG, and resulting samples were analyzed via immunoblotting for either MT-NOTCH or HA-MYC as indicated. Whole cell lysates were also analyzed directly by SDS-PAGE and immunoblotting for FLAG-FBW7, HA-MYC, or MT-NOTCH as indicated to verify expression of transfected constructs. dnCul1 was included in all transfections to block FBW7-mediated proteasomal degradation. (B) T-ALL–associated FBW7 mutants are unable to mediate MYC degradation. 293a cells were transfected as indicated. Whole cell lysates were subjected to SDS-PAGE and immunoblotting for either HA-MYC or FLAG-FBW7 as indicated. (C) Tumor-derived FBW7 mutants dominantly inhibit MYC turnover by WT-FBW7. 293a cells were transfected with a constant amount of HA-MYC with increasing amounts of either FLAG-FBW7-γ or a combination of a constant amount (500 ng) of FLAG-FBW7-γ and increasing amounts of FLAG-FBW7-γ-R465H (1–5 μg). The ratios of mutant to WT FBW7 expression are indicated above lanes 7–9. HA-cdk2, which does not affect turnover of MYC by FBW7, is included as a transfection control. Whole cell lysates were subjected to SDS-PAGE and immunoblotting for HA-MYC, HA-cdk2, or FLAG-FBW7 as indicated.
Similar articles
- The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia.
Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. Thompson BJ, et al. J Exp Med. 2007 Aug 6;204(8):1825-35. doi: 10.1084/jem.20070872. Epub 2007 Jul 23. J Exp Med. 2007. PMID: 17646408 Free PMC article. - New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia.
Hales EC, Taub JW, Matherly LH. Hales EC, et al. Cell Signal. 2014 Jan;26(1):149-61. doi: 10.1016/j.cellsig.2013.09.021. Epub 2013 Oct 16. Cell Signal. 2014. PMID: 24140475 Review. - Notch and NF-kB signaling pathways regulate miR-223/FBXW7 axis in T-cell acute lymphoblastic leukemia.
Kumar V, Palermo R, Talora C, Campese AF, Checquolo S, Bellavia D, Tottone L, Testa G, Miele E, Indraccolo S, Amadori A, Ferretti E, Gulino A, Vacca A, Screpanti I. Kumar V, et al. Leukemia. 2014 Dec;28(12):2324-35. doi: 10.1038/leu.2014.133. Epub 2014 Apr 14. Leukemia. 2014. PMID: 24727676 - Gamma-secretase inhibitors suppress the growth of leukemia and lymphoma cells.
Kogoshi H, Sato T, Koyama T, Nara N, Tohda S. Kogoshi H, et al. Oncol Rep. 2007 Jul;18(1):77-80. Oncol Rep. 2007. PMID: 17549349 - Therapeutic targeting of NOTCH1 signaling in T-cell acute lymphoblastic leukemia.
Palomero T, Ferrando A. Palomero T, et al. Clin Lymphoma Myeloma. 2009;9 Suppl 3(Suppl 3):S205-10. doi: 10.3816/CLM.2009.s.013. Clin Lymphoma Myeloma. 2009. PMID: 19778842 Free PMC article. Review.
Cited by
- SCFFBXW7 regulates G2-M progression through control of CCNL1 ubiquitination.
O'Brien S, Kelso S, Steinhart Z, Orlicky S, Mis M, Kim Y, Lin S, Sicheri F, Angers S. O'Brien S, et al. EMBO Rep. 2022 Dec 6;23(12):e55044. doi: 10.15252/embr.202255044. Epub 2022 Oct 24. EMBO Rep. 2022. PMID: 36278408 Free PMC article. - Loss of thymocyte competition underlies the tumor suppressive functions of the E2a transcription factor in T-ALL.
Parriott G, Hegermiller E, Morman RE, Frank C, Saygin C, Stock W, Bartom ET, Kee BL. Parriott G, et al. Leukemia. 2024 Mar;38(3):491-501. doi: 10.1038/s41375-023-02123-4. Epub 2023 Dec 28. Leukemia. 2024. PMID: 38155245 - Phosphorylation-dependent regulation of Notch1 signaling: the fulcrum of Notch1 signaling.
Lee HJ, Kim MY, Park HS. Lee HJ, et al. BMB Rep. 2015 Aug;48(8):431-7. doi: 10.5483/bmbrep.2015.48.8.107. BMB Rep. 2015. PMID: 26058398 Free PMC article. Review. - The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis.
Lau AW, Fukushima H, Wei W. Lau AW, et al. Front Biosci (Landmark Ed). 2012 Jun 1;17(6):2197-212. doi: 10.2741/4045. Front Biosci (Landmark Ed). 2012. PMID: 22652772 Free PMC article. Review. - FBP1 inhibits NSCLC stemness by promoting ubiquitination of Notch1 intracellular domain and accelerating degradation.
He T, Wang Y, Lv W, Wang Y, Li X, Zhang Q, Shen HM, Hu J. He T, et al. Cell Mol Life Sci. 2024 Feb 13;81(1):87. doi: 10.1007/s00018-024-05138-x. Cell Mol Life Sci. 2024. PMID: 38349431 Free PMC article.
References
- Wilson, A., and F. Radtke. 2006. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 580:2860–2868. - PubMed
- Dievart, A., N. Beaulieu, and P. Jolicoeur. 1999. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 18:5973–5981. - PubMed
- Hallahan, A.R., J.I. Pritchard, S. Hansen, M. Benson, J. Stoeck, B.A. Hatton, T.L. Russell, R.G. Ellenbogen, I.D. Bernstein, P.A. Beachy, and J.M. Olson. 2004. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 64:7794–7800. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA84069/CA/NCI NIH HHS/United States
- R01CA102742/CA/NCI NIH HHS/United States
- K08 CA109124/CA/NCI NIH HHS/United States
- R01 CA084069/CA/NCI NIH HHS/United States
- P01 HL084205/HL/NHLBI NIH HHS/United States
- R01 CA102742/CA/NCI NIH HHS/United States
- 1K08CA109124-01A2/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases